Abstract
A novel method for the detection of any alteration within a defined sequence has recently been demonstrated (A. Lishanski, N. Kurn, and E. F. Ullman, Nucleic Acids Res. 28:E42, 2000; A. Lishanski, Clin. Chem. 46:9, 2000). Essential to this method are the generation of partial duplexes that are capable of forming four-stranded structures and the ability to detect inhibition of branch migration in these structures (I. G. Panyutin and P. Hsieh, J. Mol. Biol. 230:413–424, 1993). Inhibition of branch migration indicates the presence of sequence alteration. This mutation detection method, termed branch migration inhibition (BMI), is suitable for the detection of drug resistance in M. tuberculosis, which is frequently associated with multiple mutations within known genes. We describe the genotypic determination of the rifampin (RMP) and pyrazinamide (PZA) susceptibilities of M. tuberculosis isolates, using BMI coupled with the luminescence oxygen channeling immunoassay (LOCI) (E. F. Ullman et al., Proc. Natl. Acad. Sci. USA 91:5426–5430, 1994). RMP and PZA resistances are associated with multiple mutations within the rpoB and pncA genes, respectively. M. tuberculosis genomic DNA samples prepared from 46 clinical isolates were used for genotypic determination of RMP resistance in a “blind study.” Similarly, PZA resistance was determined using genomic DNA samples prepared from 37 clinical isolates. Full agreement of the genotypic and phenotypic determinations of drug susceptibility was demonstrated. RMP susceptibility determination directly from cells of 10 clinical isolates grown in culture was also demonstrated. The genotypic result of only 1 out of 10 isolates did not agree with the phenotypic susceptibility testing result. Sequence analysis of the rpoB gene of this clinical isolate revealed a single base substitution, most likely a silent point mutation. The new BMI-LOCI mutation detection method is a rapid and accurate procedure for the genotypic determination of the RMP and PZA susceptibilities of M. tuberculosis clinical isolates. BMI can also be detected by using commercially available automated enzyme-linked immunosorbent assay plate formats (Lishanski et al., Nucleic Acids Res. 28:E42, 2000).
The incidence of tuberculosis is increasing in many countries, and control of the disease is threatened by the emergence of drug resistance. In the past few years considerable progress has been made in understanding the mechanisms of action of antimycobacterial agents and the genetic basis of resistance to some of these compounds (13). To date, there is information about 12 genes involved in resistance in Mycobacterium tuberculosis (2, 14). There is a need for rapid susceptibility tests for M. tuberculosis, particularly for first-line drugs such as rifampin (RMP), pyrazinamide (PZA), isoniazid, and ethambutol. A test method that is based on detection of mutations in the respective gene sequences is likely to be a useful adjunct to the lengthy conventional culture-based susceptibility testing and should prove useful for improving the treatment and control of the disease.
The molecular basis of M. tuberculosis resistance to RMP has been shown to be associated with deletion, insertion, or missense mutations in the 81-bp region of the rpoB gene, which encodes the β-subunit of the DNA-dependent RNA polymerase (8). More than 96% of M. tuberculosis isolates showing resistance to RMP have been reported to have one or two mutations within this region (10). Thus, the detection of a mutation in the rpoB gene can be used as an accurate marker for detection of RMP resistance in the majority of M. tuberculosis strains tested. Furthermore, RMP resistance is associated with multidrug resistance (3).
Resistance to PZA has been shown to be accompanied by loss of pyrazinamidase (PZase) activity in M. tuberculosis (10). PZase converts PZA to bactericidal pyrazinoic acid, and loss of PZase activity is associated with PZA resistance. Multiple mutations, which span the entire coding region of the pncA gene, have been shown to be associated with PZA resistance in M. tuberculosis. Mutations in the pncA gene have been identified as the cause for acquired PZA resistance in M. tuberculosis, and sequence alteration of this gene has also been reported for the naturally resistant Mycobacterium bovis strain (11, 12). The percentage of PZA-resistant M. tuberculosis strains that have alterations in the pncA gene has been reported as between 72 and 97% (1). In order to detect PZA-resistant strains, in vitro testing of the susceptibility of M. tuberculosis to PZA is highly recommended. Unfortunately, conventional agar-based testing for PZA susceptibility often leads to noninterpretable results because of insufficient growth in the acidified medium (15).
Insofar as multiple mutations in the rpoB and pncA genes have been shown to be associated with RMP and PZA resistance, respectively, molecular methods for the genotypic determination of resistance to these antibiotics should be designed to detect the presence of any and all mutations in these genes. Several methods for the detection of multiple mutations have been described in recent years. Most of these methods require gel separation and are not equally effective for the detection of all sequence alterations. We have recently described a method for rapid detection of any alteration in a test nucleic acid sequence relative to a reference sequence, which can be amplified by PCR (6).
The method is based on the detection of inhibition of branch migration in four-stranded cruciform structures. The cruciform structures are formed by annealing of partial duplexes prepared by amplification of test and reference nucleic acid sequences using specially designed PCR primers. If test and reference sequences are identical, the cruciform structures resolve into undetectable duplexes; if a mutation is present, the cruciform structures are stable and can be detected. This novel mutation detection method, termed branch migration inhibition (BMI), has been shown to be equally effective in the detection of any mutation, including single-base substitutions, insertions, and deletions. The method is particularly suitable for the genotypic determination of drug susceptibility where the resistance phenotype is associated with multiple mutations in a known genetic locus.
We describe the development and performance of a rapid assay for genotypic determination of RMP and PZA susceptibilities of M. tuberculosis, employing the BMI mutation detection method coupled with a homogeneous, light-induced luminescence detection method, the luminescence oxygen channeling immunoassay (LOCI) (16). The correct determination of drug susceptibility using either purified genomic DNA or whole bacterial cells from clinical specimens is demonstrated. The new method is simple, fast, and amenable to automation.
MATERIALS AND METHODS
Clinical isolates.
Ten samples of M. tuberculosis cells from clinical isolates were used in the study. The isolates, of known RMP susceptibility phenotypes, grown either on Lowenstein-Jensen agar or in Middlebrook 12B broth, were collected following different growth periods, spun down, and resuspended in assay buffer. Cells were harvested and heat killed prior to analysis. Three separate cell disruption methods, sonication, microwave irradiation, and heating to 98°C for 10 to 15 min, were evaluated for feasibility for direct genotypic determination of antibiotic resistance.
Purified genomic DNA samples.
A total of 64 genomic DNA samples purified from M. tuberculosis clinical isolates were used (17). These included 23 RMP-resistant isolates, 8 PZA-resistant isolates, and 33 wild-type isolates. Forty-six of these genomic DNA samples (18 RMP-resistant isolates and 28 wild-type isolates) were used in the blind study for the genotyping of RMP resistance. Thirty-seven of the genomic DNA samples (8 phenotypically PZA-resistant isolates and 29 wild-type isolates which were used in the determination of RMP resistance) were used for the genotyping of PZA resistance. Genomic DNA samples purified from four Mycobacterium other than tuberculosis (MOTT) isolates, including Mycobacterium smegmatis, Mycobacterium kansasii, M. bovis, and Mycobacterium intracellulare, were also studied. Phenotypic determination of the antibiotic susceptibilities of the isolates was carried out at a reference laboratory (Northern California State Reference Laboratory at Berkeley). RMP susceptibility testing was carried out by agar proportion methods. PZA susceptibility testing was performed by the radiometric method in 7H12 broth (2a). M. tuberculosis genomic DNA or cells from clinical isolates were provided by the Stanford Medical Center (Palo Alto, Calif.). Purified human genomic DNA was obtained from the Coriell Institute for Medical Research (Camden, N.J.).
DNA target sequences.
DNA target sequences were the 3,853-bp M. tuberculosis rpoB gene (GenBank accession no. U12565), the 561-bp M. tuberculosis pncA gene (GenBank accession no. U59967), and the human cystic fibrosis gene (CFTR), exon 11.
Oligonucleotides.
All oligonucleotides were synthesized with specific modifications by Oligos, Etc. (Wilsonville, Oreg.). The sequences of the oligonucleotide primers used for mutation analysis are listed in Table 1. All primers were 3′ modified by the addition of two ethenoadenosine residues. The added nonhybridizing modified nucleotide, ethenoadenosine, was found to provide added priming specificity when used with cloned Pfu polymerase, which was from Stratagene (San Diego, Calif.). More recent results of BMI with other DNA targets strongly suggest that the use of ethenoadenosine primers is probably not necessary as long as “hot-start” protocols are used (7).
TABLE 1.
Oligonucleotide primers
| Primera | Sequencesb |
|---|---|
| M. tuberculosis rpoB gene analysis | |
| FP1/1779 5′-biotin or digoxigenin) | 5′-GAG CGG ATG ACC ACC CAG GAC XXT-3′ |
| RP1/1963t1 | 5′-ACC ATG CTC GAG ATT ACG AGC CGG CAC GCT CAC GTG ACA XXA-3′ |
| RP1/1963t2 | 5′-GAT CCT AGG CCT CAC GTA TTC CGG CAC GCT CAC GTG ACA XXA-3′ |
| RP2/2044t1 | 5′-ACC ATG CTC GAG ATT ACG AG C AGA CCG ATG TTG GGC CCC TXX A-3′ |
| RP2/2044t2 | 5′-GAT CCT AGG CCT CAC GTA TT C AGA CCG ATG TTG GGC CCC TXX A-3′ |
| M. tuberculosis pncA gene analysis | |
| FP2/39ue (5′-biotin or digoxigenin) | 5′-TGG TGC CGC GTC GGT AGG XXT-3′ |
| RP2/5411t1 | 5′-ACC ATG CTC GAC ATT ACG AG T CAG GAG CTG CAAACCAACTCXXA-3′ |
| RP2/5411t2 | 5′-GAT CCT AGG CCT CAC GTA TT T CAG GAG CTG CAA ACC AACTCXXA-3′ |
| Human CFTR exon 11 analysis | |
| F2 (5′-digoxigenin) | 5′-TAG AAG GAA GAT GTG CCT TTC AXX T-3′ |
| R1t1 | 5′-ACC ATG CTC GACATT ACG AG G ACA TTT ACA GCA AAT GCT TGC XXT-3′ |
| R1t2 | 5′-GAT CCT AGG CCT CAC GTA TT G ACA TTT ACA GCA AAT GCT TGC XXT-3′ |
FP, forward primer; RP, reverse primer.
Underlined “tail” (t1 and t2) portions of the reverse primer sequences are not complementary to the target DNA. These are the same sequences used in the synthetic branch migration substrates of Panyutin and Hsieh (9). XX, ethenodeoxyadenosine. All primers were 3′ modified as stated in Materials and Methods.
DNA amplification.
PCR amplification of all test targets was carried out using a 5′-biotin-labeled forward primer and two-tailed reverse primers with “tail” sequences (which are not complementary to the target) as shown in Fig. 1. The reference target (wild-type M. tuberculosis) and nonrelevant target (human CFTR exon 11) were amplified using the corresponding 5′-digoxigenin-labeled forward primer and two-tailed reverse primers composed of a 3′ target-specific portion and 5′ tails which are the same as those used for the test M. tuberculosis rpoB target sequence.
FIG. 1.
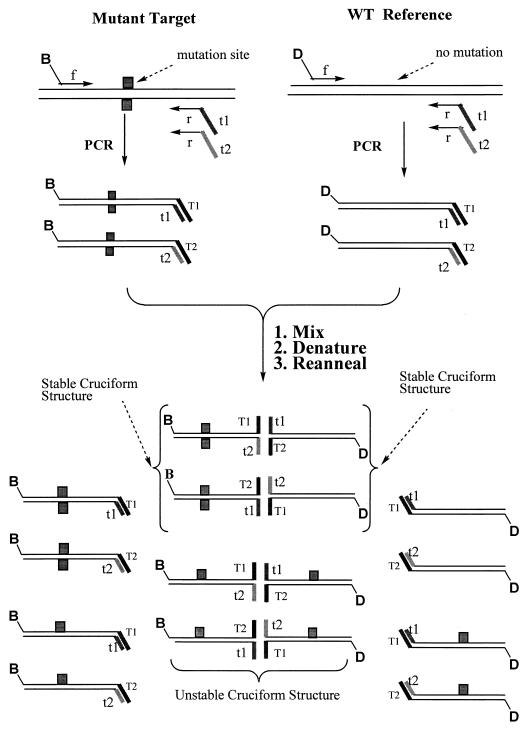
Generation of BMI-specific products. The formation of four-stranded DNA structures and their resolution depending on the presence of a mutation are described in the text. 5′-end-biotin (B)- or digoxigenin (D)-labeled oligonucleotides are used as the forward (f) primers for mutant target and wild-type reference sequences. The “tails” in the reverse (r) primers are designated t1 and t2. The BMI method is based on the detection of inhibition of branch migration in four-stranded cruciform structures formed by annealing of partial duplexes prepared by amplification of test and reference nucleic acid sequences. The stable cruciform structures are presented within braces; the rest are the unstable cruciform structures which will resolve and separate, as well as the partial duplexes.
A master reaction mixture (mix 1; 25 μl) containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mg of bovine serum albumin (BSA)/ml, 200 μM each of the four deoxynucleoside triphosphates, and 250 nM each of the primers was aliquoted to each reaction tube containing a wax gem (Perkin-Elmer) to form the wax barrier on top of the liquid reaction mixture. A second reaction mixture (mix 2; 20 μl) containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mg of BSA/ml, and 2.5 U of Pfu DNA polymerase/25 μl was also prepared and aliquoted to each reaction tube following formation of the wax barrier. Five microliters of the test or reference target was added to each tube, prepared as above. PCR amplification was carried out in a T3-Thermoblock thermocycler (Biometra Inc., Tampa, Fla.). Amplification of the M. tuberculosis rpoB gene target sequence was carried out using the following thermocycling profile: 4 min at 95°C for denaturation of target DNA, followed by 39 cycles of 45 s at 95°C and 2 min at 70°C. Amplification of the M. tuberculosis pncA gene was carried out using the following thermocycling profile: 4 min at 95°C, followed by 39 cycles of 30 s at 95°C, 1 min at 64°C, and 1 min at 72°C. Amplification of the human CFTR exon 11 gene sequence was carried out using the following thermocycling conditions: 4 min at 95°C, followed by 39 cycles of 30 s at 95°C, 1 min at 64°C, and 1 min at 72°C.
BMI mutation analysis and signal detection.
BMI analysis of test DNA amplification products was carried out by mixing reaction mixtures of test samples with amplification products of either relevant, wild-type M. tuberculosis reference genomic DNA or a nonrelevant, human cystic fibrosis exon 11 gene reference sequence. One microliter each of reference (wild-type) amplification products and test amplification products were added to PCR tubes containing 4 μl of 100 mM Tris-HCl (pH 8.3), 500 mM KCl, 15 mM MgCl2, and 2 mg of BSA/ml. The mixtures were subjected to denaturation and reannealing (2 min at 95°C followed by 30 min at 65°C). The formation of stable cruciforms, indicating the presence of an alteration(s) in the test sequence, was measured by LOCI, a homogeneous singlet oxygen chemiluminescence immunoassay (16). Fifty microliters of a LOCI bead suspension, containing 2.5 μg of streptavidin-coated sensitizer particles and 1.25 μg of anti-digoxigenin monoclonal antibody-coated chemiluminescer particles was added, and the tubes were incubated at 37°C for 30 min to allow binding of the particles. Chemiluminescence signals were read (3 cycles of 1 s of illumination (680 nm) and 1 s of read [560 to 630 nm]) with an in-house-built reader. BMI analysis using the nonrelevant reference was carried out similarly with the exception that the postamplification reaction mixture of CFTR genomic DNA was first diluted 10-fold, and 1 μl of this diluted reaction mixture was mixed with 1 μl of the post-PCR mixture of the test sample.
Normalization of BMI signals.
A normalized BMI signal, as related to the presence of sequence alterations, is obtained by calculating the ratio of the signal generated from a mixture of the test amplification product with the relevant reference amplification product (the M. tuberculosis rpoB [GenBank accession no. U12565] or pncA [U59967] gene) to the signal generated from a mixture of the test amplification product with a nonrelevant reference amplification product (the CFTR gene, exon 11 [cystic fibrosis mutation database available at http: //www.genet.sickkids.on.ca/cftr/]).
RESULTS
Experimental design.
The BMI mutation analysis method was applied to the detection of sequence alterations in M. tuberculosis. The method is based on the generation from related test and reference nucleic acid sequences of amplification products capable of forming partial duplexes, which are able to anneal to form four-stranded cruciform structures. When the test and reference sequences are identical, spontaneous branch migration occurs, and the cruciform structures formed by the association of partial duplexes dissociate to full duplexes. However, any sequence alteration in the test sequence relative to the reference sequence will result in inhibition of branch migration and the formation of stable cruciform structures (9). When suitable labels are attached to the PCR primers, the cruciform structures can be detected by various immunochemical methods.
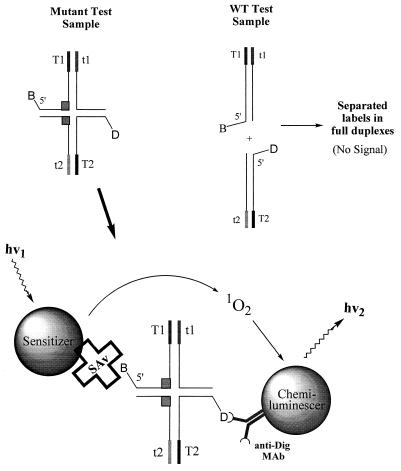
We detect the stable cruciform structures by LOCI (16) as shown in Fig. 2. The binding of sensitizer and chemiluminescer particles to the respective labels of the stable cruciform structures results in the formation of complexes capable of producing light-induced chemiluminescence. Due to the short lifetime of singlet oxygen (about 4 μs), only chemiluminescer dyed particles that are closely associated with the sensitizer particles will react to produce chemiluminescence signals. Thus, particles that are not attached to the stable cruciform structures contribute only minimally to signal generation.
FIG. 2.
LOCI detection of BMI products. The LOCI detection method employs two different dyed latex particles, a streptavidin (SAv)-labeled photosensitizer latex particle and an anti-digoxigenin monoclonal antibody (anti-Dig MAb)-coated chemiluminescer latex particle, as mentioned in the text. Illumination of the reaction mixture with a light source of defined wavelength (680 nm) results in production of singlet oxygen by the sensitizer dye, which can react with the acceptor dye to produce chemiluminescence signals (550 nm). Due to the short lifetime of singlet oxygen (4 μs), only chemiluminescent particles that are closely associated with the sensitizer particles will react to produce chemiluminescence signals. The binding of sensitizer and chemiluminescer particles to the respective labels of the cruciform structures results in the formation of complexes capable of producing light-induced chemiluminescence. WT, wild type; hv, laser light, with a defined wavelength.
BMI analysis for the detection of rpoB mutations associated with RMP resistance.
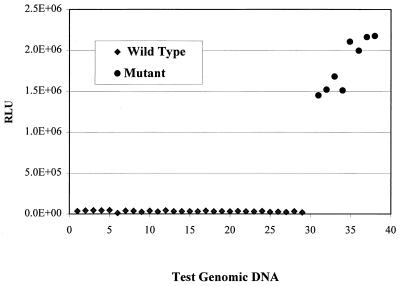
The performance of BMI for detection of sequence alteration in test DNA compared to a reference DNA is dependent on the ratio of signals obtained for the mutant and wild-type genotypes. Optimization of conditions for formation of cruciform structures and resolution of the structures when the test sequence is of a wild-type genotype revealed dependency on both the temperature and the Mg2+ ion concentration. Optimal performance was achieved at 65°C for 30 min and was the same for the various target systems tested. The detection of mutant M. tuberculosis genotypes was found to be optimal at 15 mM Mg2. Analysis of genomic DNA samples purified from 10 M. tuberculosis clinical isolates revealed clear discrimination of wild-type (n = 5) and mutant (n = 5) rpoB genotypes, with mean BMI signals for mutant genotypes (2,985,000 ± 156,000; coefficient of variation [CV], 13%) about 20-fold higher than the mean signals for the wild-type genotype (120,000 ± 17,000; CV, 5%).
Low BMI signals for mutant genotypes may result from inefficient amplification or low initial sample DNA input and could be incorrectly interpreted as wild-type genotypes. We have developed a method for normalization of BMI signals, which corrects for variations in input test DNA or in amplification efficiency and ensures correct genotype determination. Partial duplexes generated from the test and nonrelevant reference DNA amplification products can bind to each other by hybridization of the complementary tail sequences to form cruciform structures. Because the sequences of the double-stranded portions of the partial duplexes are not related, spontaneous branch migration does not occur and the four-stranded cruciform structures are stable. Signals generated from these structures are proportional to the amount of test DNA in the sample and are used for normalization of the mutation-related signals.
Genotypic determination of RMP resistance directly from cells.
The performance of BMI coupled with LOCI for detection of mutations in the rpoB gene sequence directly from cells, i.e., without prior DNA purification, was assessed. The three cell disruption methods were found to be equally efficient in terms of releasing DNA from mycobacterial cells (data not shown).
Genotypic determination of RMP susceptibility directly from heat-treated cells of 10 M. tuberculosis isolates was carried out using BMI analysis coupled with the LOCI detection method. As shown in Table 2, the genotypic determination of 9 out of the 10 isolates correlated well with phenotypic determination of RMP susceptibility. Only one discrepancy between the genotypic determination and the conventional culture-based phenotypic determination of antibiotic resistance was observed. Sequence analysis of the rpoB gene of the discrepant M. tuberculosis clinical isolate (isolate 8) revealed the presence of a mutation in a phenotypically susceptible isolate. Nucleic acid sequencing of fragments of the rpoB gene, which include the region of interest for genotypic determination of RMP resistance, was performed at the sequencing facility of Stanford University. Sequence analysis was obtained for isolate 8, a phenotypically resistant strain (isolate 7) derived from it, and a wild-type isolate. The sequence of the fragment derived from the wild-type strain was identical to that reported by Telenti (14). A point mutation at position 198 (according to the numbering of Telenti), with a replacement of C by T, was revealed for the isolate for which there is a discrepancy between the genotypic and phenotypic determinations. This new point mutation is most likely a silent mutation, which does not alter the function of the RNA polymerase. Other silent mutations in rpoB have been described previously (4). Sequence information for the rpoB fragment of the phenotypically resistant strain derived from the above isolate revealed a point mutation at position 198, as well as an additional replacement of C by T at position 248. The point mutation at position 248 was previously described as conferring RMP resistance.
TABLE 2.
Direct rpoB genotyping of M. tuberculosis clinical isolatesa
| Isolate no. | RLUb | Phenotypec | Genotyped |
|---|---|---|---|
| 1 | 8,840 | Susceptible | Wild type |
| 2 | 1,434,180 | Resistant | Mutant |
| 3 | 9,724 | Susceptible | Wild type |
| 4 | 1,495,320 | Resistant | Mutant |
| 5 | 10,980 | Susceptible | Wild type |
| 6 | 1,363,690 | Resistant | Mutant |
| 7 | 1,227,160 | Resistant | Mutant |
| 8 | 1,421,840 | Susceptible | Mutant |
| 9 | 1,047,470 | Resistant | Mutant |
| 10 | 10,208 | Susceptible | Wild type |
BMI was carried out on suspensions of heat-killed M. tuberculosis cells treated by sonication prior to analysis. One genotypically mutant isolate (isolate 8) was determined to be phenotypically susceptible.
RLU, relative luminescence units (see Materials and Methods). The RLU cutoff used for differentiation of mutant versus wild-type strains in this experiment was set at 20,000, which is above the mean plus 3 standard deviations (mean ± standard deviation, 9579 ± 2278) from a parallel experiment using five replicates of two wild-type strains.
Determined by conventional culture-based methods at a reference laboratory.
Determined by the BMI study.
Blind study of genotypic determination of RMP resistance.
A panel of 46 genomic DNA samples was tested in a “blinded” fashion for rpoB genotype. In addition to BMI analysis of the test gene relative to a wild-type reference gene, a method for the normalization of BMI signals was also employed as described above. Amplification of a CFTR sequence, exon 11, was carried out using the primer oligonucleotides specified in Table 1, and the product was employed as a nonrelevant reference nucleic acid sequence for the normalization procedure.
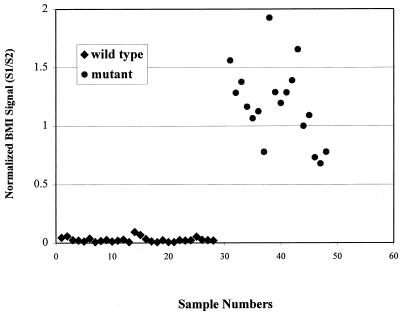
The results obtained by using this normalization scheme are summarized in Fig. 3. The normalized BMI-LOCI signal clearly differentiates between mutant and wild-type isolates. The mutant and wild-type genotype determinations by this method are in full agreement with the phenotypic results for the clinical isolates as determined by conventional culture-based methods at the Stanford Medical Center.
FIG. 3.
M. tuberculosis rpoB genotypic determination: blind study. Shown are normalized BMI signals for analysis of rpoB gene mutations. A total of 46 M. tuberculosis genomic DNA samples were analyzed by PCR-BMI-LOCI using a wild-type M. tuberculosis amplicon as the relevant reference and CFTR exon 11 as the nonrelevant reference to normalize signals. Samples 1 to 28 represent RMP-susceptible isolates, and samples 31 to 48 are RMP resistant. A normalized BMI signal is obtained by calculating the ratio of the signal generated from a mixture of the test amplification product with the relevant reference amplification product (S1) to the signal generated from a mixture of the test amplification product with the nonrelevant reference amplification product (S2).
Genotypic determination of PZA resistance.
Oligonucleotide primers (Table 1) were selected for efficient amplification of the pncA gene, followed by BMI mutation analysis coupled with LOCI detection for the genotypic determination of PZA susceptibility. Amplification of the pncA gene sequence results in the generation of a 620-bp amplicon. Thirty-seven DNA samples purified from M. tuberculosis clinical isolates were tested for genotypic determination of PZA susceptibility. These included 29 phenotypically wild-type isolates (used in the determination of RMP resistance) and 8 phenotypically PZA-resistant isolates. The results of genotypic determination of PZA susceptibility are shown in Fig. 4, demonstrating perfect correlation of the phenotypic and genotypic determinations, with very strong discrimination between susceptible and resistant genotypes.
FIG. 4.
M. tuberculosis pncA gene mutation detection by PCR-BMI-LOCI. A total of 37 genomic DNA samples (samples 1 to 29 are wild-type isolates, and samples 31 to 38 are PZA-resistant isolates) were analyzed using BMI-specific primer sets. The amplification product of wild-type M. tuberculosis was used as a reference. The amplicon size is 620 bp.
Specificity of the BMI genotypic determination of antibiotic resistance.
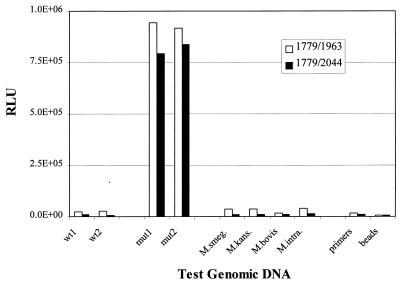
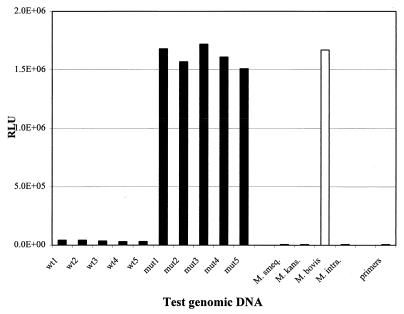
The specificity of the BMI-LOCI method for detection of M. tuberculosis mutant RMP and PZA genotypes was examined. In the event that the clinical isolates contain an additional mycobacterial species, it is possible that any sequence difference between the reference (M. tuberculosis wild-type sequence) and the nontuberculosis sequence will result in the generation of a positive BMI-LOCI signal, which may be misinterpreted as indicative of a resistant M. tuberculosis genotype. Thus, we have studied the BMI-LOCI responses obtained from genomic DNAs of a number of mycobacterial species. This study was undertaken only for the validation of the BMI-LOCI method for determination of M. tuberculosis antibiotic resistance. Genomic DNAs purified from four MOTT isolates, including isolates of M. smegmatis, M. kansasii, M. bovis, and M. intracellulare, were used in this study and tested together with wild-type and mutant genomic DNA samples of M. tuberculosis. All but M. bovis yielded low BMI signals, similar to signals obtained from wild-type M. tuberculosis DNA, as shown in Fig. 5 and 6. The BMI-LOCI analysis of M. bovis rpoB DNA showed a wild-type genotype, whereas analysis of the pncA gene sequence showed a mutant genotype. This result is in accordance with known sequence alterations resulting in the naturally PZA-resistant phenotype of M. bovis.
FIG. 5.
Species specificity of detection of mutations in the M. tuberculosis rpoB gene. BMI analysis was carried out using two PCR primer sets. The same forward primer (1779 5′-biotin or 5′-digoxigenin) was used with two sets of reverse tail primers (1963t1 and 1963t2; 2044t1 and 2044t2) with amplicon sizes of 225 and 306 bp, respectively. Samples included genomic DNA purified from two wild-type M. tuberculosis isolates (wt1 and wt2), two RMP-resistant M. tuberculosis isolates (mut1 and mut2), and four MOTT isolates (M. smegmatis, M. kansasii, M. bovis, and M. intracellulare). An amplification product from wild-type M. tuberculosis was used as a reference.
FIG. 6.
Species specificity of detection of mutations in the M. tuberculosis pncA gene. BMI analysis was carried out using a PCR primer set which generates 620-bp amplicons. Samples included genomic DNA purified from five wild-type M. tuberculosis isolates (wt1 to wt5), five PZA-resistant M. tuberculosis isolates (mut1 to mut5), and four MOTT isolates (M. smegmatis, M. kansasii, M. bovis, and M. intracellulare). An amplification product from wild-type M. tuberculosis was used as a reference.
Nonspecific PCR amplification leads to increased background signals and reduced signal-to-background ratios. Primer selection, together with a hot-start PCR amplification procedure, was found to be essential for reducing background signals. The performances of two PCR-BMI primer pairs for analysis of rpoB gene mutations are shown in Fig. 5. Although high signal-to-background (S/B) ratios were obtained with both primer pairs, this performance parameter (S/B, 37 for the primer set of 1779 [forward]–1963t1 and -t2 [reverse] versus 90 for the primer set of 1779 [forward]–2044t1 and -t2 [reverse]) can be improved with proper primer pair selection.
Reproducibility of the BMI-LOCI signals for genotypic determination of antibiotic resistance.
To assess assay reproducibility, eight independent replicate determinations of two DNA samples, one from a phenotypically wild-type and one from a phenotypically PZA-resistant M. tuberculosis isolate, were carried out as described before. Very reproducible results were obtained. The mean value of the eight wild-type signals is 22,136 ± 1,528 (CV, 6.9%), and the mean value of the eight mutant signals is 2,283,981 ± 106,972 (CV, 4.6%).
DISCUSSION
The results reported demonstrate the feasibility of the BMI-LOCI mutation detection method for genotypic testing of resistance to two antituberculosis agents, RMP and PZA. Multiple mutations in the rpoB and pncA gene sequences have previously been reported to be associated with RMP and PZA resistance (11, 13) and are present in the majority of resistant isolates. Application of a rapid DNA-based assay would be of benefit for determining drug susceptibility patterns (13). Detection of RMP resistance should also identify multidrug-resistant strains, nearly all of which are resistant to RMP (3).
Correct genotypic determination of PZA- and RMP-resistant M. tuberculosis using BMI analysis coupled with the LOCI detection method was demonstrated by employing genomic DNA purified from clinical isolates. Mutations in the rpoB and pncA genes were identified in all genomic DNA samples purified from RMP- and PZA-resistant isolates tested. Further validation of the BMI method for genotypic identification of RMP- and PZA-resistant M. tuberculosis will require testing of additional clinical isolates, preferably selected from diverse geographic settings so as to ensure that the full spectrum of resistance-conferring and silent mutations is sampled.
Nine out of 10 BMI genotypic determinations of RMP susceptibility directly from cells correlated well with phenotypic determinations by a culture-based method. Sequence analysis of the rpoB gene segment used for genotypic determination revealed a point mutation, which had not been previously described, in the single discrepant isolate. The new mutation is likely to be a silent mutation which gives a positive BMI-LOCI signal in this phenotypically susceptible isolate. Other silent rpoB mutations have been described previously (4).
The BMI method detects only the presence, not the type, of mutations. A method like BMI that is independent of the type of mutation would be very useful in antimicrobial susceptibility testing, when any of the multiple mutations within a given sequence can often give rise to drug resistance. Insofar as the new mutation detection method is capable of detecting any mutation in the gene sequence tested, it is expected to detect silent mutations as well as mutations leading to a mutant phenotype. The potential for false classification of isolates with mutant genotypes as antibiotic resistant is ever present when one is using a nucleic-acid-based method which detects any sequence alteration in the test sample relative to a reference. The practical impact of this potential discrepancy between phenotypic and genotypic determinations is yet to be determined. However, this disadvantage is likely small compared to the benefits of rapid screening of clinical M. tuberculosis isolates for drug resistance.
A simple method for the normalization of BMI-LOCI signals obtained from various test samples was also developed. This method is useful for overcoming the possible false genotypic determination of phenotypically antibiotic-resistant isolates as susceptible. This false determination could occur as a consequence of a low signal resulting from either a low input of DNA or amplification failure. Both of these causes for potential false-negative results are common for all nucleic-acid-based methods and would have significant impact on the clinical utility of the genotyping method. Another method for checking PCR success is to add ethidium bromide to the PCR mixture (7). The ability to control for these potential amplification failures will facilitate the use of the new method as a rapid screen for drug-resistant M. tuberculosis.
In conclusion, the feasibility of rapid genotypic determination of M. tuberculosis RMP and PZA resistance using the BMI mutation detection method was demonstrated. Coupled with the LOCI homogeneous detection method, the new procedure is rapid, highly reproducible, and specific. Other detection methods, such as the enzyme-linked immunosorbent assay, are also feasible, as was shown previously (6). The ability to detect mutations in specific nucleic acid sequences directly from mycobacterial cells, without prior purification of genomic DNA, should render this method suitable for the clinical microbiology laboratory.
ACKNOWLEDGMENTS
We acknowledge the contributions of A. Lishanski and E. Ullman through helpful discussions and thank S. Rose and A. Dafforn for reviewing this paper and M. Taylor for assistance in computer graphics.
REFERENCES
- 1.Alcaide F, Telenti A. Molecular techniques in the diagnosis of drug-resistant tuberculosis. Ann Acad Med Singapore. 1997;26:647–650. [PubMed] [Google Scholar]
- 2.Cole S T. Mycobacterium tuberculosis: drug resistance mechanisms. Trends Microbiol. 1994;2:411–415. doi: 10.1016/0966-842x(94)90621-1. [DOI] [PubMed] [Google Scholar]
- 2a.Heifets L B, Iseman M D. Radiometric method for testing susceptibility of mycobacteria to pyrazinamide in 7H12 broth. J Clin Microbiol. 1985;21:200–204. doi: 10.1128/jcm.21.2.200-204.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heym B, Honore N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs W R, Jr, van Embden J D, Grosset J H, Cole S T. Implication of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 4.Kim B J, Kim S Y, Park B H, Lyu M A, Park I K, Bai G H, Kim S J, Cha C Y, Kook Y H. Mutations in the rpoB gene of Mycobacterium tuberculosis that interfere with PCR-single-strand conformation polymorphism analysis for rifampin susceptibility testing. J Clin Microbiol. 1997;35:492–494. doi: 10.1128/jcm.35.2.492-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konno K, Feldmann F M, McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis. 1967;95:461–469. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- 6.Lishanski A, Kurn N, Ullman E F. Branch migration inhibition in PCR amplified DNA: homogeneous mutation detection. Nucleic Acids Res. 2000;28:E42. doi: 10.1093/nar/28.9.e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lishanski A. Screening for single-nucleotide polymorphisms using branch migration inhibition in PCR-amplified DNA. Clin Chem. 2000;46:9. [PubMed] [Google Scholar]
- 8.Miller L, Crawford J, Shinnick T. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:805–811. doi: 10.1128/aac.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panyutin I G, Hsieh P. Formation of a single base mismatch impedes spontaneous branch migration. J Mol Biol. 1993;230:413–424. doi: 10.1006/jmbi.1993.1159. [DOI] [PubMed] [Google Scholar]
- 10.Pretorius G, Sirgel F, Schaaf H, van Helden P, Victor T. Rifampin resistance in M. tuberculosis—rapid detection and implications in chemotherapy. S Afr Med J. 1996;86:50–55. [PubMed] [Google Scholar]
- 11.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculosis drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 12.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–543. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 14.Telenti A. Genetics of drug resistance in tuberculosis. Clin Chest Med. 1997;18:55–64. doi: 10.1016/s0272-5231(05)70355-5. [DOI] [PubMed] [Google Scholar]
- 15.Tummon R. Growth inhibition of Mycobacterium tuberculosis by oleate in acidified medium. Med Lab Technol. 1975;32:229–232. [PubMed] [Google Scholar]
- 16.Ullman E F, Kirakossian H, Singh S, Wu Z P, Irvin B R, Pease J S, Switchenko A C, Irvine J D, Dafforn A, Skold C N, Wagner D B. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc Natl Acad Sci USA. 1994;91:5426–5430. doi: 10.1073/pnas.91.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Soolingen D, Hermans P W, de Haas P E, Soll D R, van Embden J D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]