Abstract
Prostate cancer is featured by its heterogeneous nature, which indicates a different prognosis. Castration-resistant prostate cancer (CRPC) is a hallmark of the treatment-refractory stage, and the median survival of patients is only within two years. Neuroendocrine prostate cancer (NEPC) is an aggressive variant that arises from de novo presentation of small cell carcinoma or treatment-related transformation with a median survival of 1–2 years from the time of diagnosis. The epigenetic regulators, such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), have been proven involved in multiple pathologic mechanisms of CRPC and NEPC. LncRNAs can act as competing endogenous RNAs to sponge miRNAs that would inhibit the expression of their targets. After that, miRNAs interact with the 3’ untranslated region (UTR) of target mRNAs to repress the step of translation. These interactions may modulate gene expression and influence cancer development and progression. Otherwise, epigenetic regulators and genetic mutation also promote neuroendocrine differentiation and cancer stem-like cell formation. This step may induce neuroendocrine prostate cancer development. This review aims to provide an integrated, synthesized overview under current evidence to elucidate the crosstalk of lncRNAs with miRNAs and their influence on castration resistance or neuroendocrine differentiation of prostate cancer. Notably, we also discuss the mechanisms of lncRNA–miRNA interaction in androgen receptor-independent prostate cancer, such as growth factors, oncogenic signaling pathways, cell cycle dysregulation, and cytokines or other transmembrane proteins. Conclusively, we underscore the potential of these communications as potential therapeutic targets in the future.
Keywords: cancer, epigenetics, long non-coding RNAs, microRNAs, prostate cancer, castration-resistant, neuroendocrine
1. Introduction
Prostate cancer (PCa) is the second most commonly diagnosed malignancy, leading to 6.8% of cancer-related deaths in the male population worldwide in 2020 [1]. Its prevalence increases with age by a 1.7 odds ratio per decade and is up to 59% (48–71%) by age >79 years old [2]. At diagnosis, the treatment and the prognosis of PCa are determined by the stratified risk group on the basis of prostate-specific antigen (PSA) level, the sum of Gleason patterns, and the clinical stage. Compared with localized PCa that can be actively surveyed without management, the treatment of advanced PCa, requiring systemic therapy, is much more complicated and lethal [3,4,5]. Androgen deprivation therapy (ADT) is usually the first-line systemic treatment in advanced PCa, according to the canonical mechanism of the androgen receptor (AR) signaling pathway [6]. Nevertheless, the innate diversity of PCa cells can ultimately adapt to androgen deprivation, resulting in activation of AR signal even under a low level of serum androgen and advancing to a worse condition named castration-resistant prostate cancer (CRPC) [7,8].
2. CRPC and Its Subtypes
The diagnostic criteria of CRPC consist of serum testosterone less than 50 ng/mL or 1.7 nmol/L plus biochemical or radiologic progression [9]. Biochemical progression is defined as three continuous increases in PSA at intervals of no less than one week, leading to at least two 50% rises over the nadir and PSA more than 2 ng/mL [10]. Radiologic progression is defined as more than two new lesions detected under an imaging survey based on the rules of Response Evaluation Criteria in Solid Tumors (RECIST) [11]. The reported prevalence of CRPC within 5 years of follow-up after initial PCa treatment was 10–20%, with metastatic CRPC (mCRPC) accounting for ≥84% of all. Moreover, 33% non-metastatic CRPC (nmCRPC) advanced to metastasis within 2 years of follow-up [12]. An extensive database series in Sweden in 2020 comprised 1712 CRPC cases, stating that the median PCa-specific survival rates since diagnosis were 30.3 months and 13.3 months for nmCRPC and mCRPC, respectively [13].
The mechanism of CRPC is quite complex and alterable, and therefore even if novel therapies are constantly being proposed and used, the median PCa-specific survival in mCRPC can only increase by 2.8 to 4.8 months [14,15,16,17,18,19,20]. Sustained activation of AR signaling in cancer cells even under androgen depletion therapy is thought to be the primary mechanism of CRPC [21]. However, most CRPC are still AR-dependent. AR signaling can still be delivered through several mechanisms, including AR point mutation, increased AR expression, emersion of AR splice variants, altered intratumoral androgen biosynthesis, and co-factor activity modulation [22]. Therefore, more effective AR-targeting agents (AR signaling inhibitor, ARSI) such as abiraterone, apalutamide, and enzalutamide are recognized as first-line treatment options in CRPC [23]. However, not all castration resistance is dependent on AR signaling due to the diversity, plasticity, and heterogeneity of the tumor. In addition, the transition of tumor cells from AR-dependent to AR-independent signaling pathways may cause further resistance to ARSI [24,25]. The mechanisms of AR-independence include neuroendocrine differentiation, cell cycle dysregulation, glucocorticoid receptor upregulation, immune-mediated resistance, PI3K/AKT pathway, and MYC pathway.
Increasingly more researchers are trying to classify CRPC on the basis of histological phenotypes or by exploring the variation of AR expression, also known as AR heterogeneity in tumor cells. It helps in dissecting the development of CRPC, improving drug evolution and treatment strategy, and delivering more explicit outcomes [26,27,28,29]. Thus far, AR and neuroendocrine markers are the two most common and crucial histologic features helping to categorize CRPC into different phenotypes such as AR-high prostate cancer (ARPC); AR-low prostate cancer (ARLPC); neuroendocrine prostate cancer (NEPC); and both AR-null and NE-null CRPC, also named double-negative CRPC (DNPC) [28,30]. The study published by Bluemn et al. exhibited that through the use of ARSI in the recent two decades, the ratio of NEPC and DNPC in CRPC increased from 5.4% and 6.3% to 13.3% and 23.3%, respectively [29]. The transition from ARPC-dominant phenotype to NEPC-dominant phenotype after receiving AR-targeting therapy has also been demonstrated in animal models, cell lines, and patient-derived xenograft (PDX) studies [31,32,33,34]. In addition to the heterogeneity of cell composition of CRPC, dynamic cell selection and lineage plasticity also increase the complexity of CRPC mechanism and imply the pivotal role of microenvironment, epigenetic or transcriptional, posttranscriptional, and posttranslational regulation.
3. Non-Coding RNAs and CRPC
Non-coding RNAs (ncRNAs), which participate in diverse molecular regulatory activities including epigenetic, posttranscriptional, posttranslational regulation, and signal transduction, have been proven to be a crucial molecular substance involved in multiple pathologic mechanisms of CRPC [35,36,37]. In particular, ncRNAs are the transcripted products of genomic sequences that do not proceed to encode proteins. Advanced research and updated knowledge have altered them from useless substances to functional molecules of cellular processes in the past decades [38,39]. Recently, increasing studies have discovered the contribution of ncRNAs on cancer initiation, progression, and metastasis [40]. NcRANs are thought to be potential diagnostic and prognostic biomarkers of cancer, especially combining the detection of different ncRNAs, which can enhance the sensitivity and specificity in diagnostic and prognostic assessments [41]. According to the size, structure, transcripted source, and location, functional ncRNAs can be classified into several subsets, including microRNA (miRNA), small interfering RNA (siRNA or RNAi), small nuclear RNA (snoRNA), PIWI-interactive RNA (piRNA), and the long non-coding RNA (lncRNA).
Among the different categories of ncRNAs, miRNA and lncRNA are the two most frequently investigated in CRPC research and have been proven to involve multiple pathophysiologic pathways [35,37]. The regulatory role of miRNA in CPRC was the first to be discovered. At least 20 kinds of miRNAs have been confirmed to be related to the development of CPRC, and they participated in diverse pathogenesis pathways such as AR-related cell proliferation, cancer cell survival, apoptosis, or epithelial–mesenchymal transition (EMT) [36].
On the other hand, lncRNA is a larger and more complex ncRNA, and it has been gradually explored to be related to CPRC in the past decade due to advances in detection technology [35]. Moreover, because lncRNA carries more information and affinity to biomolecules in the human body, a single type of lncRNA can have multi-pathway regulation. Therefore, lncRNA is also a key regulator in PCa progression from androgen-dependent status to CRPC [42]. For example, AR-regulated long noncoding RNA 1 (ARLNC1, a lncRNA) could be upregulated by the AR protein and stabilize the AR transcript via RNA–RNA interaction [43]. Knockdown of ARLNC1 caused the suppression of AR expression and inhibition of AR-dependent PCa growth in vitro and in vivo [43]. Moreover, prostate cancer-associated non-coding RNA1 (PRNCR1, a lncRNA) and prostate cancer gene expression marker 1 (PCGEM1, a lncRNA) were highly expressed in CRPC cells and reported to be involved in AR signaling pathway, while knockdown of either PRNCR1 or PCGEM1 would suppress in vivo tumor growth of CRPC [44,45,46].
An alternative mechanism for lncRNAs to influence the AR is through interactions with miRNAs. Although the contribution of numerous miRNAs and lncRNA in CRPC development has been summarized and reviewed in the literature, these authors only elaborated on the impact of these two ncRNAs separately without sorting out the interaction of lncRNAs and miRNAs [47,48]. There is accumulating evidence proposed that cancer development is highly correlated with the interaction patterns between lncRNAs and miRNAs [49,50]. LncRNAs and miRNAs are proven to interact in many ways, including the “sponge” effect that is most frequently described and utilized [51,52]. In the last five years, an increasing number of studies have also described the interaction and regulation between lncRNAs and miRNAs in different pathologic pathways of CRPC. Therefore, in this literature review, we comprehensively summarize the currently known interaction between lncRNA and miRNA on the development of CRPC. Further, our collation is classified according to the different molecular mechanisms and histology types of CRPC.
4. LncRNA and Its Interaction with miRNA
LncRNAs are defined as non-coding transcripts usually longer than 200 nucleotides. They are one of the most abundant classes within ncRNAs. They outnumber protein-coding transcripts in the ratio of about three to one [53]. In the past few decades, researchers mainly studied the protein-coding genes and their transcripts, yet there are still many unraveled fields in whole transcriptome data. LncRNAs are now found to demonstrate variable effects when binding to DNA, RNA, or proteins. According to their locations corresponding to protein-coding genes, lncRNA can be classified into several groups, including (1) intronic lncRNAs: transcribed from introns of protein-coding genes, (2) intergenic lncRNAs: transcribed between two protein-coding genes, (3) antisense lncRNAs: transcribed in the opposite direction of protein-coding genes, and (4) overlapping lncRNAs: transcripts that cover protein-coding genes [54].
LncRNAs can fold themselves into structures to interact with other genomes, transcripts, or proteins to regulate chromatin activities, transcription, protein chromatin assembly, splicing, and telomere biology. They are capable of chromatin remodeling, histone methylation, acting as miRNA “sponges” or transcription factor decoys, and affecting the proteome by mediating complex formation [42]. If lncRNAs are localized in the nucleus, they usually vise proteins to regulate target gene expression at the transcriptional levels. However, if lncRNAs are distributed mainly in the cytoplasm, they are likely to enhance target gene expression at the posttranscriptional level by binding with miRNA.
MiRNAs are small single-stranded ncRNAs with about 22 nucleotides. Most of the time, miRNAs interact with the 3’ untranslated region (UTR) of target mRNAs to repress their expression via the promotion of RNA degradation or inhibition of protein translation. Previously, it has been revealed that 3’UTR of oncogene such as CCND1 can be shortened to escape miRNA-mediated repression in cis and lead to its overexpression in cancer [55]. However, a recent study has demonstrated that 3’UTR shortening of competing for endogenous RNA (ceRNA), an RNA transcript that regulates the levels of other RNA transcripts by competing for shared microRNAs, has a trans effect on the repression of tumor suppressor gene via release of sequestered miRNA and results in tumorigenesis [56]. Taken together, 3’UTR shortening might not only induce proto-oncogene expression in cis but also repress tumor suppressor gene expression in trans via miRNA-dependent manners. This concept of ceRNAs may also be applied to the interactions of lncRNAs and miRNAs and has raised much attention because they can promote or inhibit cancers depending on the roles of the genes they modulate.
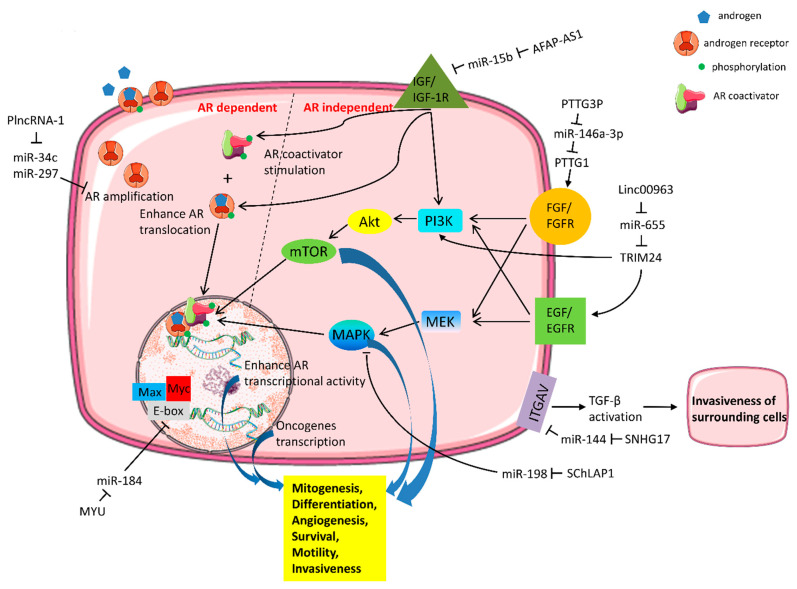
In addition, miRNAs have been recognized as well-known effectors concerning castration resistance. The mechanisms of castration resistance related to miRNAs mainly include (1) modulation of AR amplification; (2) interfering AR transcriptional activity; (3) signal cascades independent of AR and crosstalk with AR, or regulation of AR coactivator/corepressor; and (4) AR-targeted miRNAs [35]. Since lncRNAs and miRNAs are two major families of the non-protein-coding transcripts, lncRNAs are prone to act as ceRNAs and competitively bind to miRNAs and create the lncRNA–miRNA–mRNA networks that extensively exist in CRPC cell lines or specimens. Table 1 demonstrates the current studies about the interaction of lncRNAs and miRNAs and their related mechanisms to castration resistance. Among them, signal cascades independent of AR and crosstalk with AR are the most frequently seen and dissected pathways [28]. The AR bypass/crosstalk mechanisms mainly consist of growth factors, cytokines, MAPK, PI3K/AKT, MYC pathway, or cell cycle dysregulation (Table 1, Figure 1) [28].
Table 1.
Content curation of long-non-coding RNA (lncRNA) and its targeting microRNA (miRNA) and related mechanism of castration resistance prostate cancer.
| Mechanism of Castration Resistance | Subject Investigated (Cell Lines/Tissue) | lncRNA | miRNA | Interactions | Reference | |
|---|---|---|---|---|---|---|
| AR amplification | 22RV1/human PCa | PlncRNA-1 | miR-34c/miR-297 | PlncRNA-1 promotes AR expression via competitive inhibition of miRNA-34c/miR-297 targeting AR. | [57] | |
| Increased AR transcriptional activity | PC3, DU145/human PCa | CCAT1 | miR-28-5P | In cytoplasm: competing for miR-28-5P to promote cell proliferation and colony formation. | [58] | |
| In nucleus: CCAT1 acts as a scaffold for DDX5(P68) and AR transcriptional complex to facilitate expression of AR-regulated castration resistance gene (UBE2C). | ||||||
| Signal cascades independent of AR and crosstalk with AR | Growth factors | C4-2, PC3, DU145 | AFAP1-AS1 | miR-15b | AFAP1-AS1 upregulates IGF1R by competitively binding with miR-15b to de-repress IGF1R. | [59] |
| human PCa | PTTG3P | miR-146a-3p | PTTG3P upregulates PTTG1 to stimulate FGF expression by competing for miR-146a-3p. | [60] | ||
| Other oncogenic signal pathways | PC3 | SChLAP1 | miR-198 | SChLAP1 regulates the miR-198 expression and influences cancer progression by the MAPK1 pathway. | [61] | |
| human PCa | Linc00963 | miR-655 | Linc00963 competitively binds with miR-655 and upregulates TRIM24 expression to activate the PI3K/AKT pathway. | [62] | ||
| PC3, DU145/human PCa | MYU(VPS9D1-AS1) | miR-184 | MYU upregulates the MYC expression by competitively binding with miR-184. | [63] | ||
| Cell cycle dysregulation | DU145/human PCa | SNHG7 | miR-503 | MiR-503 targets 3′-UTR of SNHG and inhibits cell cycle proteins (CDK4, CDK6, Cyclin D), inducing G0/G1 cell cycle arrest. | [64] | |
| PC3, DU145 | LOXL1-AS1 | miR-541-3p | LOXL1-AS1 interferes with miR-541-3p targeting cell cycle regulator Cyclin D and promotes cell proliferation. | [65] | ||
| DU145/human PCa | TTTY15 | let-7 | FOXA1, acting as a transcription factor of TTTY15, promotes PCa progression by sponging let-7 and upregulating CDK6 and FN1. | [66] | ||
| Cytokine | C4-2, PC3, DU145 | SNHG17 | miR-144 | SNHG17 acts as a ceRNA to upregulate CD51 (integrin alpha-V) expression through competitively sponging miR-144. | [67] | |
| Unidentified mechanisms | PC3, DU145 | HOTAIR | miR-34a | MiR-34a directly targets HOTAIR and inhibits cell growth. | [68] | |
| PC3 | HOTAIR | miR-193a | HOTAIR couples with EZH2 to repress miR-193a by trimethylation of H3K27me3; miR-193a directly targets HOTAIR to reduce HOTAIR level in miR-193a overexpressed cells. | [69] | ||
| PC3 | PCGEM-1 | miR-148a | Putative PCGEM1 binding site is identified in the 5′-UTR of miR-148a; PCGEM-1 expression represses miR-148a and cell apoptosis. | [70] | ||
| PC3, DU145/human PCa | PCSEAT | miR-143-3p-/miR-24-2-5p | PCSEAT competitively sponges miR-143-3p/miR-24-2-5p and decreases PCSEAT-mediated cell proliferation. | [71] | ||
| PC3, DU145/human PCa | Linc00308 | miR-137 | LncRNA-miRNA-mRNA networks regulate tumor suppressor gene PTGS2 and DUSP2 and affect survival. | [72] | ||
| Linc00355 | miR-122/miR-506 | |||||
| OSTN-AS1 | miR-137/miR-506 | |||||
AR: Androgen receptor; FGF: fibroblast growth factor; FN1: fibronectin; IGF1R: insulin-like growth factor 1 receptor; PCa: prostate cancer; ceRNA: competing endogenous RNA.
Figure 1.
The mechanisms of actions for long-non-coding RNA (lncRNA) and microRNA (miRNA) in androgen receptor (AR) independence/crosstalk signaling in castration-resistant prostate cancer cells. A number of miRNAs that are regulated by lncRNAs and involve the activation of AR crosstalk (AR dependent) and AR bypass (AR independent) signaling axis are demonstrated and further discussed in the present manuscript.
5. LncRNA–miRNA Interaction and Their Roles in AR-Independent/Crosstalk Mechanisms
5.1. Growth Factors
Growth factors include insulin-like growth factor-1 (IGF-1), fibroblast growth factor (FGF), and epidermal growth factor (EGF). IGF-1 and insulin-like growth factor 1 receptor (IGF1R) is the most extensively studied growth factor pathway involving AR signaling [73]. IGF1R can interact with AR and modulate the translocation of AR from the cytoplasm to the nucleus and thus alter AR activity [74]. On the other hand, IGF1R can promote AR signaling by stimulating AR coactivators such as transcriptional intermediary factor 2 (TIF2) [74]. Moreover, EGF signal transduction can also activate AR via a ligand-independent pathway [73]. In contrast, FGF activates MAPK signaling and bypasses the AR in promoting CRPC tumor growth by inducing the expression of inhibitor of differentiation 1 (ID1) [29].
Since IGF1R, EGFR, and FGFR are receptor tyrosine kinases (RTKs), the role of RTKs and their downstream signal cascade activation may take part in shifting phenotypic and molecular landscapes of metastatic CRPC. A substantial body of evidence has documented that lncRNA–miRNA interactions are involved in these signaling pathways due to their distinctive multifaceted features. Liu et al. assessed the expression of lncRNA AFAP1-AS1 in castration-resistant C4-2, PC3, and DU145 cell lines and found that AFAP1-AS1 level was significantly elevated compared with the androgen-sensitive LNCaP cells. AFAP1-AS1 could bind to miR-15b and disturb its tumor suppressor role to repress the IGF1R [59]. Huang et al. detected that lncRNA PTTG3P was remarkably increased in the castration-resistant cell lines and CRPC tissues. PTTG3P could upregulate PTTG1 expression by competing for miR-146a-3p [60]. In vitro experiments have shown that the PTTG-binding factor directly binds PTTG to promote PTTG nuclear translocation and subsequently increase the expression of FGF mRNA [75].
5.2. Oncogenic Signaling Pathways
LncRNA SChLAP1 was found highly expressed in PCa and closely related to tumor progression. SChLAP1 could downregulate the expression of miR-198, and miR-198 had a targeted correlation with 3’ UTR of MAPK1. The miR-198 inhibition significantly increased the expression of MAPK1, and its major target genes such as ELK-1, F-actin, and PAK1 are related to cancer progression [61].
Upregulated lncRNA Linc00963 was detected in C4-2 cells compared to LNCaP cells, indicating its role in transitioning from androgen-dependent PCa to androgen-independent CRPC [76]. Linc00963 directly bound to miR-655 in CRPC cells and inhibited its interaction with TRIM24 mRNA, promoting cell proliferation by enhancing TRIM24 expression [62]. TRIM24 is a well-known oncogene in advanced CRPC [77]. TRIM24 expression could promote CRPC by activating AR through PI3K/AKT pathway at shallow androgen levels. Additionally, TRIM24 was found to be a transcriptional regulator of EGFR and PIK3CA genes, while PIK3CA and EGFR demonstrated synergetic effects on the PI3K/AKT pathway activation in PCa [62].
LncRNA MYU(VPS9D1-AS1) upregulated the c-Myc transcription level by competitively binding with miR-184 [63]. MYC protein forms a heterodimer with the related transcription factor MAX. This complex acts as transcription factors that activate the expression of multiple proliferative genes through binding to enhancer box sequences and recruiting histone acetyltransferases [78]. Interestingly, MYC is activated by various mitogenic signals such as serum stimulation or by Wnt, Shh, and EGF through the MAPK/ERK pathway [79]. This phenomenon implies the close relationship of the cell surface receptor and intracellular signal networks in promoting PCa cell survival in the absence of androgen or being independent of AR activation.
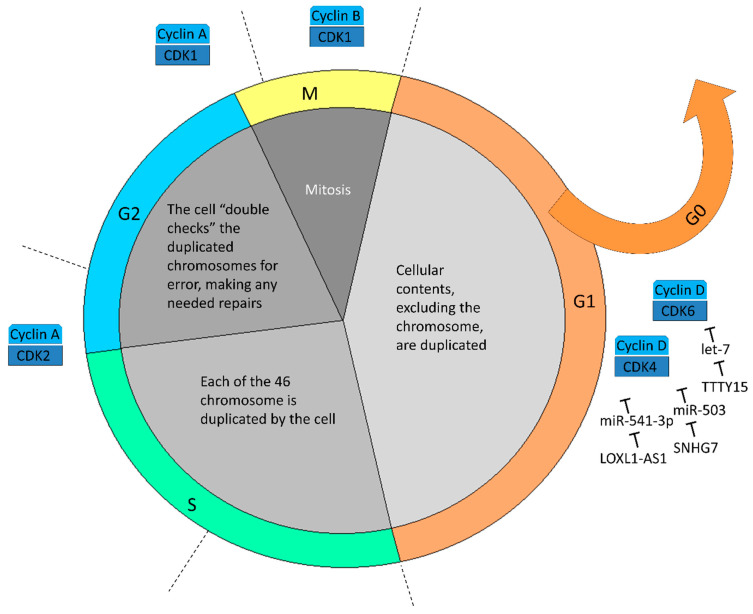
5.3. Cell Cycle Dysregulation
The lncRNA SNHG7 level is also over-expressed in PCa tissue and cell lines, being closely correlated with poor prognosis. Knockdown of SNHG7 significantly decreased the level of cell-cycle regulators, including Cyclin D, CDK4, and CDK6. In vivo and in vitro experiments confirmed the cell cycle arrest at G0/G1 phase after silencing of SNHG7 [64]. In addition, MiR-503 was found to have complementary binding sites with 3’UTR of SNHG7 and Cyclin D. Transfection with miR-503 mimics could downregulate the SNHG7 and Cyclin D mRNA expression and suppress tumor growth [64].
LncRNA LOXL1-AS1 was predominantly located in the PCa cell cytoplasm and functioned as a miRNA sponge to interact with miR-541-3p [65]. MiR-541-3p could inhibit Cyclin D expression by binding with its 3’UTR and induce G0/G1 phase cell cycle arrest. LOXL1-AS1 induced upregulated genes significantly enriched in cell cycle checkpoint and nuclear division. Collectively, LOXL1-AS1 regulated cell cycle progression through modulating the expression of miR-541-3p and, subsequently, the expression of Cyclin D [65].
Y chromosome-transcribed lncRNA TTTY15 was also mainly located in the cytoplasm and acted as a ceRNA to absorb the let-7 family members let-7a, let-7b, let-7c, and let-7f. Among the let-7 targets, CDK6 and fibronectin (FN1) were of clinical interest. CDK6 regulates cell proliferation, and FN1 plays a vital role in cancer metastasis [80]. Furthermore, FN1 has been shown to stimulate the gonadal steroids that interact with vertebrate ARs, capable of controlling the expression of Cyclin D and related genes involved in cell cycle control. Taken together, TTTY15 may promote PCa progression by sponging let-7 family members and enhancing CDK6 and FN1 expression [66]. As a monoallele on the Y chromosome, the TTTY15 gene has a unique advantage for gene editing therapy and thus may possess more therapeutic potential. Lastly, Figure 2 summarizes current molecular shreds of evidence of lncRNAs and miRNAs in the cell cycle progression of CRPC.
Figure 2.
The mechanisms of actions for lncRNAs and miRNAs in cell cycle progression in castration-resistant prostate cancer cells. Most of the miRNAs in contemporary pieces of evidence function in the canonical checkpoint that prevents G1 phase cells from entering the S phase. Cancer cells overwhelm pRb growth suppression through inactivation of pRb by Cyclin D-CDK4/6. MiRNAs inhibit Cyclin D-CDK4/6, but their inhibitory ability is abolished by lncRNAs indicated above.
5.4. Cytokines and Other Transmembrane Proteins
Integrins are transmembrane proteins that facilitate cell-to-cell and cell-to-extracellular matrix adhesion [81]. CD51, otherwise named integrin subunit alpha V (ITGAV), is a member of the integrin alpha chain family [82]. CD51 inhibited E-cadherin expression and activated transforming growth factor-beta (TGF-β), a well-known multifunctional cytokine [83]. Mechanical tension of the cells alongside binding of the large latent TGF-β complex exposes the active TGF-β through the latency-associated peptide (LAP) opening. By binding to TGF-β receptor on neighboring cells, activated TGF-β acts on the surrounding stromal, immune, endothelial, and smooth muscle cells. It results in immunosuppression and angiogenesis, finally enhancing the invasiveness of cancer cells [84]. LncRNA SNHG17 level is elevated in CRPC cell lines and tissues and is positively correlated with CD51 expression. Mechanically, SNHG17 directly bound with miR-144 and inhibited miR-144 targeting downstream CD51. Herein, SNHG17 enhanced cell proliferation and invasion in vitro and promoted tumor growth and metastasis in vivo [67].
6. The Interplay between lncRNA and miRNA/Epigenetic Regulators in NEPC
6.1. General Introduction of NEPC
NEPC is a rare subtype of prostate cancer. From 2004 to 2013, only 0.06% of patients were classified to de novo NEPC among patients diagnosed with prostate cancer in the Surveillance, Epidemiology, and End Results (SEER) database [85]. However, with the advancement of AR-targeting therapy, NEPC can be detected more and more nowadays. Compared to de novo NEPC, patients who received ARSI may develop NEPC, called treatment-emergent neuroendocrine prostate cancer (t-NEPC). The t-NEPC is aggressive and lethal. Clinically, t-NEPC was found to have a higher visceral organs metastatic rate, a higher tumor cell proliferation rate, a higher resistance rate of ADT, and a poor prognosis [86]. Moreover, t-NEPC is AR-independent with negativity for PSA, and its prevalence rate is about 10~20% among patients who developed mCRPC [87]. Therefore, the way in which to treat t-NEPC has become an important issue in the era of ARSI. The mechanism of NEPC development is still limited knowledge today. There are two possible proposals as follows: (1) genomic mutation, (2) epigenetic/non-coding RNA interactome (ENI). We describe them separately here.
6.2. Genomic Mutation and NEPC
In recent evidence, retinoblastoma (Rb), p53, PTEN, AURKA, MYCN, and SRRM4 were found to be associated with NEPC. Loss of Rb gene, p53, and PTEN was present in more than 50% of NEPC [32,88,89,90]. Inactivation of Rb and p53 in PCa may increase the expression of SOX2, which is a reprogramming transcription factor, thus inducing stem cell-like status and promoting lineage plasticity [33]. Moreover, amplification and overexpression of AURKA and MYCN were present above 70% in NEPC but only 5% in ordinary PCa [91]. Moreover, N-Myc and AKT1 induce adenocarcinoma from prostate epithelial cells into neuroendocrine tumors [92]. Notably, overexpression of SRRM4 was also found in NEPC and might induce NEPC development by SOX2 expression, which promotes stem cell-like status and lineage plasticity [93,94]. Moreover, SRRM4 targets repressor element-1 silencing transcription factor (REST), which drives neuroendocrine transdifferentiation [95]. In brief, loss of tumor suppressor genes (such as Rb, p53, and PTEN) or protooncogene activation (such as AURKA, MYCN, and SRRM4) may induce lineage plasticity and drive NEPC development.
6.3. ENI and NEPC
Emerging evidence has revealed t-NEPC development from PCa by transdifferentiation of luminal cells into neuroendocrine cells. It was described in many studies that epigenetic alterations might drive NEPC formation. Moreover, EZH2, SOX2, REST, FOXA1, FOXA2, BRN2, ONECUT2, E2F1, and ASCL1 were found to be associated with NEPC progression. In NEPC, Rb inactive or N-Myc activation can induce EZH2 overexpression, promoting t-NEPC development [96,97]. In addition, ADT can induce CREB (cAMP response element-binding protein)-EZH2-TSP1 (thrombospondin-1) pathway, which promotes neuroendocrine differentiation and angiogenesis in advanced PCa [98]. Moreover, N-Myc interacts with EZH2 and promotes the reprogramming of cells into a stem-like cell status. This step may speed up neuroendocrine differentiation in advanced PCa [97]. As mentioned above, SOX2 overexpression promotes cancer stem-like cell formation and lineage plasticity and NEPC progression, regulated by Rb, p53, and SRRM4 [33,94]. Moreover, REST, also known as neuron-restrictive silencer factor (NRSF), is a transcriptional repressor of neuronal differentiation. REST influences neuroendocrine transdifferentiation, associated with SRRM4 expression and epithelial–mesenchymal transition (EMT) [95,99]. The association between EMT and cancer stem-like cells was demonstrated in previous reports [100]. The crosstalk between the WNT–β-catenin and IL-6-STAT3 pathways was found in EMT/cancer stem-like cells in cancer cells [101]. It is reported that activation of the EMT may induce cellular-plasticity, stem-like cell status, and neuroendocrine differentiation in advanced Pca [102]. The FOXA family, including FOXA1 and FOXA2, is also associated with neuroendocrine differentiation in PCa. The high expression of FOXA2 was found in NEPC [103,104]. By contrast, FOXA1 inhibits neuroendocrine differentiation via IL-8 and the MAPK/ERK pathway in PCa [105]. In short conclusion, the overexpression of EZH2, SOX2, FOXA2, BRN2, ONECUT2, E2F1, and ASCL1 but the downregulation of FOXA1 and REST may promote NEPC development [106,107,108,109,110].
6.4. The Interplay between lncRNA and miRNA in NEPC
The interplay between lncRNA and miRNA/epigenetic regulators in NEPC is described in Table 2. According to previous studies, lncRNA including HOTAIR, H19, LINC00261, FENDRR, and MALAT1 could crosstalk with miRNA in NEPC. For example, HOTAIR and miR-31-5, LINC00261 and miR-8485, H19 and miR-675, FENDRR and miR-301b-3p/miR-18a-5p, and MALAT1 and miR-1 have been reported [111,112,113,114,115,116,117].
Table 2.
The interplay between lncRNA and miRNA/epigenetic regulators in NEPC.
| The Interplay between lncRNA and miRNA | ||||
|---|---|---|---|---|
| lncRNA | miRNA | Target | Pathway and Influence | Reference |
| HOTAIR | miR-31-5p | REST, EZH2 | A direct interaction network among AR, HOTAIR, ESR1, and miR-31-5p was proposed. MiR-31-5p inhibits the expression of AR, and then AR transcriptionally inhibits HOTAIR. | [111,112] |
| LINC00261 | miR-8485 | CBX2, FOXA2 | In the cytoplasm, LINC00261 binds to miR-8485, which reduces the inhibition of CBX2. In the nucleus, LINC00261 activates FOXA2 expression via the SMAD2/3 transcriptional complex. |
[113] |
| H19 | miR-675 | TGF-β1 | H19 and miR-675 negatively regulate the expression of TGFβ1, inhibiting prostate cancer migration. | [114] |
| FENDRR (FOXF1-AS1) | miR-301b-3p | CSNK1E, PRC2 | Reduce tumor invasion by targeting CSNK1E. | [115] |
| miR-18a-5p | RUNX1 | FENDRR inhibits tumor cell proliferation by binding to miR-18a-5p with RUNX1. | [116] | |
| MALAT1 | miR-1 | MALAT1 acts as a sponge of miR-1, resulting in downregulating KRAS in AR independent prostate cancer. | [117] | |
| The interplay between lncRNA and epigenetic regulators | ||||
| LncRNA-p21 | EZH2 | LncRNA-p21 promotes EZH2 to enhance STAT3 methylation and drives neuroendocrine transdifferentiation. | [118] | |
| MIAT | Polycomb genes | MIAT interacts with polycomb genes and is positively associated with Rb mutation and CBX2 expression. | [111,119] | |
| LINC00514 | TADA3 | LINC00514 interacts with TADA3 and reduces p53 activity. | [120] | |
| SSTR5-AS1 | KDM4B | SSTR5-AS1 binds with KDM4B. KDM4B interacts with N-Myc, which drives the progression of NEPC. | [120] | |
Notably, it was proposed that there was a direct interaction network among AR, HOTAIR, ESR1, and miR-31-5p. It was found in NEPC that miR-31-5p inhibits the expression of AR, while AR transcriptionally inhibits HOTAIR [111]. Otherwise, HOTAIR is a REST-repressed lncRNA that drives NEPC development [112]. Interestingly, AR itself, in turn, can repress miR-31 transcription, and HOTAIR can enhance both AR and ESR1 [111]. The four molecules form a mutual regulatory model in NEPC.
Mather et al. reported that a highly conserved lncRNA, LINC00261, would bind with miR-8485 to reduce its inhibition to chromobox 2 (CBX2) in the cytoplasm [113]. CBX2 overexpression can promote tumor proliferation, while the overexpression of miR-8485 can inhibit the expression of CBX2. On the other hand, LINC00261 actives forkhead box A2 (FOXA2) expression via SMAD2/3 transcriptional complex in the nucleus, while FOXA2 overexpression may induce neuroendocrine differentiation.
Zhu et al. demonstrated lncRNA H19 suppresses tumor cell migration via miR-675. MiR-675 directly binds to the 3’UTR of TGFβ1 mRNA that inhibits its translocation to inhibit tumor migration [114]. In addition, FENDRR, also known as FOXF1-AS1, can reduce tumor invasion by targeting CSNK1E in PCa. Meanwhile, miR-301b-3p plays a role in upregulatory tumor progression, although the interaction is not clear between FENDRR and miR-301b-3p [115]. Moreover, it was proposed that FENDRR inhibits tumor cell proliferation and migration by binding to miR-18a-5p with RUNX1 [116].
Recently, the association between MALAT1 and NEPC was demonstrated by Ostano et al. [111]. Although the mechanism between MALAT1 and miRNA in NEPC remains unclear, the relation between MALAT1 and miR-1 in AR-independent PCa cell lines was shown. In PCa cell lines, MALAT1 expression was upregulated, while miR-1 expression was downregulated. Chang et al. proposed the idea that MALAT1 acts as a molecular sponge of miR-1, resulting in downregulating KRAS in AR-independent PCa cell lines [117]. In this way, tumor cell proliferation and migration are suppressed by silencing MALA1.
In addition to the above, there were other lncRNAs interacting with epigenetic regulators in NEPC, including lncRNA-p21, MIAT, LINC00514, and SSTR5-AS1. For example, lncRNA-p21 interacts with EZH2 and disrupts the PRC2 complex. Then, lncRNA-p21 promotes EZH2 to enhance STAT3 methylation and drive neuroendocrine transdifferentiation. This phenomenon was observed when patients were treated with enzalutamide [118]. Crea et al. demonstrated that MIAT interacts with polycomb genes and is positively associated with higher metastatic potential, Rb mutation, and CBX2 expression [119]. LINC00514 links with TADA3 and inhibits its activity in NEPC, while TADA3 can bind and stabilize p53 [120,121]. Regarding SSTR5-AS1, it is the highest expressed lncRNA in NEPC and binds with KDM4B, a histone demethylase and a key molecule in AR signaling [120]. In addition, KDM4B influences AR transcriptional activity in prostate adenocarcinoma and interacts with N-Myc in neuroblastoma [122,123]. Because N-Myc plays a vital role in neuroendocrine transdifferentiation, KDM4B may drive NEPC via N-Myc [120].
7. Conclusions
In recent years, ncRNA biology has been regarded as an important role in our understanding of PCa progression, especially in the era of AR-targeting agents. In order to deal with a complete diminishment of direct AR activation, cancer cells have started learning to “survive” in the environment poverty of AR signaling. As a member of the steroid hormone nuclear receptor family, AR can act as a nuclear transcription factor with its coactivators and directly binds to androgen response elements (AREs) to mediate its target gene function. However, cancer cells can still bypass AR-binding-independent pathways via activation of second messengers such as MAPK, ERK, or AKT. Growth factors or cytokines are also crucial for PCa to develop castration resistance.
On the other hand, due to the widespread use of AR-targeting agents, the prevalence of NEPC also increased in decades. LncRNA–miRNA–mRNA networks appear to be widespread in PCa cells and tissues. LncRNAs are also recognized as master epigenetic regulators of the genome in the development of NEPC. Furthermore, an investigation into the mechanisms of ncRNA actions is a rising issue due to the relative accessibility to construct their mimics and put them into therapeutic potential. Methods are now available for interfering lncRNAs, but the application of these approaches is still immature. However, our current literature review rendered a curation primarily focused on the interaction of lncRNA and miRNA in CRPC and NEPC. More attention and research in this field are warranted.
Author Contributions
Conceptualization, T.-Y.L. and C.-C.C.; methodology, C.-Y.H. and K.-Y.W.; formal analysis, C.-Y.H. and K.-Y.W.; investigation, all authors; data curation, T.-Y.L. and C.-C.C.; writing—original draft preparation, C.-Y.H. and K.-Y.W.; writing–review and editing, T.-Y.L. and C.-C.C.; visualization, C.-Y.H. and T.-Y.L.; supervision, T.-Y.L. and C.-C.C.; project administration, T.-Y.L. and C.-C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Bell K.J., Del Mar C., Wright G., Dickinson J., Glasziou P. Prevalence of incidental prostate cancer: A systematic review of autopsy studies. Int. J. Cancer. 2015;137:1749–1757. doi: 10.1002/ijc.29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Litwin M.S., Tan H.J. The diagnosis and treatment of prostate cancer: A review. JAMA. 2017;317:2532–2542. doi: 10.1001/jama.2017.7248. [DOI] [PubMed] [Google Scholar]
- 4.Mottet N., Bellmunt J., Bolla M., Briers E., Cumberbatch M.G., De Santis M., Fossati N., Gross T., Henry A.M., Joniau S., et al. EAU-ESTRO-SIOG Guidelines on prostate cancer. Part 1: Screening, diagnosis, and local treatment with curative intent. Eur. Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Cornford P., van den Bergh R.C.N., Briers E., Van den Broeck T., Cumberbatch M.G., De Santis M., Fanti S., Fossati N., Gandaglia G., Gillessen S., et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on prostate cancer. Part II-2020 update: Treatment of relapsing and metastatic prostate cancer. Eur. Urol. 2021;79:263–282. doi: 10.1016/j.eururo.2020.09.046. [DOI] [PubMed] [Google Scholar]
- 6.Gillessen S., Omlin A., Attard G., de Bono J.S., Efstathiou E., Fizazi K., Halabi S., Nelson P.S., Sartor O., Smith M.R., et al. Management of patients with advanced prostate cancer: Recommendations of the St Gallen Advanced Prostate Cancer Consensus Conference (APCCC) 2015. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015;26:1589–1604. doi: 10.1093/annonc/mdv257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C.D., Welsbie D.S., Tran C., Baek S.H., Chen R., Vessella R., Rosenfeld M.G., Sawyers C.L. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 8.Watson P.A., Arora V.K., Sawyers C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015;15:701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornford P., Bellmunt J., Bolla M., Briers E., De Santis M., Gross T., Henry A.M., Joniau S., Lam T.B., Mason M.D., et al. EAU-ESTRO-SIOG Guidelines on prostate cancer. Part II: Treatment of relapsing, metastatic, and castration-resistant prostate cancer. Eur. Urol. 2017;71:630–642. doi: 10.1016/j.eururo.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Trock B.J., Han M., Freedland S.J., Humphreys E.B., DeWeese T.L., Partin A.W., Walsh P.C. Prostate cancer-specific survival following salvage radiotherapy vs observation in men with biochemical recurrence after radical prostatectomy. JAMA. 2008;299:2760–2769. doi: 10.1001/jama.299.23.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Kirby M., Hirst C., Crawford E.D. Characterising the castration-resistant prostate cancer population: A systematic review. Int. J. Clin. Pract. 2011;65:1180–1192. doi: 10.1111/j.1742-1241.2011.02799.x. [DOI] [PubMed] [Google Scholar]
- 13.Aly M., Leval A., Schain F., Liwing J., Lawson J., Vágó E., Nordström T., Andersson T.M., Sjöland E., Wang C., et al. Survival in patients diagnosed with castration-resistant prostate cancer: A population-based observational study in Sweden. Scand. J. Urol. 2020;54:115–121. doi: 10.1080/21681805.2020.1739139. [DOI] [PubMed] [Google Scholar]
- 14.De Bono J.S., Oudard S., Ozguroglu M., Hansen S., Machiels J.P., Kocak I., Gravis G., Bodrogi I., Mackenzie M.J., Shen L., et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: A randomised open-label trial. Lancet. 2010;376:1147–1154. doi: 10.1016/S0140-6736(10)61389-X. [DOI] [PubMed] [Google Scholar]
- 15.Kantoff P.W., Higano C.S., Shore N.D., Berger E.R., Small E.J., Penson D.F., Redfern C.H., Ferrari A.C., Dreicer R., Sims R.B., et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 16.Fizazi K., Scher H.I., Molina A., Logothetis C.J., Chi K.N., Jones R.J., Staffurth J.N., North S., Vogelzang N.J., Saad F., et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: Final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012;13:983–992. doi: 10.1016/S1470-2045(12)70379-0. [DOI] [PubMed] [Google Scholar]
- 17.Scher H.I., Fizazi K., Saad F., Taplin M.E., Sternberg C.N., Miller K., de Wit R., Mulders P., Chi K.N., Shore N.D., et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 18.Parker C., Nilsson S., Heinrich D., Helle S.I., O’Sullivan J.M., Fosså S.D., Chodacki A., Wiechno P., Logue J., Seke M., et al. Alpha emitter radium-223 and survival in metastatic prostate cancer. N. Engl. J. Med. 2013;369:213–223. doi: 10.1056/NEJMoa1213755. [DOI] [PubMed] [Google Scholar]
- 19.Ryan C.J., Smith M.R., Fizazi K., Saad F., Mulders P.F., Sternberg C.N., Miller K., Logothetis C.J., Shore N.D., Small E.J., et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): Final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2015;16:152–160. doi: 10.1016/S1470-2045(14)71205-7. [DOI] [PubMed] [Google Scholar]
- 20.Beer T.M., Armstrong A.J., Rathkopf D., Loriot Y., Sternberg C.N., Higano C.S., Iversen P., Evans C.P., Kim C.S., Kimura G., et al. Enzalutamide in men with chemotherapy-naïve metastatic castration-resistant prostate cancer: Extended analysis of the phase 3 PREVAIL study. Eur. Urol. 2017;71:151–154. doi: 10.1016/j.eururo.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scher H.I., Sawyers C.L. Biology of progressive, castration-resistant prostate cancer: Directed therapies targeting the androgen-receptor signaling axis. J. Clin. Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 22.Hoang D.T., Iczkowski K.A., Kilari D., See W., Nevalainen M.T. Androgen receptor-dependent and -independent mechanisms driving prostate cancer progression: Opportunities for therapeutic targeting from multiple angles. Oncotarget. 2017;8:3724–3745. doi: 10.18632/oncotarget.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Qi M., Zhang J., Sun X., Guo H., Pang Y., Zhang Q., Chen X., Zhang R., Liu Z., et al. Differential response to neoadjuvant hormonal therapy in prostate cancer: Predictive morphological parameters and molecular markers. Prostate. 2019;79:709–719. doi: 10.1002/pros.23777. [DOI] [PubMed] [Google Scholar]
- 24.De Winter J.A., Trapman J., Brinkmann A.O., Boersma W.J., Mulder E., Schroeder F.H., Claassen E., van der Kwast T.H. Androgen receptor heterogeneity in human prostatic carcinomas visualized by immunohistochemistry. J. Pathol. 1990;160:329–332. doi: 10.1002/path.1711600409. [DOI] [PubMed] [Google Scholar]
- 25.Choucair K., Ejdelman J., Brimo F., Aprikian A., Chevalier S., Lapointe J. PTEN genomic deletion predicts prostate cancer recurrence and is associated with low AR expression and transcriptional activity. BMC Cancer. 2012;12:543. doi: 10.1186/1471-2407-12-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q., Deng Q., Chao H.P., Liu X., Lu Y., Lin K., Liu B., Tang G.W., Zhang D., Tracz A., et al. Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses. Nat. Commun. 2018;9:3600. doi: 10.1038/s41467-018-06067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vellky J.E., Ricke W.A. Development and prevalence of castration-resistant prostate cancer subtypes. Neoplasia. 2020;22:566–575. doi: 10.1016/j.neo.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makino T., Izumi K., Mizokami A. Undesirable status of prostate cancer cells after intensive inhibition of AR signaling: Post-AR era of CRPC treatment. Biomedicines. 2021;9:414. doi: 10.3390/biomedicines9040414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bluemn E.G., Coleman I.M., Lucas J.M., Coleman R.T., Hernandez-Lopez S., Tharakan R., Bianchi-Frias D., Dumpit R.F., Kaipainen A., Corella A.N., et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–489.e6. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng Q., Tang D.G. Androgen receptor and prostate cancer stem cells: Biological mechanisms and clinical implications. Endocr.-Relat. Cancer. 2015;22:T209–T220. doi: 10.1530/ERC-15-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin D., Wyatt A.W., Xue H., Wang Y., Dong X., Haegert A., Wu R., Brahmbhatt S., Mo F., Jong L., et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014;74:1272–1283. doi: 10.1158/0008-5472.CAN-13-2921-T. [DOI] [PubMed] [Google Scholar]
- 32.Ku S.Y., Rosario S., Wang Y., Mu P., Seshadri M., Goodrich Z.W., Goodrich M.M., Labbé D.P., Gomez E.C., Wang J., et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mu P., Zhang Z., Benelli M., Karthaus W.R., Hoover E., Chen C.C., Wongvipat J., Ku S.Y., Gao D., Cao Z., et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou M., Toivanen R., Mitrofanova A., Floch N., Hayati S., Sun Y., Le Magnen C., Chester D., Mostaghel E.A., Califano A., et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 2017;7:736–749. doi: 10.1158/2159-8290.CD-16-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shih J.W., Wang L.Y., Hung C.L., Kung H.J., Hsieh C.L. Non-coding RNAs in castration-resistant prostate cancer: Regulation of androgen receptor signaling and cancer metabolism. Int. J. Mol. Sci. 2015;16:28943–28978. doi: 10.3390/ijms161226138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kojima S., Goto Y., Naya Y. The roles of microRNAs in the progression of castration-resistant prostate cancer. J. Hum. Genet. 2017;62:25–31. doi: 10.1038/jhg.2016.69. [DOI] [PubMed] [Google Scholar]
- 37.Ding L., Wang R., Shen D., Cheng S., Wang H., Lu Z., Zheng Q., Wang L., Xia L., Li G. Role of noncoding RNA in drug resistance of prostate cancer. Cell Death Dis. 2021;12:590. doi: 10.1038/s41419-021-03854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santosh B., Varshney A., Yadava P.K. Non-coding RNAs: Biological functions and applications. Cell Biochem. Funct. 2015;33:14–22. doi: 10.1002/cbf.3079. [DOI] [PubMed] [Google Scholar]
- 39.Panni S., Lovering R.C., Porras P., Orchard S. Non-coding RNA regulatory networks. Biochim. Biophys. Acta—Gene Regul. Mech. 2020;1863:194417. doi: 10.1016/j.bbagrm.2019.194417. [DOI] [PubMed] [Google Scholar]
- 40.Hu W., Liu C., Bi Z.Y., Zhou Q., Zhang H., Li L.L., Zhang J., Zhu W., Song Y.Y., Zhang F., et al. Comprehensive landscape of extracellular vesicle-derived RNAs in cancer initiation, progression, metastasis and cancer immunology. Mol. Cancer. 2020;19:102. doi: 10.1186/s12943-020-01199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim M., Baird A.M., Aird J., Greene J., Kapoor D., Gray S.G., McDermott R., Finn S.P. RNAs as candidate diagnostic and prognostic markers of prostate cancer-from cell line models to liquid biopsies. Diagnostics. 2018;8:60. doi: 10.3390/diagnostics8030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramnarine V.R., Kobelev M., Gibb E.A., Nouri M., Lin D., Wang Y., Buttyan R., Davicioni E., Zoubeidi A., Collins C.C. The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur. Urol. 2019;76:546–559. doi: 10.1016/j.eururo.2019.07.040. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y., Pitchiaya S., Cieślik M., Niknafs Y.S., Tien J.C., Hosono Y., Iyer M.K., Yazdani S., Subramaniam S., Shukla S.K., et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat. Genet. 2018;50:814–824. doi: 10.1038/s41588-018-0120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., Merkurjev D., Ohgi K.A., Meng D., Zhang J., et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bardhan A., Banerjee A., Basu K., Pal D.K., Ghosh A. PRNCR1: A long non-coding RNA with a pivotal oncogenic role in cancer. Hum. Genet. 2021:1–15. doi: 10.1007/s00439-021-02396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aird J., Baird A.M., Lim M., McDermott R., Finn S.P., Gray S.G. Carcinogenesis in prostate cancer: The role of long non-coding RNAs. Noncoding RNA Res. 2018;3:29–38. doi: 10.1016/j.ncrna.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma G., Tang M., Wu Y., Xu X., Pan F., Xu R. LncRNAs and miRNAs: Potential biomarkers and therapeutic targets for prostate cancer. Am. J. Transl. Res. 2016;8:5141–5150. [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y., Liu K.Y., Liu Q., Cao Q. Androgen receptor-related non-coding RNAs in prostate cancer. Front. Cell Dev. Biol. 2021;9:660853. doi: 10.3389/fcell.2021.660853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam C., Wong J.H., Tsui S.K.W., Zuo T., Chan T.F., Ng T.B. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: Updates in recent years. Appl. Microbiol. Biotechnol. 2019;103:4649–4677. doi: 10.1007/s00253-019-09837-5. [DOI] [PubMed] [Google Scholar]
- 50.Sun B., Liu C., Li H., Zhang L., Luo G., Liang S., Lü M. Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol. Lett. 2020;19:595–605. doi: 10.3892/ol.2019.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J., Xu T., Liu L., Zhang W., Zhao C., Li S., Li J., Rao N., Le T.D. LMSM: A modular approach for identifying lncRNA related miRNA sponge modules in breast cancer. PLoS Comput. Biol. 2020;16:e1007851. doi: 10.1371/journal.pcbi.1007851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.López-Urrutia E., Bustamante Montes L.P., Ladrón de Guevara Cervantes D., Pérez-Plasencia C., Campos-Parra A.D. Crosstalk between long non-coding RNAs, micro-RNAs and mRNAs: Deciphering molecular mechanisms of master regulators in cancer. Front. Oncol. 2019;9:669. doi: 10.3389/fonc.2019.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S., et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin W., Zhou Q., Wang C.Q., Zhu L., Bi C., Zhang S., Wang X., Jin H. LncRNAs regulate metabolism in cancer. Int. J. Biol. Sci. 2020;16:1194–1206. doi: 10.7150/ijbs.40769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayr C., Bartel D.P. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park H.J., Ji P., Kim S., Xia Z., Rodriguez B., Li L., Su J., Chen K., Masamha C.P., Baillat D., et al. 3’ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat. Genet. 2018;50:783–789. doi: 10.1038/s41588-018-0118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang Z., Xu C., Li Y., Cai X., Ren S., Liu H., Wang Y., Wang F., Chen R., Qu M., et al. A feed-forward regulatory loop between androgen receptor and PlncRNA-1 promotes prostate cancer progression. Cancer Lett. 2016;374:62–74. doi: 10.1016/j.canlet.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 58.You Z., Liu C., Wang C., Ling Z., Wang Y., Wang Y., Zhang M., Chen S., Xu B., Guan H., et al. LncRNA CCAT1 promotes prostate cancer cell proliferation by interacting with DDX5 and MIR-28-5P. Mol. Cancer Ther. 2019;18:2469–2479. doi: 10.1158/1535-7163.MCT-19-0095. [DOI] [PubMed] [Google Scholar]
- 59.Liu B., Jiang H.Y., Yuan T., Zhou W.D., Xiang Z.D., Jiang Q.Q., Wu D.L. Long non-coding RNA AFAP1-AS1 facilitates prostate cancer progression by regulating miR-15b/IGF1R axis. Curr. Pharm. Des. 2021;27:4261–4269. doi: 10.2174/1381612827666210612052317. [DOI] [PubMed] [Google Scholar]
- 60.Huang S., Liao Q., Li W., Deng G., Jia M., Fang Q., Ji H., Meng M. The lncRNA PTTG3P promotes the progression of CRPC via upregulating PTTG1. Bull. Cancer. 2021;108:359–368. doi: 10.1016/j.bulcan.2020.11.022. [DOI] [PubMed] [Google Scholar]
- 61.Li Y., Luo H., Xiao N., Duan J., Wang Z., Wang S. Long noncoding RNA SChLAP1 accelerates the proliferation and metastasis of prostate cancer via targeting miR-198 and promoting the MAPK1 pathway. Oncol. Res. 2018;26:131–143. doi: 10.3727/096504017X14944585873631. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Bai M., He C., Shi S., Wang M., Ma J., Yang P., Dong Y., Mou X., Han S. Linc00963 promote cell proliferation and tumor growth in castration-resistant prostate cancer by modulating miR-655/TRIM24 axis. Front. Oncol. 2021;11:636965. doi: 10.3389/fonc.2021.636965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J., Yang X., Li R., Wang L., Gu Y., Zhao Y., Huang K.H., Cheng T., Yuan Y., Gao S. Long non-coding RNA MYU promotes prostate cancer proliferation by mediating the miR-184/c-Myc axis. Oncol. Rep. 2018;40:2814–2825. doi: 10.3892/or.2018.6661. [DOI] [PubMed] [Google Scholar]
- 64.Qi H., Wen B., Wu Q., Cheng W., Lou J., Wei J., Huang J., Yao X., Weng G. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed. Pharmacother. 2018;102:326–332. doi: 10.1016/j.biopha.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 65.Long B., Li N., Xu X.X., Li X.X., Xu X.J., Liu J.Y., Wu Z.H. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem. Biophys. Res. Commun. 2018;505:561–568. doi: 10.1016/j.bbrc.2018.09.160. [DOI] [PubMed] [Google Scholar]
- 66.Xiao G., Yao J., Kong D., Ye C., Chen R., Li L., Zeng T., Wang L., Zhang W., Shi X., et al. The long noncoding RNA TTTY15, which is located on the Y chromosome, promotes prostate cancer progression by sponging let-7. Eur. Urol. 2019;76:315–326. doi: 10.1016/j.eururo.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 67.Bai M., Lei Y., Wang M., Ma J., Yang P., Mou X., Dong Y., Han S. Long non-coding RNA SNHG17 promotes cell proliferation and invasion in castration-resistant prostate cancer by targeting the miR-144/CD51 axis. Front. Genet. 2020;11:274. doi: 10.3389/fgene.2020.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiyomaru T., Yamamura S., Fukuhara S., Yoshino H., Kinoshita T., Majid S., Saini S., Chang I., Tanaka Y., Enokida H., et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS ONE. 2013;8:e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ling Z., Wang X., Tao T., Zhang L., Guan H., You Z., Lu K., Zhang G., Chen S., Wu J., et al. Involvement of aberrantly activated HOTAIR/EZH2/miR-193a feedback loop in progression of prostate cancer. J. Exp. Clin. Cancer Res. 2017;36:159. doi: 10.1186/s13046-017-0629-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang S., Li Z., Zhang L., Xu Z. MEF2activated long noncoding RNA PCGEM1 promotes cell proliferation in hormonerefractory prostate cancer through downregulation of miR148a. Mol. Med. Rep. 2018;18:202–208. doi: 10.3892/mmr.2018.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang X., Wang L., Li R., Zhao Y., Gu Y., Liu S., Cheng T., Huang K., Yuan Y., Song D., et al. The long non-coding RNA PCSEAT exhibits an oncogenic property in prostate cancer and functions as a competing endogenous RNA that associates with EZH2. Biochem. Biophys. Res. Commun. 2018;502:262–268. doi: 10.1016/j.bbrc.2018.05.157. [DOI] [PubMed] [Google Scholar]
- 72.Jiang T., Guo J., Hu Z., Zhao M., Gu Z., Miao S. Identification of potential prostate cancer-related pseudogenes based on competitive endogenous RNA network hypothesis. Med. Sci. Monit. 2018;24:4213–4239. doi: 10.12659/MSM.910886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Culig Z., Hobisch A., Cronauer M.V., Radmayr C., Trapman J., Hittmair A., Bartsch G., Klocker H. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res. 1994;54:5474–5478. doi: 10.1159/000475232. [DOI] [PubMed] [Google Scholar]
- 74.Wu J.D., Haugk K., Woodke L., Nelson P., Coleman I., Plymate S.R. Interaction of IGF signaling and the androgen receptor in prostate cancer progression. J. Cell Biochem. 2006;99:392–401. doi: 10.1002/jcb.20929. [DOI] [PubMed] [Google Scholar]
- 75.Chien W., Pei L. A novel binding factor facilitates nuclear translocation and transcriptional activation function of the pituitary tumor-transforming gene product. J. Biol. Chem. 2000;275:19422–19427. doi: 10.1074/jbc.M910105199. [DOI] [PubMed] [Google Scholar]
- 76.Wang L., Han S., Jin G., Zhou X., Li M., Ying X., Wang L., Wu H., Zhu Q. Linc00963: A novel, long non-coding RNA involved in the transition of prostate cancer from androgen-dependence to androgen-independence. Int. J. Oncol. 2014;44:2041–2049. doi: 10.3892/ijo.2014.2363. [DOI] [PubMed] [Google Scholar]
- 77.Groner A.C., Cato L., de Tribolet-Hardy J., Bernasocchi T., Janouskova H., Melchers D., Houtman R., Cato A.C.B., Tschopp P., Gu L., et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29:846–858. doi: 10.1016/j.ccell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rahl P.B., Young R.A. MYC and transcription elongation. Cold Spring Harb. Perspect. Med. 2014;4:a020990. doi: 10.1101/cshperspect.a020990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campisi J., Gray H.E., Pardee A.B., Dean M., Sonenshein G.E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984;36:241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- 80.Jackstadt R., Roh S., Neumann J., Jung P., Hoffmann R., Horst D., Berens C., Bornkamm G.W., Kirchner T., Menssen A., et al. AP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancer. J. Exp. Med. 2013;210:1331–1350. doi: 10.1084/jem.20120812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hynes R.O. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 82.Conroy K.P., Kitto L.J., Henderson N.C. Alphav integrins: Key regulators of tissue fibrosis. Cell Tissue Res. 2016;365:511–519. doi: 10.1007/s00441-016-2407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hinz B. The extracellular matrix and transforming growth factor-beta1: Tale of a strained relationship. Matrix Biol. 2015;47:54–65. doi: 10.1016/j.matbio.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 84.Blobe G.C., Schiemann W.P., Lodish H.F. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 85.Zaffuto E., Pompe R., Zanaty M., Bondarenko H.D., Leyh-Bannurah S.R., Moschini M., Dell’Oglio P., Gandaglia G., Fossati N., Stabile A., et al. Contemporary incidence and cancer control outcomes of primary neuroendocrine prostate cancer: A SEER database analysis. Clin. Genitourin. Cancer. 2017;15:e793–e800. doi: 10.1016/j.clgc.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 86.Aggarwal R., Zhang T., Small E.J., Armstrong A.J. Neuroendocrine prostate cancer: Subtypes, biology, and clinical outcomes. J. Natl. Compr. Cancer Netw. JNCCN. 2014;12:719–726. doi: 10.6004/jnccn.2014.0073. [DOI] [PubMed] [Google Scholar]
- 87.Aggarwal R., Huang J., Alumkal J.J., Zhang L., Feng F.Y., Thomas G.V., Weinstein A.S., Friedl V., Zhang C., Witte O.N., et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J. Clin. Oncol. 2018;36:2492–2503. doi: 10.1200/JCO.2017.77.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tan H.L., Sood A., Rahimi H.A., Wang W., Gupta N., Hicks J., Mosier S., Gocke C.D., Epstein J.I., Netto G.J., et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin. Cancer Res. 2014;20:890–903. doi: 10.1158/1078-0432.CCR-13-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen H., Sun Y., Wu C., Magyar C.E., Li X., Cheng L., Yao J.L., Shen S., Osunkoya A.O., Liang C., et al. Pathogenesis of prostatic small cell carcinoma involves the inactivation of the P53 pathway. Endocr.-Relat. Cancer. 2012;19:321–331. doi: 10.1530/ERC-11-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng L., MacLennan G.T., Lopez-Beltran A., Montironi R. Anatomic, morphologic and genetic heterogeneity of prostate cancer: Implications for clinical practice. Expert Rev. Anticancer Ther. 2012;12:1371–1374. doi: 10.1586/era.12.127. [DOI] [PubMed] [Google Scholar]
- 91.Mosquera J.M., Beltran H., Park K., MacDonald T.Y., Robinson B.D., Tagawa S.T., Perner S., Bismar T.A., Erbersdobler A., Dhir R., et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia. 2013;15:1–10. doi: 10.1593/neo.121550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee J.K., Phillips J.W., Smith B.A., Park J.W., Stoyanova T., McCaffrey E.F., Baertsch R., Sokolov A., Meyerowitz J.G., Mathis C., et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29:536–547. doi: 10.1016/j.ccell.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang X., Coleman I.M., Brown L.G., True L.D., Kollath L., Lucas J.M., Lam H.M., Dumpit R., Corey E., Chéry L., et al. SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin. Cancer Res. 2015;21:4698–4708. doi: 10.1158/1078-0432.CCR-15-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee A.R., Gan Y., Tang Y., Dong X. A novel mechanism of SRRM4 in promoting neuroendocrine prostate cancer development via a pluripotency gene network. EBioMedicine. 2018;35:167–177. doi: 10.1016/j.ebiom.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y., Donmez N., Sahinalp C., Xie N., Wang Y., Xue H., Mo F., Beltran H., Gleave M., Wang Y., et al. SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur. Urol. 2017;71:68–78. doi: 10.1016/j.eururo.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 96.Bohrer L.R., Chen S., Hallstrom T.C., Huang H. Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: A potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology. 2010;151:5136–5145. doi: 10.1210/en.2010-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dardenne E., Beltran H., Benelli M., Gayvert K., Berger A., Puca L., Cyrta J., Sboner A., Noorzad Z., MacDonald T., et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30:563–577. doi: 10.1016/j.ccell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y., Zheng D., Zhou T., Song H., Hulsurkar M., Su N., Liu Y., Wang Z., Shao L., Ittmann M., et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 2018;9:4080. doi: 10.1038/s41467-018-06177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang Y.T., Lin T.P., Campbell M., Pan C.C., Lee S.H., Lee H.C., Yang M.H., Kung H.J., Chang P.C. REST is a crucial regulator for acquiring EMT-like and stemness phenotypes in hormone-refractory prostate cancer. Sci. Rep. 2017;7:42795. doi: 10.1038/srep42795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hao J., Li T.G., Qi X., Zhao D.F., Zhao G.Q. WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev. Biol. 2006;290:81–91. doi: 10.1016/j.ydbio.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 102.Soundararajan R., Paranjape A.N., Maity S., Aparicio A., Mani S.A. EMT, stemness and tumor plasticity in aggressive variant neuroendocrine prostate cancers. Biochim. Biophys. Acta—Rev. Cancer. 2018;1870:229–238. doi: 10.1016/j.bbcan.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chiaverotti T., Couto S.S., Donjacour A., Mao J.H., Nagase H., Cardiff R.D., Cunha G.R., Balmain A. Dissociation of epithelial and neuroendocrine carcinoma lineages in the transgenic adenocarcinoma of mouse prostate model of prostate cancer. Am. J. Pathol. 2008;172:236–246. doi: 10.2353/ajpath.2008.070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park J.W., Lee J.K., Witte O.N., Huang J. FOXA2 is a sensitive and specific marker for small cell neuroendocrine carcinoma of the prostate. Mod. Pathol. 2017;30:1262–1272. doi: 10.1038/modpathol.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim J., Jin H., Zhao J.C., Yang Y.A., Li Y., Yang X., Dong X., Yu J. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene. 2017;36:4072–4080. doi: 10.1038/onc.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rotinen M., You S., Yang J., Coetzee S.G., Reis-Sobreiro M., Huang W.C., Huang F., Pan X., Yáñez A., Hazelett D.J., et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat. Med. 2018;24:1887–1898. doi: 10.1038/s41591-018-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guo H., Ci X., Ahmed M., Hua J.T., Soares F., Lin D., Puca L., Vosoughi A., Xue H., Li E., et al. ONECUT2 is a driver of neuroendocrine prostate cancer. Nat. Commun. 2019;10:278. doi: 10.1038/s41467-018-08133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fenner A. BRN2 is a neuroendocrine driver. Nat. Rev. Urol. 2017;14:10. doi: 10.1038/nrurol.2016.237. [DOI] [PubMed] [Google Scholar]
- 109.Aggarwal R.R., Quigley D.A., Huang J., Zhang L., Beer T.M., Rettig M.B., Reiter R.E., Gleave M.E., Thomas G.V., Foye A., et al. Whole-genome and transcriptional analysis of treatment-emergent small-cell neuroendocrine prostate cancer demonstrates intraclass heterogeneity. Mol. Cancer Res. MCR. 2019;17:1235–1240. doi: 10.1158/1541-7786.MCR-18-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clegg N., Ferguson C., True L.D., Arnold H., Moorman A., Quinn J.E., Vessella R.L., Nelson P.S. Molecular characterization of prostatic small-cell neuroendocrine carcinoma. Prostate. 2003;55:55–64. doi: 10.1002/pros.10217. [DOI] [PubMed] [Google Scholar]
- 111.Ostano P., Mello-Grand M., Sesia D., Gregnanin I., Peraldo-Neia C., Guana F., Jachetti E., Farsetti A., Chiorino G. Gene expression signature predictive of neuroendocrine transformation in prostate adenocarcinoma. Int. J. Mol. Sci. 2020;21:1078. doi: 10.3390/ijms21031078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chang Y.T., Lin T.P., Tang J.T., Campbell M., Luo Y.L., Lu S.Y., Yang C.P., Cheng T.Y., Chang C.H., Liu T.T., et al. HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Lett. 2018;433:43–52. doi: 10.1016/j.canlet.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 113.Mather R.L., Parolia A., Carson S.E., Venalainen E., Roig-Carles D., Jaber M., Chu S.C., Alborelli I., Wu R., Lin D., et al. The evolutionarily conserved long non-coding RNA LINC00261 drives neuroendocrine prostate cancer proliferation and metastasis via distinct nuclear and cytoplasmic mechanisms. Mol. Oncol. 2021;15:1921–1941. doi: 10.1002/1878-0261.12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu M., Chen Q., Liu X., Sun Q., Zhao X., Deng R., Wang Y., Huang J., Xu M., Yan J., et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 115.Zhang Y.Q., Chen X., Fu C.L., Zhang W., Zhang D.L., Pang C., Liu M., Wang J.Y. FENDRR reduces tumor invasiveness in prostate cancer PC-3 cells by targeting CSNK1E. Eur. Rev. Med. Pharmacol. Sci. 2019;23:7327–7337. doi: 10.26355/eurrev_201909_18838. [DOI] [PubMed] [Google Scholar]
- 116.Zhang G., Han G., Zhang X., Yu Q., Li Z., Li Z., Li J. Long non-coding RNA FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. Biomarkers. 2018;23:435–445. doi: 10.1080/1354750X.2018.1443509. [DOI] [PubMed] [Google Scholar]
- 117.Chang J., Xu W., Du X., Hou J. MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS. OncoTargets Ther. 2018;11:3461–3473. doi: 10.2147/OTT.S164131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luo J., Wang K., Yeh S., Sun Y., Liang L., Xiao Y., Xu W., Niu Y., Cheng L., Maity S.N., et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat. Commun. 2019;10:2571. doi: 10.1038/s41467-019-09784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Crea F., Venalainen E., Ci X., Cheng H., Pikor L., Parolia A., Xue H., Nur Saidy N.R., Lin D., Lam W., et al. The role of epigenetics and long noncoding RNA MIAT in neuroendocrine prostate cancer. Epigenomics. 2016;8:721–731. doi: 10.2217/epi.16.6. [DOI] [PubMed] [Google Scholar]
- 120.Ramnarine V.R., Alshalalfa M., Mo F., Nabavi N., Erho N., Takhar M., Shukin R., Brahmbhatt S., Gawronski A., Kobelev M., et al. The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. GigaScience. 2018;7:giy050. doi: 10.1093/gigascience/giy050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mirza S., Katafiasz B.J., Kumar R., Wang J., Mohibi S., Jain S., Gurumurthy C.B., Pandita T.K., Dave B.J., Band H., et al. Alteration/deficiency in activation-3 (Ada3) plays a critical role in maintaining genomic stability. Cell Cycle. 2012;11:4266–4274. doi: 10.4161/cc.22613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coffey K., Rogerson L., Ryan-Munden C., Alkharaif D., Stockley J., Heer R., Sahadevan K., O’Neill D., Jones D., Darby S., et al. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res. 2013;41:4433–4446. doi: 10.1093/nar/gkt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang J., AlTahan A.M., Hu D., Wang Y., Cheng P.H., Morton C.L., Qu C., Nathwani A.C., Shohet J.M., Fotsis T., et al. The role of histone demethylase KDM4B in Myc signaling in neuroblastoma. J. Natl. Cancer Inst. 2015;107:djv080. doi: 10.1093/jnci/djv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.