Abstract
Autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) is an early-onset neurodegenerative disease that was originally discovered in the population from the Charlevoix-Saguenay-Lac-Saint-Jean (CSLSJ) region in Quebec. Although the disease progression of ARSACS may start in early childhood, cases with later onset have also been observed. Spasticity and ataxia could be common phenotypes, and retinal optic nerve hypermyelination is detected in the majority of patients. Other symptoms, such as pes cavus, ataxia and limb deformities, are also frequently observed in affected individuals. More than 200 mutations have been discovered in the SACS gene around the world. Besides French Canadians, SACS genetics have been extensively studied in Tunisia or Japan. Recently, emerging studies discovered SACS mutations in several other countries. SACS mutations could be associated with pathogenicity either in the homozygous or compound heterozygous stages. Sacsin has been confirmed to be involved in chaperon activities, controlling the microtubule balance or cell migration. Additionally, sacsin may also play a crucial role in regulating the mitochondrial functions. Through these mechanisms, it may share common mechanisms with other neurodegenerative diseases. Further studies are needed to define the exact functions of sacsin. This review introduces the genetic mutations discovered in the SACS gene and discusses its pathomechanisms and its possible involvement in other neurodegenerative diseases.
Keywords: ARSACS, sacsin, neurodegeneration, ataxia, mutation
1. Introduction: Sacsin (SACS) Gene and ARSACS
The sacsin gene (SACS) is located on chromosome 13 (13q12.12: chr13:23,288,689-23,433,763, GRCh38/hg38), with 145,075 bases, and is oriented in the minus strand of DNA (https://www.genecards.org/cgi-bin/carddisp.pl?gene=SACS accessed on 24 December 2021). SACS comprises 10 exons, with nine coding exons, and the 10th exon contains 11,487 base pairs, notable as the longest exon among vertebrates [1,2]. The SACS gene encodes a large 520-kDa multidomain protein of 4579 amino acids, called sacsin. It contains several different domains, including the ubiquitin-like (UBL) domain in the N-terminal region, three sacsin internal repeat (SIRPT or SRR) domains, the helical XPC-binding domain, a sacsin J-domain and the higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain in the C-terminal region [3]. Sacsin is expressed in several different tissues, with higher expression in the central nervous system or skin and lower expression in the pancreas and skeletal muscle. In the brain, sacsin expression is the highest in the motor system, including the cerebellum, granular system and in Purkinje cells [2].
SACS is associated with early-onset cerebellar ataxia due to mutation, called the spastic ataxia of Charlevoix-Saguenay (ARSACS), in an autosomal recessive pattern; it was first discovered in a population by the linkage disequilibrium studies conducted in the Charlevoix-Saguenay-Lac-Saint-Jean (CSLSJ) region in Quebec [4,5]. Based on several relatives with ataxia syndrome, the founder effect was present in the French Canadian population. Since this family immigrated to French Canada in the 17th century from the Perche region, it is possible that the disease was present in France, although similar cases may remain unrecognized. Other cases of SACS ataxia in non-Quebec populations, such as in Italy, Japan, Spain, Tunisia or Turkey, were discovered [6,7,8].
ARSACS is associated with ataxia, dysarthria, nystagmus, spasticity, distal muscle wasting and deformities in fingers or feet [6,7]. Affected patients show slow progression of spastic ataxia, which may affect all four limbs. Patients also experience a loss of muscle tissue (amyotrophy) and language impairment (their speech became slurred). Ocular movements may also be impaired [6,7]. Disease may occur during childhood (lower limb ataxia), but their intelligence may not be impaired. Cerebellar signs and pes cavus appear until patients reach their 20 s. Other characteristics may also be possible, such as retinal nerve fiber hypermyelination [8,9,10]. The first characteristics of the disease may be gait initiation, which can be noticed when children start to walk (around 12–18 months of age). Childhood ataxia and spasticity may be prominent as well. In early adulthood (20 s or 30 s), disease progression may be accelerated, and patients may lose the ability to walk by the age of 50 [9]. Bladder and bowel dysfunctions could appear in patients in their 50 s [10]. In addition, patients may show biochemical dysfunctions, such as impaired pyruvate oxidation, hyperbilirubinemia and low serum beta- or HDL lipoproteins [11]. In patients with ARSACS, non-phosphorylated neurofilaments (NFs) may occur in different neurons, including Purkinje cells or motor neurons in the cortex. The cultured motor neurons of SACS knockout mouse embryos presented abnormal neurofilament rearrangements [12,13]. Abnormal mitochondrial functions were also detected, with lower mitochondrial motility and elongation. Since NFs could impact cytoskeletal organization, the alteration of NFs in patients with ARSACS may present mitochondrial impairments, resulting in elevated cellular vulnerability [12,13]. In this review, mutations in the SACS gene in patients with ARSACS and the potential involvement of sacsin in other forms of neurodegeneration are discussed.
2. Sacsin Functions and Cell/Animal Models of ARSACS
One of the limitations in understanding the role of SACS in ARSACS was the unavailability of brain tissues from the affected patients. Ideal cell/animal models should provide insights and reduce the time and costs of analysis, which could be used in screening drug candidates in preclinical studies [14]. To date, several cell and mouse models (hiPSC, SH-SY5Y, knockout mouse models) have been developed with high priority in ARSACS to reflect the accurate human conditions for the discovery of the disease mechanisms. However, the differences in genome, anatomy and metabolisms between humans and mice could represent limitations of mismatch in the ideal replication of the disease [2,13,15]. SACS knockout mice, investigated by Larivière (2015), revealed typical ARSACS symptoms, loss of Purkinje cells in the cerebellum and abnormal aggregation on non-phosphorylated neurofilaments in the somatodendritic brain region with altered mitochondrial dynamics [12]. An additional study on SACS –/– mouse Purkinje cells by Ady et al. showed that SACS knockout cells could reduce the firing rate, lower the excitatory synaptic rate and lead to the loss of Purkinje cells in the anterior lobes, but not in the posterior ones [15]. Loss of Purkinje cells was prominent in the deep cellular nuclei, and the disturbances appeared in the cerebellar circuit [15]. Human-derived cell models may mimic more accurately the human-related pathways and could be more effective in studying disease-related mechanisms and therapies [16]. Human-induced pluripotent stem cells (hiPSC) were suggested to open new avenues in disease modeling and also drug development. They may mimic more correctly the disease mechanisms, even from the earliest cellular dysfunctions. However, the challenge with hiPSC could be that they may not reflect the complexity of different brain areas (cerebellum in ARSACS). An additional issue could be the need for protocols to be developed for an appropriate ARSACS model, such as creating more humanized cell lines, providing nutrients to cells. With hiPSC cells, it may be difficult to model aging-related diseases [16]. SH-SY5Y cells with sacsin knockout may also be a promising approach to monitor the alterations of gene/protein expression or cellular changes [17,18]. Since SH-SY5Y are widely used cell lines in neuroscience and are low-cost human-derived cell lines, these can be used to differentiate various neuronal phenotypes [18]. However, the issues with SH-SY5Y cells are that no standardized protocols are available on them to maintain the culture and to grow the neuron of interest [14]. The studies by Duncan et al. (2017), Crisuolo et al. (2015) and Girard et al. (2012) on human dermal fibroblasts (HDFs) from ARSACS patients revealed reduced sacsin expression. Lower sacsin expression was associated with impaired mitochondrial functions, IF (vimentin) aggregation, abnormal chaperon activity and increased autophagy [19,20,21]. HDFs could be a convenient disease model without genetic engineering to study the disease mechanisms of several human diseases. HDFs have been used to study several neuropsychiatric disorders [22,23]. However, isolation and culture techniques in HDFs are time-consuming and require materials and extensive studies to improve the optimization of the protocols [24]. Taken together, promising studies have been performed to analyze sacsin functions and ARSACS disease mechanisms in cell or mouse models, but further research is needed on the ideal disease model organism.
3. Sacsin Protein Domains
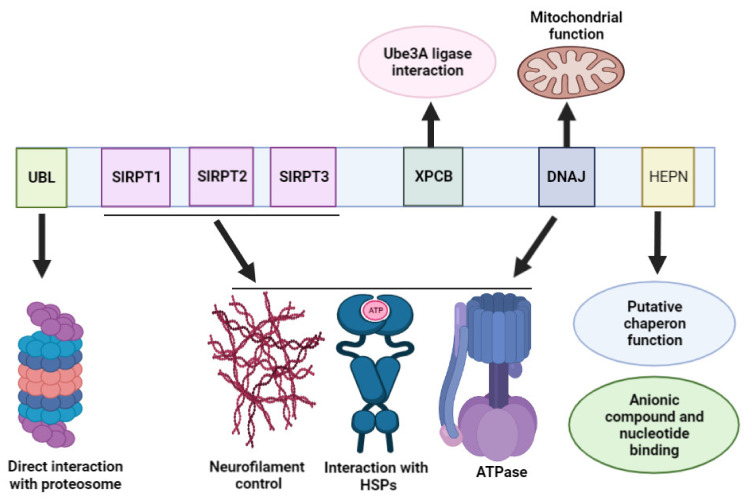
Sacsin has five domains: ubiquitin-like domain (Ubl), three large sacsin internal repeats (SIRPT1, SIRPT2, SIRPT3), xeroderma pigmentosum C-binding domain (XPCB), J-domain (DNAJ) and higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domain. These domains could have different functions and interacting partners (Figure 1). However, they also could also cooperate in different pathways [25].
Figure 1.
Schematic showing the domain structure of the of sacsin protein and the potential functions of different domains.
The N-terminal region of sacsin could interact with proteasomes. Additionally, it may play a role in regulating protein folding by interacting with heat shock proteins. The Ubl domain could interact directly with the proteasome system through the 19S cap and 26S proteasomes and be involved in the degradation pathway [2,26]. The SIRPT domains contain a homologous region, the Hsp90 chaperone [5,13,27,28], which was suggested to be involved in ATPase activities. ATP hydrolysis is important for proper sacsin function. Dysfunctions of the SIRPT domain were associated with reduced or lost ATP hydrolysis. Along with the J-domain, SIRPT domains could stimulate the ATPase activity of Hsp70 [28,29]. The XPC-binding domain (XPCB) could bind the Ube3A ubiquitin ligase. It may be possible that dysfunctions in Ube3A–sacsin interaction could impact the onset of Angelman syndrome-related ataxia [30].
The J-domains can enhance the protein–protein interactions and regulate the activity of heat shock proteins, including Hsp70. A J-domain contains Hsp40 homologous sequences and may impact homeostasis. An interaction between sacsin, Hsp70 and the ubiquitin proteosome system may be involved in defensive mechanisms against abnormal protein aggregation [2]. Sacsin was suggested to play a role in controlling the homeostasis between intermediate filaments (IFs) and neurofilaments (NFs) [3]. If sacsin is missing, nerve cells could contain abnormal bundles of NFs. Patient-derived HDF cells showed abnormal IF (vimentin) distribution and broken microtubule organization. In ARSACS cells, the misfolded IFs aggregated and formed a cage-like structure around the microtubule organization center. These aggregates may result in abnormal clearance and autophagy [20]. Adding the SIPRT and J-domain into motor neurons from sacsin knockout mice resulted in a reduced amount of NF bundles. Both domains could prevent the assembly of NFs [3]. Treatment with SacsJ-myc-TAT in SACS+/+ motor neurons resulted in the induction of IF and NF disassembly. In Sacs–/– motor neurons, the NF bundles were resolved, and the NF network was restored [21]. These data suggest that the J-domain could impact the regulation of IF-NF assembly and disassembly directly [3,20,31].
The function of the HEPN domain was initially unclear [27], but it was found in different eukaryotes and prokaryotes. In bacteria, the HEPN region could impact antibiotic resistance (for example, kanamycin). In eukaryotes, the HEPN domain may have nucleotide-binding activity, but it could also bind anionic compounds in neurons [22,23,32,33]. HEPN domains could dimerize and form a high-affinity site for GTP binding, and potentially impact the chaperon activity of the sacsin protein. The HEPN domain may be involved in the elevation of the ATP/GTP concentration, and in the sacsin–Hsp70 interaction. The HEPN domain may also co-operate with the J-domain, and they contribute to nucleotide binding. Mutations in the HEPN region were suggested to disrupt the nucleotide-binding activity and result in abnormal folding/oligomerization of sacsin [3,27,32,33].
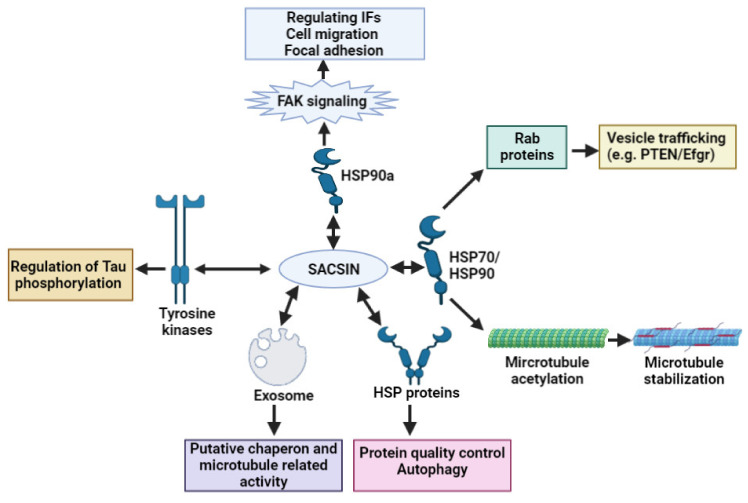
A recent study by Romano et al. [17] revealed that, besides controlling the filament architecture, sacsin could play a crucial role in cell adhesion, microtubule organization and trafficking proteins. The mentioned authors provided an extensive study on the possible functions of sacsin. Knockout of the SACS gene in SH-SY5Y cell lines revealed several disease-associated changes. Microtubule organization and dynamics were altered in KO cells, microtubule polymerization was enhanced, and the movement of tubules became abnormal. Sacsin may also regulate Tau phosphorylation by interacting with tyrosine kinase enzymes. Several kinesin proteins were hyperphosphorylated, resulting in disturbances in mitochondrial movements. Tau and STMN1 pS16 were also hyperphosphorylated. These findings revealed that sacsin could possibly interact with tyrosine kinases. Additionally, sacsin interactions with HSP proteins could be critical in microtubule organization and stabilization. Non-functional sacsin also resulted in disrupted focal adhesion and dynamics, which could result in disturbances in axonal growth, synaptic formation and balance in the brain. Focal adhesion kinases were suppressed in SACS KO cells, and modulation of FAK-PTEN pathways may be beneficial in the case of cellular deficits. Disturbances were found in the adhesion mechanisms, since the membrane-bound adhesion molecules and cell adhesion molecules were both mis-localized. These findings suggest that, besides abnormal trafficking and cellular interactions, localizations could contribute to the reduced interaction between heat shock proteins and sacsin, leading to ARSACS pathology. Additionally, sacsin may interact with exosomes, which could also impact several chaperons or microtubule-related mechanisms. Figure 2 demonstrates that sacsin may serve as a critical regulator of different cellular processes, including chaperon activity, the transport of vesicles and microtubule and filament organization [17].
Figure 2.
Possible impact of sacsin in different cellular processes, including chaperon functions, microtubule organization and vesicle trafficking.
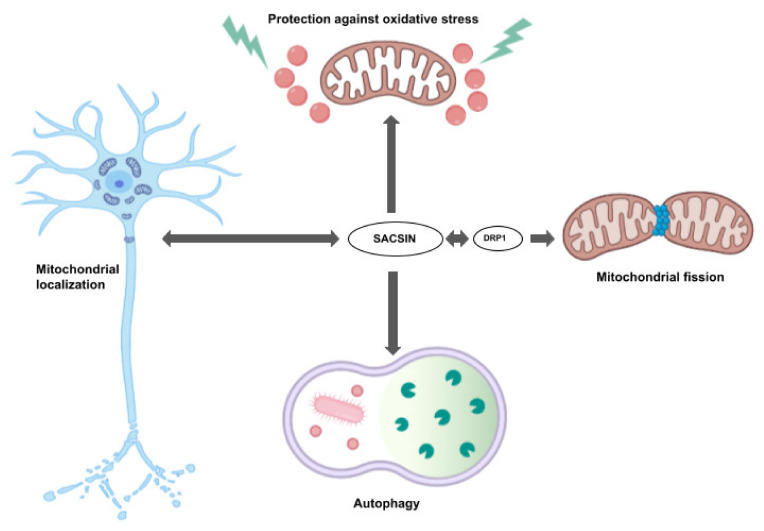
Sacsin is also localized in mitochondria and was suggested to play a key role in different mitochondrial functions (Figure 3). It was suggested to be involved in mitochondrial network regulation and connection and morphology. Sacsin was verified to bind the dynamin-related protein 1 (Drp1) [21]. Lower sacsin levels resulted in failure to recruit appropriate amounts of Drp1, leading to abnormalities in mitochondrial quality control [34]. Cellular and animal model studies were investigated to define the pathways related to sacsin dysfunctions. Girard et al. (2012) found that fibroblasts from ARSACS patients presented abnormal mitochondrial functions [31]. Later, Pilliod et al. suggested that monitoring mitochondrial abnormalities could be a possible diagnostic biomarker in ARSACS patients [35,36]. Sacsin also plays an important role in controlling the localization of mitochondria in neurons, and in appropriate dendrite development and morphology. Knockdown of SACS could result in several mitochondrial dysfunctions, such as abnormal hyperfused/balloon-like/bulbed mitochondria, or an abnormally interconnected network through lower fission. Additional dysfunctions could be disturbances of mitochondrial transport into neurons. Mitochondria accumulated in soma and proximal dendrites instead of the inner neural cell bodies and along the full length of dendrites. In nerve cells with non-functional sacsin, the number of dendrites was reduced. In addition, the remaining dendrites became thicker compared to the normal ones [21]. HGF studies by Criscuolo et al. revealed that sacsin dysfunctions could reduce the mitochondrial respiratory rate and ATP synthesis, resulting in oxidative stress [19]. Morani et al. used Crispr/Cas9 technology to knockout sacsin in SH-SY5Y cell lines, which showed that cells with SACS knockout obtained reduced oxygen consumption and higher DNA damage by reactive oxygen species (ROS). The gene expression profile was also changed: differently expressed genes were observed in several pathways, such as autophagy, mitochondrial dynamics and apoptosis [36]. There is a possibility that sacsin may also influence autophagy. Morani et al. (2019) analyzed differentially expressed genes (DEGs) in sacsin knockout SH-SY5Y neuroblastoma cells, and, besides oxidative phosphorylation and mitochondrial dynamics, the autophagy- and cell-death-related genes were included among the most significant DEGs. Cells without functional sacsin showed a reduced degree of autophagosome aggregation and its fusion for lysosomes. These findings also reveal that ARSACS dysfunctions could result in reduced clearance of damaged cellular organelles [36].
Figure 3.
Sacsin involvement in mitochondrial functions.
4. SACS Genetics and Mutations
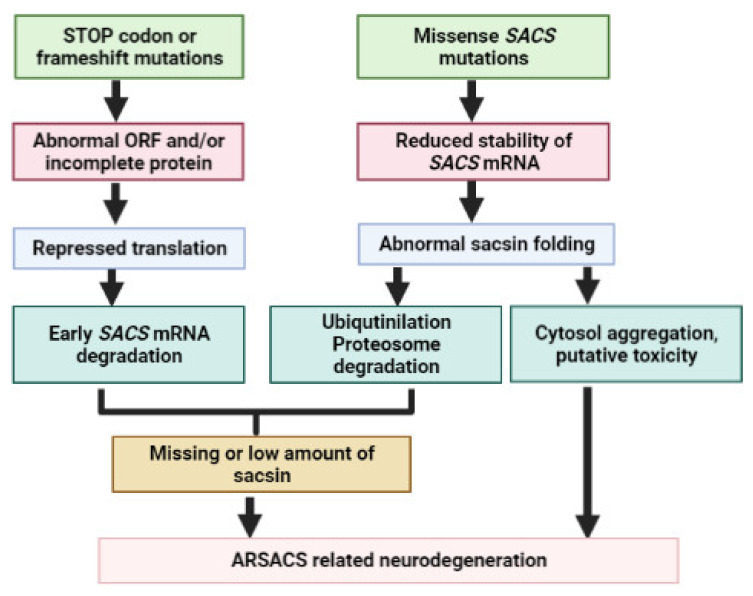
More than 200 pathogenic mutations have been reported in the SACS gene all around the world (Table 1, Table 2, Table 3 and Table 4). The majority of mutations were found in exon 10, which was verified as the longest exon among vertebrates [37]. The majority of mutations may result in non-functional SACS or reduced sacsin expression. Disease-associated variants can be either homozygous or compound heterozygous. The phenotypes of mutations may be diverse; besides the classical phenotypes (ataxia, spasticity), additional atypical symptoms may also be present (mental retardation, memory dysfunctions). Mutations can be either missense mutations, STOP codon mutations or frameshift variants (Figure 4). The missense mutations could result in lower stability and abnormal conformation of sacsin. Normally, the misfolded sacsin goes through co-translational ubiquitination and degradation by the proteosome system. If degradation does not happen, mutant sacsin could potentially aggregate in cytosol. Based on other neurodegenerative diseases, the potential aggregation of sacsin may result in putative additional gain-of-function toxicity. The frameshifts or nonsense variants may repress the translation, resulting in the degradation of the SACS transcript. All of the above could be associated with missing sacsin expression or insufficient amounts of sacsin protein [38].
Table 1.
SACS mutations detected in French Canadian and Tunisian patients. “hm” means mutation carried the homozygous form, “c het” means mutation had compound heterozygous allele. “rs” means respectively.
| Mutation | Exon | Domain | AOO | Clinical Symptoms | Neurological Changes | Population (Ethnicity) |
Refs. |
|---|---|---|---|---|---|---|---|
| c.6594delT (I2949Ffs*4,) or c.5254C > T (p.Q1752X), hm or c-het |
10 | SIRPT1 + SIRPT3 rs | 12–18 mths |
|
|
Canada (French Canadian) |
[5,38] |
| several variants, c.8844delT (p.I2949FfsX2952): most common, hm or c-het |
8 or 10 |
SIRPT1, 2 or 3, XPBC or DNAJ | NA |
|
NA |
[42] | |
| c.9284dupC 9p.Ala3096Cysfs*2), hm | 10 | SIRPT3 | early child- hood |
|
|
[41] | |
| c.10046G > C (p.A3324P), hm | 10 | Between SIRPT3 and XPBC | 2–4 yrs |
|
|
Tunisian (Tunisian) |
[43] |
| c.1411delT, hm | 8 | SIRPT1 | 5 yrs |
|
|||
| c.1155insA, hm | 8 | SIRPT1 | 1–9 yrs |
|
|||
| c.3662T > C (p.W1196R), hm | 10 | SIRPT1 | 10–14 yrs |
|
|||
| c.12846_12850delAGAG, hm | 10 | Between XPBC and DNAJ | 1–3 yrs |
|
|
[44] | |
| c.2439-2440delAT(V815Gfs*4), hm | 10 | SIRPT1 | 13–19 yrs |
|
|
[45] |
Table 2.
SACS mutation cases discovered in Europe. “hm” means mutation carried the homozygous form, “c het” means mutation had compound heterozygous allele. “rs” means respectively.
| Mutation | Exon | Domain | AOO | Clinical Symptoms | Neurological Changes | Country (Ethnicity) |
Refs. |
|---|---|---|---|---|---|---|---|
| c.1859insC, hm |
8 | SIRPT1 | 2 yrs |
|
|
Italy (Italian) |
[48] |
| c.4999del CAGAA5000-(p.C1679X), hm |
10 | SIRPT2 | before 10 yrs |
|
|
[47] | |
| c.1858C>T (p.Q620X), c.4585insA:V1528→p.fsX1540), c.het | 8 & 10, rs | SIRPT1 & SIRPT2, rs | before 2 yrs |
|
|
||
| c.563G>A (p.G188E) + c.7394C>T (S2465L), c.het | 7 & 10, rs | SIRPT1 & SIRPT3, rs | 15 yrs |
|
|
[49] | |
| c.962G>A; p.R321Q + c.8330G>A, (R2777L), c.het | 8 & 10, rs | SIRPT1 & SIRPT3, rs | 15 mths |
|
|
||
| 16 novel mutations, including a large deletion, hm or c-het | 8 & 10 | SIRPT 1-2-3 or XPCB or DNAJ | 1–32 yrs |
|
|
[50] | |
| c.4198T>A/c.5719C>T, c.het | 10 | SIRPT2 | 15–16 yrs |
|
|
[51] | |
| c.13132C >T (p.R4378X), hm | 10 | DNAJ | 2 yrs |
|
|
[52] | |
| c.5629C>T- (p.R1877X), hm | 10 | SIRPT2 | 26 mths |
|
|
[13] | |
| c.600_604+1delAACAGG (p.I202fsX6), hm; 1.5 MB large deletion | 8 | SIRPT1 | 42 yrs |
|
|
[53] | |
| c.6680T>C (p.L2374S), hm | 10 | SIRPT2 | early childhood |
|
|
[13] | |
| c.10743C>T (p.Q3582X), hm | 10 | XPCPB | early childhood |
|
|
[54] | |
| c.11471A>G, (p.N3799D), hm | 10 | SIRPT3 | 2.5–3.5 yrs |
|
|
Turkey (Turkish) |
[55,56] |
| c.9655_9658 delAGTT), truncation of I3199, hm | 10 | SIRPT3 | 1.5 yrs |
|
|
||
| c. 2018T>C (p.C648R), hm | 8 | SIRPT1 | 2.5 yrs |
|
|
||
| c. 8124delC-truncation of p.A2708, hm |
10 | SIRPT3 | 2 yrs |
|
|
||
| c.1160A>G (p.F4011S), hm |
10 | between SIRPT3 & DNAJ | 1 yr |
|
|
[57] | |
| c. 6945A>G (p.V3369A), hm |
10 | SIRPT3 | 3–7 yrs |
|
|
||
| c.12841T>A (p.K1404), c.6945A>G (p.V3369A), c.het | 10 | SIRPT2 & SIRPT3,rs | 2 yrs |
|
|
||
| c.6945A>G (p.V3369A), c.12020C (p.S4007F), c.het | 10 | SIRPT3 & loop between XPCB-DNAJ | 4 yrs |
|
|
||
| c.8346–8347insT (p.G2902V), hm | 10 | SIRPT2 | 2 yrs |
|
|
||
| c.5677G>A (p.R3801X), hm |
10 | SIRPT3 | 3 yrs |
|
|
||
| c. c.13485delC (p.K4495N), hm |
10 | DNAJ | 3 yrs |
|
|
||
| p.G2772A, hm | 10 | SIRPT3 | early childhood |
|
|
[58] | |
| c.2182C>T (p.R728), hm | 8 | SIRPT1 | 4 yrs |
|
|
[59] | |
| c.12461delC (p.P4154QfsX20), hm | 10 | between XPCB & DNAJ | After 1 yr |
|
|
[60] | |
| Several mutations, hm or c het | 8 or 10 | SIRPT2 or 3 or between SIRPT3 & DNAJ | 1–13 yrs |
patients had similar symptoms:
|
|
Netherlands (Dutch, British, Turkish) |
[61] |
| c.3491T>A, p.M1164K, hm | 10 | SIRPT1 | 12–13 yrs |
|
|
Belgium (Belgian) |
[62] |
| Several mutations, hm or c -het | 8 or 10, deletion of exon 3–5 | SIRPT1 or 2 or 3 or HEPN | 1–24 yrs |
|
|
Belgium (Belgian, Moroccan, Serbian, Hungarian) |
[63] |
| c.10517T>C (p.F3506S) + chromosomal deletion, c-het | 10 | SIRPT3 | 16 yrs |
|
|
[76] | |
| NA | NA | NA | 1 yr |
|
|
Spain (Spanish) |
[77] |
| c.7848C>T (p.R2556C), hm | 10 | SIRPT3 | before 1 yr |
|
|
[64] | |
| c.826C>T (p.R276C) + c.3904C>T (p.P1302S), c-het | 8 & 10, rs | SIRPT1+ between SIRPT3 & DNAJ, rs | early childhood |
|
|
[65] | |
| c.9695delC (p.T3232KfsX24),hm | 10 | SIRPT3 | 8–10 yrs |
|
|
Greece (Greek) |
[66] |
| c.9804_9805insC, (p.S3268 _Ifs), hm | 10 | SIRPT3 | 1.5–5 yrs |
|
|
Poland (Polish) |
[67] |
| c.72276C>T (p.R2426X) | 10 | SIRPT3 | 32 yrs |
|
|
Russia (Russian) |
[68] |
| c.13352T>C (p.L4451P) + c.6890T>G (p.L2297W), c.het | 10 | HEPN+ SIRPT2, rs | before 10 yrs |
|
|
Norway (Norwegian) |
[69] |
| 17 novel SACS mutations, hm or c-het | 8 + 9 + 10 |
SIRT1 or 2 or 3 or DNAJ or HEPN | 1–30 yrs |
|
|
Germany (German, Turkish, Greek, Macedonian) |
[4] |
| 9 different mutations in 6 families, hm or c-het | 8 or 10 | SIRT1 or 2 or 3 or DNAJ or HEPN | 2–15 yrs |
|
|
Germany (German, Italian, Romanian, Turkish, Arabic) | [70] |
| c.2076delG (p.T692) + c.3965_3966del (p.G1322Vfs*1343), c-het | 8 & 10 | SIRT1+ SIRPT2, rs | 19–26 yrs |
|
|
UK (British) |
[71] |
| 20 mutations, 11 novel mutations, hm or c-het | 10 | SIRT1 or 2 or 3 or DNAJ or HEPN | 1–46 yrs |
|
|
[72] | |
| c.5744_5745delAT (p.H1915Rfs*19),hm | 10 | SIRPT2 | 9–19 yrs |
|
|
France (French) |
[73] |
| p.E1100K + p.N1489S + p.M1359T, c-het | 10 | SIRT1+ SIRPT2, rs | 6–15 yrs |
|
|
Finland (Finnish) |
[74] |
| c.13721T/G (p.F4574C), hm | 10 | HEPN | 37 yrs |
|
|
Macedonia (Macedonian) |
[75] |
Table 3.
SACS mutations discovered in Asia. “hm” means mutation carried the homozygous form, “c het” means mutation had compound heterozygous allele. “rs” means respectively.
| Mutation | Exon | Domain | AOO | Clinical Symptoms | Neurological Changes | Country (Ethnicity) |
References |
|---|---|---|---|---|---|---|---|
| p.3774C>T, p.Q1198X, c.het | 10 | XPCB & SIRPT1, rs | 9 yrs |
|
|
Japan (Japanese) |
[78] |
| c.2951_2952delAG(p.Q984GfsX986)+3922delT(p.1308LfsX1326), c. het | 10 | SIRPT1 + between SIRPT3 & XPBC, rs | 15–20 yrs |
|
|
[81] | |
| c.32627-32636delACACTGTTAC(p.W395-fsX407), c.31760delT (p.V687-fsX713), c.het | 8 | SIRPT1 | under 10 yrs |
|
|
[79] | |
| c.6543delA, (p.R2002X), hm | 10 | SIRPT2 | early childhood |
|
|
[80] | |
| c.5988-9del CT, hm | 10 | SIRPT2 | early childhood |
|
|
[81] | |
| c. 987C>T (p.F304S), hm | 8 | SIRPT1 | before 10 yrs |
|
|
[82] | |
| c. 6355C>T (p.R2119X), hm | 10 | SIRPT2 | 20s |
|
|
[83] | |
| c.482delA (p.L802P), c.2405T>C (p.N161fsX175), c-het | 10 & 7 rs | SIRPT1 | late 10s- early 20s |
|
|
[85] | |
| c.12976A/G (p.K4326Q), c.4233-4236 delACTT (p.L1412Kfs*16), c-het | 10 | DNAJ + SIRPT2, rs | ~22 yrs |
|
|
[85] | |
| c.3769 G>T (p.G1257X)+11361–2insT(p.R3788SfsX3820), c-het | 10 | SIRPT2 & SIRPT3, rs | 12 yrs |
|
|
[86] | |
| c.414 C>G (p.Y138X) +5263–4delAA (p.K1755VfsX1775), c-het | 7 & 10, rs | SIRPT1 & SIRPT2, rs | 12–19 yrs |
|
|
[87,88] | |
| c.4756_4760del (p.N1586Yfs*3)+ putative noncoding mutation, c-het | 10 | SIRPT2 | early childhood |
|
|
Korea (Korean) |
[89] |
| c.8844delT (p.I2949Ffs*4) + c.11781_11782dupGC (p.P3928Rfs*17), c-het | 10 | SIRPT3 +between XPCB & DNAJ, rs | ~20 yrs |
|
|
[90] | |
| c.7272C>A (p.C2424X), c.11319_11321del (p.R3774del), c-het | 10 | SIRPT3 +XPCB, rs | ~10 yrs |
|
|
[91] | |
| c.11803C>T (p.Q3935X)+ 1.33Mb deletion, c-het | 10 | between SIRPT3 & XPCB | 6 yrs |
|
|
China (Chinese) |
[92] |
| c.12637 _12638delGA (p.Q4213Rfs*3)+ c.11274_11276delAAC (p.I3758_ TdelinsM), c-het | 10 | between XPCB & DNAJ + XPCB, rs | 10’s |
|
|
[93] | |
| c. 8000T>C (p.F2667S), c. 10685_10689del (p.F3562X), c-het | 10 | SIRPT3 + between XPCB & DNAJ + XPCB, rs | early childhood |
|
|
[94] | |
| c.5236dupA (p.T1746fs)+ c.13085T/G (p.I4362R), c-het | 10 | SIRPT2 +DNAJ, rs | NA |
|
|
[96] | |
| c.9019C>T, p.P3007S and c.10174_10183del, p.H3392fs | 10 | SIRPT2 + between XPCB & DNAJ | early childhood |
|
|
[95] | |
| c.1773C>A (p.S578X) + c.8088_8089 insCA (p.M2697Q fs*43), c-het | 8 & 10, rs | SIRPT1 + SIRPT3, rs | 6 yrs |
|
|
||
| c.5692 G>T, p.E1898X; c.12673-12677 del TATCA, p.Y4225D fs*6-c-het | 10 | SIRPT1 +DNAJ, rs | Early childhood |
|
|
[97] | |
| c.1773C>A, p.S578X; c.8088-8089 in. CA, p.M2697Q fs*4 | 10 | SIRPT1 + SIRPT3, rs | 6 yrs |
|
|
||
| c.382_383del (p.Q128Sfs*2), hm | 7 | SIRPT1 | 2 yrs |
|
|
Thailand (Thai) |
[98] |
| c.5824_5827delTACT (p.Y1942Mfs*9), hm | 10 | SIRPT2 | early teens |
|
|
Kuwait (Kuwait) |
[99] |
| c.429_430delTT (p.W144VfsX39), hm | 7 | SIRPT1 | 3 yrs |
|
|
Iran (Iranian) |
[100] |
| c.4117_4118delGCinsAG (p.A1373R), hm | 10 | SIRPT2 | early childhood |
|
|
[101] | |
| c.14329fs*2725 (p.R707Kfs*6), hm | 10 | SIRPT2SIRPT1 | 9–15 yrs |
|
|
India (Indian) |
[102] |
| c.11690_11693dupGTGA (p.N3898QfsX2),hm | 10 | XPCB | 4 yrs |
|
|
[103] | |
| c.8605delT (p.C2869VfsX15), hm | 10 | SIRPT3 | 14 mths |
|
|
[104] | |
| c.8793 delA, hm | 10 | SIRPT1 | early childhood |
|
|
[105] | |
| c. 4232T>G + c.8132C>T, c-het | 10 | SIRPT2 + SIRPT3, rs | 3 yrs |
|
|
[106] | |
| c.2656C>T (p.Q886*), hm | 10 | SIRPT1 | 11–12 yrs |
|
|
Pakistan (Pakistani) | [107] |
| c.4756_4760delAATCA (p.N1586Yfs*3), hm | 10 | SIRPT2 | 9–10 yrs |
|
|
||
| c.9119dupA (p.N3040Kfs*4), hm | 10 | SIRPT3 | 1.5 yrs |
|
|
[108] | |
| p.N2380K & p.D3269N, c-het | 10 | SIRPT2 + SIRPT3, rs | 16 mths |
|
|
Israel (Ashkenazi Jews) |
[109] |
Table 4.
SACS mutations discovered in USA, South America and New Zealand. “hm” means mutation carried the homozygous form, “c het” means mutation had compound heterozygous allele. “rs” means respectively.
| Mutation | Exon | Domain | AOO | Clinical Symptoms | Neurological Changes | Country (Ethnicity) |
Refs |
|---|---|---|---|---|---|---|---|
| c.3484G>T (p.E1162X), & c. 11707C>T (p.R3903X), c-het | 10 | SIRPT1+ between SIRPT3 & XPBC, rs | 2.5–3.5 yrs |
|
|
USA (American) | [110] |
| c.11637_11638delAG (p.R3879fs), hm | 10 | SIRPT3 & XPBC, rs | 4 yrs |
|
|
[111] | |
| Chr13 duplication | NA | NA | 13 yrs |
|
|
[112] | |
| c.4744G>A (p.D1582N) + c.7205_7206delTT (p.L2402Rfs*6), c-het | 10 | SIRPT2 + SIRPT3, rs |
2 yrs |
|
|
USA (American or mixed European) | [113] |
| c.11824dup (p.M3942Nfs*4), hm | 10 | XPCB | 11 yrs |
|
|
USA (African American) |
[114] |
| 11 SACS variants in 39 patients, hm or c-het | 8 or 10 | SIRPT 1-3, XPBC or HEPN | NA |
|
|
USA (NA) |
[115] |
| NA | NA | NA | early childhood |
|
|
Brazil (Brazilian) | [116] |
| c.5150_5151insA | 10 | SIRPT2 | early childhood |
|
|
Brazil (German) |
[117] |
| Several variants, including p.L393Cfs*17 & p.N2760Mfs*6 | 8 or 10, rs |
SIRPT 1-3 or XPBC | 1–44 yrs |
|
|
Brazil (Brazilian) | [118] |
| c.8107C>T (p.R2703C) +c.922C>T-(p.L308F), hm | 10 + 8, rs | SIRPT3 + SIRPT1, rs | 8–9 yrs |
|
|
[119] | |
| c.7962T>G (p.Y2654X), hm | 10 | SIRPT3 | 20’s |
|
|
New Zealand (Maori & English) |
[120] |
Figure 4.
Potential effects of mutations in SACS gene.
The first SACS mutations were reported in the CSLSJ family. In this population, more than 300 patients were observed who could have been the descendants of a single founder [5,37]. Several mutations were found in the affected population in the Quebec region. Initially, two pathogenic mutations were identified, c.6594delT (mutation location on transcript) or p.I2949Ffs*4 (mutation location on protein, 94% of mutant alleles) and c.5254C>T (p.Q1752X, 3%). Both mutations could be related to the truncation of the sacsin protein [5,37,38]. Later, additional variants were observed in the Quebec population, including five missense variants, five indels, one nonsense and one large genome deletion. The mutation c.8844delT (p.I2949FfsX2952) seemed to be relatively common among ARSACS patients. However, additional variants also appeared in several ARSACS cases, such as c.4744G>A (p.D1582N), c.814C>T (p.R272C), c.237insAfsX (p.S80IfsX98) and c.5836T>C (p.W1946R). This study also suggested that large deletions in French Canadians (Table 1) may not be either common or a lethal variant. The diverse phenotypes of the disease may appear due to the partial loss of function in the sacsin protein [39,40,41].
Besides French Canadians, several SACS mutations and ARSACS cases have been detected around the world. In Tunisian families, patients with ARSACS were late-onset cases with similar clinical symptoms in comparison to the French Canadians [43,44]. In 2003, 18 ARSACS patients from four families in Tunisia were analyzed and displayed two STOP codon and two missense mutations: c.10046G>C (A3324P), c.1411delT (frameshift, resulting in premature STOP codon of residue 456), c.1155insA (frameshift, resulting in premature STOP codon of residue 360) and c.3662T>C (W1196R) (Table 2). The STOP codon variants were predicted to be involved in the loss of DNAJ domain of the sacsin protein and reduced or loss of chaperon activity. The missense mutations could result in abnormal folding of the protein secondary structure. Although these families were not related to each other, inbreeding was found to be high in Tunisia, suggesting the possibility of a founder effect [43]. Furthermore, in 2009, an additional mutation was found in a Tunisian male patient with a familial case of ARSACS who developed dysarthria and gait dysfunction between one and two years of age, followed by language impairment and ataxia. A four-base deletion was found in proband c.112846_12850delAGAG, resulting in a STOP codon at residue 4305. Parents of patients were first cousins and carried the heterozygous indel. The mutation was associated with a loss of 274 amino acids and missing C-terminal region, including the DNAJ domain [44].
In Europe, several ARSACS cases were discovered in different countries, including Italy, Spain, Turkey and Belgium (Table 2) [46]. In Italy, ARSACS has been quite extensively studied. The first cases of SACS mutations were discovered by Grieco et al. (2004), and this study reported three novel variants in patients. The first patient carried a 5-base deletion (del4999cagaa5003) at residue 4999, which could result in the absence of the most protein sequence, including the DNAJ and HSP90 domains. The second patient carried two possibly pathogenic variants: 1858C>T (Q620X) and an insertion (4585insA: V1528fsX1540) [47]. Another Italian mutation was discovered in SACS, 1859insC, resulting in a premature STOP codon at residue 599, where most of the protein was lost [48]. Bi-allelic cases also appeared in Italian patients, such as c.563G>A (p.G188Q) + c.7394C>T (p.S2465L) double mutations and c.962G>A (p.R321Q) + c.8330G>A (p.R2777K) double mutation in two different Italian patients, and both were predicted to be damaging in silico. Both patients presented elevated levels of mitochondrial fragmentation. The patient with c.563G>A (p.G188Q) + c.7394C>T (p.S2465L) mutations also showed reduced sacsin levels in fibroblasts [49]. In 2013, Prodi et al. found several homozygous or compound heterozygous SACS mutations, and one of them was a large in-frame deletion. The majority of patients developed the first disease phenotypes at early childhood, but one of them presented late onset (32 years). These patients had early gait instability, Babinski sign and pes cavus, but spasticity and elevated muscle tone were not among the early symptoms. Imaging showed cerebellar superior vermis atrophy, pontine changes and thinning corpus callosum in all patients [50]. Additional homozygous variants also appeared in Italian patients, such as c.4198T>A + c.5719C>T [51], c. 1859insC [48], c.13132C>T [52], c.5629 >T [13], c.600_604+1delAACAGG [53], c.6680T>C [13] or c.10743C>T [54], c.11471A>G [54] and a large deletion at the SACS region. The majority of these cases were related to young-onset ataxia, and hypermyelinated retina was common among them. The patient with the homozygous c.600_604+1delAACAGG variant with the large 1.5 MB large deletion developed an ataxic phenotype at an atypically late age of 42 years [53]. Another compound heterozygous case of ARSACS mutations was found by Pensabene et al. (2020) in two siblings who developed ataxic symptoms before the age of 20, and it worsened in their 30 s [51]. Several SACS mutations were reported in Turkish cases. The first mutations were observed in 2004: this study reported two missense (W1196R and N3799D) mutations and two indels (L3193-fsX3199 and T2683-fsX2708)[55,56]. Later, additional mutations were also discovered: Oguz et al. analyzed nine unrelated families and revealed eight novel mutations in the SACS gene (5019A>G/F4011S, 14370G>T/S894X, 12841T>A/K1404X, 5031 G>A/ S4007F, 12660A>G/ I1464T, 8346–8347insT, 5677G>A/ R3801, 5566delC), and one variant (6945A>G/V3369A) [57]. Kurt et al. reported a homozygous G2772A mutation in a Turkish female patient with ataxia and spondyloepiphyseal dysplasia, but it was heterozygous in unaffected family members. The mutation co-existed with two homozygous mutations in the ACAN gene, which may have been responsible for skeletal deformations [58]. An additional mutation, c.2182C>T, appeared in a Turkish child with low progressive difficulty in walking and slurred speech. This child was negative for mutations responsible for any kind of spinocerebellar ataxia [59]. Recently, an indel (p.P4154QfsX20) was observed in a family with progressive spastic ataxia and dysarthria, with uncommon symptoms such as intellectual disability, hearing dysfunctions and epileptic seizures [60].
Besides Italy and Turkey, additional patients have been examined in other European countries. In the Dutch population, SACS mutations seemed to be quite prominent in patients with early-onset cerebellar ataxia. Vermeer et al. (2008) analyzed 43 families with ataxia. Patients developed a disease phenotype under 25 years of age. Among them, 16 patients showed novel mutations in the SACS gene in either the homozygous or compound heterozygous stage. Among these variants, there were eight STOP codon mutations, three were missense, two splice site variants and three deletions (in-frame or frameshift). Symptoms of mutations were very similar in each family: all of them developed early-onset ataxia (before 13 years of age) with lower limp spasticity, neuropathy in semi-motor axons and atrophy in the cerebellum [61]. The first Belgian case of SACS mutation was a missense variant, c.3491T>A (p.M1164K), detected in a family with similar symptoms to Quebec patients, but additional phenotypes also appeared, such as no retinal nerve hypermyelination or teenage onset of disease [62]. Baets et al. (2010) examined several ARSACS patients; the majority of them were Belgian descendants. In 11 families (17 patients), 18 mutations were observed, including an intragenic SACS exon 3-5 deletion. In 12 individuals, classic ARSACS phenotypes were found. Several patients had later onset of disease, since they developed gait difficulties or distal weakness after 20 years of age, which may not be associated with classical ARSACS [63]. The first mutation in Spain, reported by Criscuolo in 2005, involved c.7848C>T(p.R2556C), which presented a similar classical phenotype to the Quebec population. The mutation spared the DNAJ domain, co-segregating in the family, and was missing in controls. In addition, this variant was reported to affect a conservative residue (R2556), which also confirms its role in the disease [64]. Another Spanish female patient with a compound heterozygous p.R276C and p.P1302S was reported in 2015. She developed gait instability and slurred speech in her teenage years [65]. A Greek family carried a deletion, p.T3232KfsX24 (c.9695delC), where the siblings were homozygous for this variant, but their unaffected parents were both heterozygous. Affected siblings developed gait disturbances and ataxia in their first decade of life [66]. The first Polish case of ARSACS was described in 2017, in a patient who carried two novel mutations in the compound heterozygote stage: p.S3268_I3269fs/c.9804_9805insC and p.D4192N/c.12574 G>A. The mutation was missing in the healthy population, while unaffected siblings and the mother carried only one of these variants. Besides the classical ARSACS symptoms, affected patients presented cognitive or behavioral dysfunctions too [67]. A Russian case of the disease was discovered with atypically late onset of ARSACS and c.72276C>T (p.R2426X) mutation. The patient developed typical ataxia symptoms at the age of 32 years [68]. A Norwegian case of a compound heterozygous mutations c.13352T>C, p.L4451P; c.6890T>G, p.L2297W were found in a family with a typical form of spastic ataxia and other dysfunctions, such as cognitive decline and epilepsy [69]. In Germany, two studies were performed on ARSACS patients [4,70]. Synofzik et al. (2013) sequenced 22 patients with early-onset ataxia, and 17 novel homozygous or compound heterozygous variants were reported. All variants were missing in unaffected individuals and located in conserved domains. While some patients presented classical cases of ARSACS, some atypical symptoms were also apparent, such as pure neuropathy (which characterizes the Charcot–Marie–Tooth disease/CMT), missing neuropathy, absence of spasticity or cerebellar ataxia. Interestingly, besides the cerebellum and pons, MRI revealed abnormalities in other brain areas, such as atrophy of the cerebral parietal region or thinning of the corpus callosum [4]. Vill et al. (2018) identified seven SACS variants in nine patients who were diagnosed with non-syndromic hereditary motor and sensory neuropathy (HMSN). These mutations were associated with atypical phenotypes, such as sensory ataxia only, absence of spasticity and pyramidal signs. MRI also did not present any typical ARSACS characteristics, since it was either normal or presented isolated atrophy of the superior cerebellar vermis [70]. In the UK, a case was detected with unexplained multisystem neurological disorder in two siblings, who presented two heterozygous SACS variants (c.2076delG, p.Thr692Thr fs*713 and c.3965_3966delAC, p.Gly1322Val fs*1343). Patients had pyramidal signs, later onset (19–26 years) and no retinal nerve thickening or hypermyelination [71]. An additional study in the UK analyzed 191 patients with different types of genetic ataxia and 101 controls. Among them, 17 patients had ARSACS and carried 20 variants. Among these, 11 were unknown mutations. The majority of variants were compound heterozygous, but one deletion was found to be homozygous. The age of onset was variable (1–46 years). All of them had ataxia and nystagmus. The majority of them presented spasticity (expect one). Additional possible symptoms were sensory loss, limb weakness and dysarthria. Patients did not have visual complaints, but 12 out of 17 presented thickened retinal nerve fibers [72]. In France, a homozygous variant was found in two siblings with demyelinating motor-sensitive neuropathy (c.5744_5745delAT). Patients were also heterozygous for a larger SACS exon 10 deletion [73]. In a Finnish family, three heterozygous mutations were detected (p.E1100K, p.N1489S and p.M1359T). Patients presented spastic ataxia and distal weakness, and some family members were initially diagnosed with CMT. Sensorimotor demyelinating and cerebellar ataxia were common among them, and disease onset was between 6 and 25 years [74]. In Macedonia, one patient presented ataxia, gait, speech issues and mood swings at the age of 53, but depression appeared in her 30 s. She carried a homozygous SACS mutation, c.13721T>G;p. (F4574C), and developed cognitive decline [75].
In Japan, SACS mutations were widely investigated (Table 3). Several Japanese cases presented similar phenotypes to the Quebec patients, as well as atypical cases, such as late-onset ataxia, ataxia without spasticity or retinal optic nerve hypermyelination. In 2004, a compound heterozygous sacsin mutation (C3774T, Q1198X) was discovered in a woman who developed spastic gait at the age of 9, which deteriorated in her 30s (clumsiness in hands, unsteadiness) [78]. Another compound heterozygous case of mutation was found in monozygotic twin sisters (c.2951_2952delAG+3922delT), who were unable to run fast and fell easily during their childhood. Their menstruation cycle became scanty and irregular, and pollakiuria appeared in them. No additional family member presented any neurological symptoms [79]. In 2005, two mutations, W395-fsX407 and V687-fsX713A, were found in a 25-year-old woman. The patient had slow gait progression, and she needed assistance with walking in her 20s. In addition, her speech became slurred. No myelinated retinal fiber was notable in her, and her motor nerve conduction became lower in several brain regions, including the median and ulnar nerves [80]. A homozygous deletion, c.6543delA, was discovered in two siblings with unique clinical symptoms, such as dementia, ophthalmoplegia, spasticity and amyotrophy. However, myelinated retinal fibers were absent [81]. Another homozygous two-base deletion, c.5988–9 del CT, was detected in a 29-year-old patient with spastic ataxic gait, distal muscle weakness and myelinated retinal nerve fibers in the retina [82]. Later, a sacsin T987C mutation was found in two patients without spasticity, but with a mild degree of ataxia in the form of slurred speech [83]. Additional atypical cases revealing an R2119* nonsense mutation were found in two Japanese siblings with spasticity and without retinal optic nerve hypermyelination [84].
In 2008, a compound heterozygous case of SACS N161fsX175 and L802P from two siblings with typical Japanese ARSACS patients was reported. Several family members with only one separate mutation were not affected [85]. In 2012, two novel mutations of compound heterozygous (c.[3769 G>T]+[11361–2insT] and c.[414 C>G]+[5263–4delAA]) were reported by Shizamaki et al. (2012) with age of onset at 12 years, and imaging analyses revealed atrophy in the cerebellum and pons [86]. In 2021, a compound heterozygous case with p.K4326E and p.L1412Kfs*16 mutations was discovered in a woman in her 40s, who had exhibited a slow progressive gait since childhood. Although she revealed the typical phenotype, it appeared to be milder than the typical ARSACS phenotypes. She did not present spasticity upon neurological examination, and the polyneuropathy was also missing [87,88].
Cases of ARSACS have also been discovered in other Asian countries, including China, Iran and Korea. The first Korean case of ARSACS was discovered in 2015 with a heterozygous deletion c.[4756_4760del] (p.N1586Yfx*3) in a 17-year-old female patient with classical early-onset spastic ataxia with sensorimotor polyneuropathy and distal amyotrophy. Since the patient was heterozygous with the novel deletion, it was possible that other mutations could be identified from the noncoding region (for example, splice site) or larger deletions [89]. In 2018, the second Korean case was found, in a patient who carried the compound heterozygous mutations c.8844delT (p.I2949Ffs*4) and c.11781_11782dupGC (p.P3928Rfs*17). The patient presented early-onset cerebellar ataxia, gait disturbances and weakness in the lower extremities. Neuroimaging and ophthalmologic analysis also supported the ARSACS diagnosis [90]. A recent case of another compound heterozygous SACS mutation, c.7272C>A:p.(C2424*) and c.11319_11321del:p.(R3774del), was found in a patient with cerebellar ataxia with migraine [91]. The patient developed gait disturbances in his teenage years, and his younger sister also developed similar symptoms [91].
In China, the first ARSACS case was discovered in 2016 in a patient with a compound heterozygous mutation, a c.11803C>T (p.Q3935X) variant, and a 1.33 megabase deletion of SACS [92]. The patient was diagnosed with an atypical case of ARSACS without spasticity in the legs or cerebellar ataxia. Instead, the clinical presentation involved early-onset peripheral neuropathy, mild spastic gait and horizontal gaze nystagmus [93]. Another case of spastic ataxia was discovered in 2018 in a patient with two novel variants, c.12637_12638delGA (p.E4213Rfs*3) and c.11274_11276delAAC (p.I3758_T3759delinsM) [93]. Each mutation was inherited from each unaffected parent. The patient presented cerebellar ataxia from his teenage years, followed by sensory–motor neuropathy, finger deformities and thickened retinal nerve fibers. Nerve conduction studies revealed that sensory action potentials were missing in all of his limbs, with reduced motor conductions [93]. Recently, a compound heterozygous mutation, c.8000T>C, p.F2667S and c.10685_10689del, p.F3562* mutations, were reported in a 30-year-old male patient with progressive ataxia without lower limb spasticity [94]. Additional Chinese cases of SACS mutations were also detected in ARSACS patients, such as p.P3007S + p.H3392fs; p.W1367X [95], p.T1746fs)+ p.I4362R [96], E1898X+ Y4225D or p.S578X + p.M2697Q fs*4 [97].
In Thailand, the first ARSACS patient was found at the age of 2, with progressive spastic ataxia from a homozygous mutation, c.382_383del (p.E128Sfs*2). [98] The patient presented several cerebellar dysfunctions and hypermyelination in the nerves of the optic disc. Cognitive functions remained normal [98].
In 2020, a homozygous frameshift mutation (c.5824_5827delTAC, a premature termination at residue 1942, p.Y1942Mfs*9) was found in an ARSACS patient from the Arabic peninsula (Kuwait) [99]. The patient presented a typical form of cerebellar ataxia, which also appeared in several family members (nephews, nieces). Balance disorder started from his teenage years. He also had a history of diabetes mellitus, retinal hemorrhage and transient ischemic attack prior to ARSACS [99].
An Iranian family presented atypical ARSACS with mirror movements, hypokinesia, bradykinesia and rigidity. Thickening in retinal axons was present in the affected patients. Affected family (female proband and her brother) members carried a novel homozygous mutation c.429_430delTT: p. W144VfsTer39 [100]. Another Iranian case revealed a homozygous A1373R mutation in a child, who was initially diagnosed with spinal muscular atrophy II (SMA-II). The significance of this mutation is currently unclear [101].
In recent years, a few ARSACS cases have been reported in Indian patients. The first case was found in 2014 in a patient with suspected Friedrich ataxia without spasticity and retinal fiber abnormalities initially, prior to a finding of a homozygous frameshift (c.14329fs*2725, p.R707Kfs*6) [102]. The second case was observed in 2017 with a 4-base-pair duplication in exon 10 (c.11690_11693dupGTGA; p.D3898EfsX2), in a patient with typical ARSACS symptoms: ataxia, motor dysfunctions, language impairment and hypermyelinated nerve fibers [103]. In 2019, another ARSACS case was found in a patient in his 20s from the Remote Tribal Jammu and Kashmir region, who had mild intellectual disabilities [104]. A homozygous frameshift mutation (C2869VfsX15) was detected in this patient. Imaging analyses revealed a hyperintense rim (“bithalamic stripes”) near the thalamic region [104]. In 2020, another homozygous deletion (c.8793 del A) was reported in a female patient from Kerala, who experienced difficulties in walking from her early childhood, followed by tremors and difficulties in holding objects [105]. One of her sisters also developed similar phenotypes [105]. Next, a compound heterozygous mutation, c.4232T C>G nonsense mutation and c.8132C>T missense variant, was reported in 2020 in a patient with progressive gait ataxia, dysarthria and lower limb stiffness [106].
Three ARSACS cases with homozygous SACS mutations c.2656C>T, c.4756_4760delAATCA and c.9119dupA were discovered in Pakistan [107]. The c.2656C>T and c.4756_4760delAATCA were associated with later-onset disease (11–12 years and 9–10 years, respectively), followed by cognitive decline with mental retardation [107]. Another patient with the c.9119dupA mutation presented typical ARSACS symptoms at the age of 1.5 years old [108].
In Israel, compound heterozygous mutations (sacsin D3269N and N2380K) were found in an ARSACS patient with ataxia and hearing impairment. A study with the patient’s fibroblasts revealed mitochondrial abnormalities, such as reduced numbers of mitochondria and an impaired mitochondrial network. An atypical symptom of retinal degeneration also appeared in this patient [109]. These mutations are shown in Table 3.
The first case of ARSACS in the USA was reported in 2011, which was a compound heterozygous case (c. 3484 G>T, p. E1162X; and c.11707C>T, p. R3903X) detected in two Caucasian siblings (Table 4). Both siblings experienced clumsiness in walking, spasticity, and they were suggested to have spastic paraparesis initially. Due to the cerebellar atrophy, neuropathy in sensory motor neurons and possible autosomal recessive inheritance pattern, they were diagnosed with suspected ARSACS [110].
In 2013, a four-year-old child developed ataxia with delays in gross motor development and polyneuropathy. Whole-exome analyses revealed a homozygous mutation in SACS, 11637_11638delAG (p.R3879fs). In this family, consanguinity was present without a clear phenotype for ARSACS. Three of the patient’s cousins died with severe neonatal aspartylglucosaminuria [111]. Although a copy number variant (CNV) was found in one patient with multiple sclerosis (MS), features of ARSACS were prominent. This patient developed tremors and a mild balance disorder in their teenage years, with a history of learning difficulty and dyslexia. It could be possible that this individual carried both disorders [112].
Another case of ARSACS was detected in a male teenager with European heritage, who experienced stiffness in his legs and slow movement. At the age of 2, he was diagnosed with cerebral palsy and developed rigidity and bradykinesia later in life. He was also diagnosed with Parkinsonism. Although his speech production became slow, his language and intellectual abilities remained normal. The patient revealed a compound heterozygous known D1582N mutation and a frameshift c.7205_7206delTT (p.L2402Rfs*6). He also carried the A2510T variant with uncertain significance [113].
The first ARSACS case in the African American population was discovered in 2018, and the patient developed gait abnormalities and motor delay with low IQ. The patient also displayed headache and blurry vision. Array analysis revealed a 1.422 megabase loss in the chromosome 13q12.12 region, which contained the SACS gene. Another variant (c.11824dup) was detected in this patient. The maternal great grandfather of the patient also presented gait problems. Visual symptoms and seizures were prominent in several family members [114]. Fogel et al. (2012) analyzed several patients with sporadic ataxia, and discovered 11 SACS mutations in 39 patients, including p.N4573H, p.N4549D. A3927V or p.E174X. Majority of these patients presented spastic ataxia, but other disease phenotype also appeared, such as spinocerebellar ataxia, pure cerebellar ataxia or spastic paraplegia [115]
In Brazil, the first case appeared in a family with typical symptoms and neuroimaging features of ARSACS. Three siblings were affected, but the mutation was not found due to the absence of molecular genetic testing at that time [116]. In 2017, a case of homozygous c.5150_5151insA appeared in two female cousins of Germanic descent. Patients presented early-onset and slow progressive spastic ataxia. Retinal and nerve conduction abnormalities were also prominent [117]. In 2019, 13 Brazilian patients were investigated with ARSACS, who presented ataxia, spasticity and retinal nerve fiber thickening. Neuropathy and retinal abnormalities (peripapillary striations) or papillomacular fold were common in all 13 patients. Genetic analysis revealed 14 variants, among which two variants (p.L393Cfs*17 and N2760Mfs*6) were suggested to be novel [118].Two homozygous cases of p.R2703C and p.L308F were reported in 2017, in two unrelated patients. Both of these cases were associated with axonal CMT [119].
One mutation, c.7962T/G or p.(Y2654X), was reported in two Maori siblings with English ancestry from New Zealand (Table 4). They were initially suspected to have Friedrich ataxia, but no mutation was found in the FXN gene. Patients had lower limb weakness with upper limb ataxia. Symptoms appeared in their 20s and progressed further into their 40s [120].
5. Potential Involvement of SACS in Other Neurodegenerative Diseases
Currently, SACS has only been related to ARSACS. According to several studies on non-functional sacsin or sacsin knockout in cell lines or mice, sacsin is involved in various cellular roles, such as chaperon functions, mitochondrial mechanisms, microtubule filament control and cell adhesion [17]. It may not be ruled out that sacsin could exert an impact, directly or indirectly, in other types of neurodegenerative diseases (especially ataxias) than ARSACS. Since sacsin could interact with Hsp70 and ubiquitin proteasomes, its association may be involved in the defensive mechanisms against abnormal protein aggregations. When sacsin expression was attenuated, higher toxicity of repeat expansion was observed in comparison to normal sacsin expression. With Hsp70, sacsin may regulate the processing of ataxin-1 with polyglutamate expansion, especially the aberrant ataxin-1 degradation for protection against spinocerebellar ataxia-1 [2]. A putative association between sacsin and ataxin-3 was reported, where the N-terminal UbL domain of sacsin could directly interact with proteasomes. Since ataxin-3 could also interact with proteasomes, sacsin could affect the pathogenic mechanisms of ataxin-3 dysfunctions [121].
Interestingly, newly discovered SACS mutations in suspected patients with CMT-like neuropathy and atypical disease phenotypes suggest a potential pathological overlap between ARSACS and CMT [4].
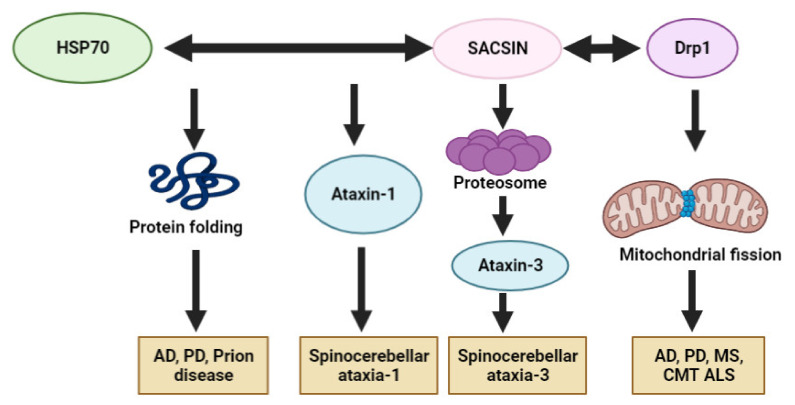
The involvement of the SACS gene in other neurodegenerative diseases, such as AD, PD, ALS or CJD, has not been reported yet. Sacsin is closely involved in controlling the mitochondrial functions and dynamics. Sacsin dysfunctions have been associated with impairment of mitochondrial morphology, dynamics, organization and dysfunctions in Drp1, and could cause synaptic dysfunctions and loss of Purkinje cells [21,26]. In addition, the interactions between Drp1 and sacsin for proper mitochondrial functions, and Drp dysfunctions in the imbalance of mitochondrial fusion or fission, could also indicate the involvement of SACS mutations in AD, ALS, MS or PD. Altered Drp1 expression could cause the overexpression of amyloid beta, huntingtin or alpha synuclein. By interacting with Drp1, sacsin may influence the expression of different neurodegenerative-disease-related proteins indirectly. Defects in mitochondrial dynamics in other neurodegenerative disorders could be a common pathway associated with ARSACS and with SACS mutations. Hence, the contribution of sacsin in different neurodegenerative diseases could be hypothesized [122,123,124].
An additional putative common pathway between sacsin and other neurodegenerative diseases could be through its chaperon function. Sacsin contains homologous sequences with Hsp90. In addition, sacsin could interact with Hsp70 to exert several neuroprotective mechanisms [125]. Hsp proteins may play as a key role in protein folding and protect neurons against various protein-folding-related diseases. Impairment of Hsps and other chaperons could result in elevated oxidative stress and mitochondrial dysfunctions. Overexpressed Hsp70 was reported in AD mouse models, suggesting enhanced protective mechanisms by reducing APP cleavage and amyloid peptide production. Hsp70 could also enhance the transport of Tau protein and amyloid oligomers into proteasomes [126]. Hsp70 seemed to play an essential role in protecting against prion misfolding and aggregation [127]. In Hsp70 knockout mice, prion propagation and toxicity was accelerated [128]. Lastly, Hsp70 could block alpha synuclein oligomerization through a noncanonical site in the C-terminal domain to exert a protective function against PD and other synucleopathies [128]. These studies suggest that sacsin may contribute either directly or indirectly to neuroprotection against non-ataxia-related diseases. Figure 5 summarizes the possible common pathways between sacsin and other neurodegenerative diseases.
Figure 5.
Possible disease mechanisms of sacsin protein in other neurodegenerative diseases: AD, PD, MS, CMT, ALS, SCA and CJD.
Additional evidence of an association involving common pathways between sacsin and other forms of neurodegenerative diseases was published by Morani et al. (2020) [36]. Their study analyzed proteomic data from ARSACS mouse models and isolated cells from ARSACS patients using SomaLogic technology. Several dysregulated pathways and differentially expressed proteins (DEPs) were found to be associated with neuroinflammation, synaptogenesis or cell engulfment. Several DEPs were found, which were involved in other neurodegenerative diseases: AD, PD, dementia with Lewy bodies and spastic paraplegia. Significant DEPs in ARSACS models were ephrins (EFNB2, EPHA3 and EPHB2), SNCA, APOE, ICAM5, SPHK1 and/or STUB1. This result points to the possibility of shared pathways between ARSACS and other neurodegenerative diseases. Hence, DEPs may act as risk factors or risk modifiers for ARSACS disease progression. Nevertheless, sacsin dysfunctions may alter the expression of ARSACS-related genes/proteins and may impact the pathological mechanisms of other neurodegenerative diseases, including AD, PD, ALS and CJD [36].
6. ARSACS Diagnosis and Potential Therapeutics
Genetic testing is required for the specific diagnosis of ARSACS. The standard Sanger sequencing would be challenging due to the size of the SACS gene. Hence, next-generation sequencing techniques would be the optimal approach in the discovery of novel causative genes or mutations in the SACS gene [129]. Additional biomarkers would be needed to enhance the disease’s diagnosis in combination with imaging biomarkers, such as brain and retinal imaging, in ARSACS diagnosis. Interestingly, “bithalamic stripes” detected by MRI imaging or retinal nerve fiber layer thickening could be an additional useful diagnostic biomarker of the disease [104].
Retinal nerve thickening was reported in several ARSACS patients. Recently, optical coherence tomography (OCT) was suggested to represent a significant diagnostic tool for investigating visual impairments and diseases in retina or neuropathy, as a non-invasive and cost-effective test. Hence, OCT could observe retinal nerve fiber thickening in patients with ARSACS [72,91]. Parkinson et al. (2018) performed OCT on patients with different types of ataxia (191) and controls (101). Retinal nerve fiber thickening was present only in ARSACS cases, and not in controls or other kinds of ataxia, such as Friedrich ataxia or spinocerebellar ataxia. This study proposed a cut-off value of 119 μm for the average retinal nerve fiber thickness. This value provided high specificity and sensitivity (100% and 99.4%, respectively) among ataxia patients [72]. Since the retina may be an excellent source of potential surrogate biomarkers in ARSACS, Rezende Filho et al. (2021) performed fundoscopy (another simple cost-effective technique) and OCT on patients with ARSACS and other forms of ataxia (spinocerebellar ataxia, autosomal recessive cerebellar ataxia, hereditary spastic paraplegia). The investigated retinal nerve fiber thickening in ARSACS by fundoscopy provided false negative data, suggesting that this method has lower sensitivity than OCT [130].
ARSACS patients presented other types of retinal abnormalities in comparison to control patients or those with other forms of ataxia. All ARSACS patients presented foveal hypoplasia, in addition to other impairments, such as retinal hyperplasia, sawtooth appearance or papillomacular fold. The above results suggest that monitoring neurophysiological abnormalities could yield promising biomarkers for ARSACS diagnosis [72,130], such as nerve conduction or nerve ultrasonography. Nerve enlargement and peripheral demyelination may be useful biomarkers in ARSACS [35,36]. Since the typical imaging biomarkers could be missing in some cases of ARSACS, Pilliod et al. analyzed 321 diagnosed patients with spinocerebellar degeneration from the SPATAX (http://spatax.wordpress.com accessed on 23 September 2021) database. They also collected fibroblast samples from skin biopsies in 11 ARSACS patients and 8 controls in order to perform mitochondrial morphology analyses, which indicated that mitochondrial abnormalities, such as bulbed mitochondria, were common among ARSACS patients. In addition, mitochondrial mass, oxygen consumption and the ratio of mitochondrial DNA/nuclear DNA were reduced among ARSACS patients. These anomalies in the mitochondrial network may be a useful diagnostic and prognostic biomarker to predict the pathogenicity of ARSACS [35]. Since spasticity is one of the typical key features, with reduced movement and coordination in patients with ARSACS, Lessard et al. performed the Lower Extremity Motor Coordination Test (LEMOCOT) for its possible usage in attempting diagnosis [131]. Analyzing pendulum oscillation amplitudes and their ratios using a wireless electro-goniometer provided information on the degree of spasticity in ARSACS patients. This device could effectively measure the evolution of spasticity in patients and provide an easy tool to compare the data from the pendulum between patients and unaffected individuals. Hence, this method would be an easy, rapid and cost-effective test for ARSACS diagnosis [132].
Since no therapy is currently available for ARSACS, the best disease management strategy is to mitigate the disease symptoms and improve the quality of the patient’s life. Positive associations were confirmed between fitness activities and the symptoms of neuromuscular/neurological diseases. An eight-week workout program, used by Audet et al. (2018), suggested that physical training may be beneficial for the fitness and functional capacity of ARSACS patients. Strengthened muscular and functional capacity was observed in patients who participated in the program. In addition, the regular workout improved patients’ ability to perform their daily activities. Furthermore, the reduced frequency of falls was notable. Regular training in patients with ARSACS could help them to retain and even regain their independence of movement [133]. Speech training was also effective to treat language impairments. Vogel et al. (2019) recruited ARSACS patients into a 4-week program with rater-blinded assessment of intelligibility. Although this was a preliminary study, the speech treatment improved the intelligibility of patients and enhanced the spontaneity of their speech [134].
Although no effective drug is available for ARSACS yet, certain candidate drugs may improve the quality of life of ARSACS patients. The main criterion for drug development is their capacity to cross the blood–brain barrier [135]. Docosahexaenoic acid (DHA), a dietary supplement, was investigated for its possible neuroprotective functions—namely, its anti-apoptosis, anti-inflammatory and anti-autophagic properties. Since DHA contains phospholipids, similar lipids to brain phospholipids, it was proposed in patients with ataxias with cerebellar and pyramidal involvement. Ricca et al. (2020) carried out a small clinical study in two ARSACS patients with SACS mutations. DHA was administrated orally for 20 months. Afterwards, investigators noticed stalled or slowed disease progression or the deterioration of clinical symptoms, with slight improvements in the functioning of the lower limbs and speech being reported, suggesting DHA as a safe and promising add-on therapy in patients with ARSACS and SACS mutations [136].
In addition, Idebenone (IDE), an analogue of coenzyme Q10, was successfully used to treat brain disorders with mitochondrial etiology by protecting against free radical toxicity. Since the solubility of IDE is low, many researchers have investigated it for the best drug delivery systems. Nanostructured lipid carriers (NLCs) loaded with IDE, with stability in water or cell media, and with successful penetration of the blood–brain barrier, were invented, representing a promising future approach in ARSACS therapy [135].
In sacsin knockout cells, the modulation of PTEN-FAK signaling could improve the ARSACS-related cellular dysfunctions, including microtubule organization, cell migration or adhesion [17]. PTEN has been verified as a negative regulator of focal adhesion. Hence, PTEN signal modulation has been suggested as a possible therapeutic target in ARSACS.
Since higher inductions of Hsp90 are observed in neurodegenerative diseases, a Hsp90 inhibitor was investigated for a possible reduction in disease progression and toxicity. A potential therapeutic candidate was KU-32, a Hsp90 inhibitor [137]. Hence, inhibiting Hsp90 was contemplated for its possible benefits in patients with ARSACS and different neurodegenerative disorders [138]. Ku-32 treatment improved the mitochondrial functions (electron transport, mitochondrial membrane potential) in cells among ARSACS patients [137].
7. Discussion and Future Insights
ARSACS appears to be one of the most common forms of ataxia besides Friedrich ataxia. Besides the French Canadian population, emerging cases of SACS mutations have been reported worldwide, including in Tunisia, Japan and Turkey. However, several cases of ARSACS may be unreported [34]. The typical disease phenotype of ARSACS includes the early onset of the disease, slow progression, cerebellar ataxia, spasticity, cerebellar atrophy, neuropathy, axonal demyelination and retinal nerve thickening. Additional symptoms, such as mental retardation, later disease onset and cognitive dysfunction, were also reported [19,68]. Interestingly, atypical cases of disease may also be possible (for example, lack of spasticity or retinal optic nerve hypermyelination), apart from the typical phenotype [4,83,85]. Additional atypical cases include patients with epilepsy, a CMT-like phenotype or hearing loss [4].
Homozygous or compound heterozygous mutations in SACS were associated with ARSACS. Noticeably, since clinical heterogeneities were observed, even from the same family, it may be possible that other genetic or environmental factors could impact the disease phenotype, besides the SACS mutations. Recently, the disease diagnosis became easier with the development of next-generation sequencing techniques, i.e., genome-wide and/or transcriptome-wide analyses, which could provide valuable insights into additional disease-modifying factors in ARSACS and or with SACS [38,129]. These genetic data could be correlated with imaging analyses, such as MRI [104] and OCT [72], to improve the disease’s diagnosis. Proteomic analysis would be a promising future investigation in understanding ARSACS and its progression and diagnosis [36].
Further studies are required to understand the functions of sacsin, especially in other neurodegenerative diseases. Although sacsin has been confirmed to be the causative factor of ARSACS, its impact on other diseases should be investigated, especially given the limited reports on its functions, which focus on chaperon interactions with Hsp70 and mitochondrial homeostasis [3,21]. Since the pathomechanisms of ARSACS may share similar pathways with other neurodegenerative diseases, such as AD, PD, ALS and CJD, it will be crucial to investigate the consequences of the gain or loss of functions [125,126,127,128,129]. The study by Morani et al. suggested that multi-omic (proteomic, genomic, transcriptomic) analysis in ARSACS models could be promising in the disease’s diagnosis, as well as in discovering the specific disease-causing pathways and risk-modifying factors, especially in the development of therapeutics [36].
Author Contributions
Conceptualization, all authors.; collection the literature, writing—review and editing the manuscript, E.B. and J.B.; substantive supervision and revision over the article, all authors; project administration, S.S.A.A.; funding acquisition, S.S.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Research Foundation of Korea (NRF) by the Korean government (2020R1A2B5B01002463 & 2021R1A6A1A03038996).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Takiyama Y. Autosomal recessive spastic ataxia of Charlevoix-Saguenay. Neuropathology. 2006;26:368–375. doi: 10.1111/j.1440-1789.2006.00664.x. [DOI] [PubMed] [Google Scholar]
- 2.Parfitt D.A., Michael G.J., Vermeulen E.G., Prodromou N.V., Webb T.R., Gallo J.-M., Cheetham M.E., Nicoll W.S., Blatch G.L., Chapple J.P. The ataxia protein sacsin is a functional co-chaperone that protects against polyglutamine-expanded ataxin-1. Hum. Mol. Genet. 2009;18:1556–1565. doi: 10.1093/hmg/ddp067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gentil B.J., Lai G.-T., Menade M., Larivière R., Minotti S., Gehring K., Chapple J.-P., Brais B., Durham H.D. Sacsin, mutated in the ataxia ARSACS, regulates intermediate filament assembly and dynamics. FASEB J. 2019;33:2982–2994. doi: 10.1096/fj.201801556R. [DOI] [PubMed] [Google Scholar]
- 4.Synofzik M., Soehn A.S., Gburek-Augustat J., Schicks J., Karle K.N., Schüle R., Haack T.B., Schöning M., Biskup S., Rudnik-Schöneborn S., et al. Autosomal recessive spastic ataxia of Charlevoix Saguenay (ARSACS): Expanding the genetic, clinical and imaging spectrum. Orphanet J. Rare Dis. 2013;8:41. doi: 10.1186/1750-1172-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engert J., Bérubé P., Mercier J., Doré C., Lepage P., Ge B., Bouchard J.-P., Mathieu J., Melançon S.B., Schalling M., et al. ARSACS, a spastic ataxia common in northeastern Québec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat. Genet. 2000;24:120–125. doi: 10.1038/72769. [DOI] [PubMed] [Google Scholar]
- 6.De Braekeleer M., Giasson F., Mathieu J., Roy M., Bouchard J.P., Morgan K. Genetic epidemiology of autosomal recessive spastic ataxia of Charlevoix-Saguenay in northeastern Quebec. Genet. Epidemiol. 1993;10:17–25. doi: 10.1002/gepi.1370100103. [DOI] [PubMed] [Google Scholar]
- 7.Heyer E. Genetic consequences of differential demographic behaviour in the Saguenay region, Québec. Am. J. Phys. Anthr. 1995;98:1–11. doi: 10.1002/ajpa.1330980102. [DOI] [PubMed] [Google Scholar]
- 8.Takiyama Y. Sacsinopathies: Sacsin-related ataxia. Cerebellum. 2007;6:353–359. doi: 10.1080/14734220701230466. [DOI] [PubMed] [Google Scholar]
- 9.Bouchard J.-P., Richter A., Mathieu J., Brunet D., Hudson T.J., Morgan K., Melançon S.B. Autosomal recessive spastic ataxia of Charlevoix–Saguenay. Neuromuscul. Disord. 1998;8:474–479. doi: 10.1016/S0960-8966(98)00055-8. [DOI] [PubMed] [Google Scholar]
- 10.Laberge A.-M., Michaud J., Richter A., Lemyre E., Lambert M., Brais B., Mitchell G. Population history and its impact on medical genetics in Quebec. Clin. Genet. 2005;68:287–301. doi: 10.1111/j.1399-0004.2005.00497.x. [DOI] [PubMed] [Google Scholar]
- 11.Bouhlal Y., Amouri R., El Euch-Fayeche G., Hentati F. Autosomal recessive spastic ataxia of Charlevoix–Saguenay: An overview. Park. Relat. Disord. 2011;17:418–422. doi: 10.1016/j.parkreldis.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Larivière R., Gaudet R., Gentil B.J., Girard M., Conte T.C., Minotti S., Leclerc-Desaulniers K., Gehring K., McKinney R.A., Shoubridge E.A., et al. Sacs knockout mice present pathophysiological defects underlying autosomal recessive spastic ataxia of Charlevoix-Saguenay. Hum. Mol. Genet. 2014;24:727–739. doi: 10.1093/hmg/ddu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romano A., Tessa A., Barca A., Fattori F., Fulvia de Leva M., Terracciano A., Storelli C., Santorelli F.M., Verri T. Comparative analysis and functional mapping of SACS mutations reveal novel insights into sacsin repeated architecture. Hum. Mutat. 2013;34:525–537. doi: 10.1002/humu.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slanzi A., Iannoto G., Rossi B., Zenaro E., Constantin G. In vitro Models of Neurodegenerative Diseases. Front. Cell Dev. Biol. 2020;8:328. doi: 10.3389/fcell.2020.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ady V., Toscano-Márquez B., Nath M., Chang P.K., Hui J., Cook A., Charron F., Larivière R., Brais B., McKinney R.A., et al. Altered synaptic and firing properties of cerebellar Purkinje cells in a mouse model of ARSACS. J. Physiol. 2018;596:4253–4267. doi: 10.1113/JP275902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro A.A., Machuca C., Jimenez F.J.R., Jendelova P., Erceg S., Arellano C.M. Short Review: Investigating ARSACS: Models for understanding cerebellar degeneration. Neuropathol. Appl. Neurobiol. 2019;45:531–537. doi: 10.1111/nan.12540. [DOI] [PubMed] [Google Scholar]
- 17.Romano L.E., Aw W.Y., Hixson K.M., Novoselova T.V., Havener T.M., Howell S., Hall C.L., Xing L., Beri J., Nethisinghe S., et al. The ataxia protein sacsin is required for integrin trafficking and synaptic organization. bioRxiv. 2021 doi: 10.1101/2021.08.20.456807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovalevich J., Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 2013;1078:9–21. doi: 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Criscuolo C., Procaccini C., Meschini M.C., Cianflone A., Carbone R., Doccini S., Devos D.A.V.I.D., Nesti C., Vuillaume I., Pellegrino M., et al. Powerhouse failure and oxidative damage in autosomal recessive spastic ataxia of Charlevoix-Saguenay. J. Neurol. 2015;262:2755–2763. doi: 10.1007/s00415-015-7911-4. [DOI] [PubMed] [Google Scholar]
- 20.Duncan E.J., Larivière R., Bradshaw T.Y., Longo F., Sgarioto N., Hayes M.J., Romano L.E., Nethisinghe S., Giunti P., Bruntraeger M.B., et al. Altered organization of the intermediate filament cytoskeleton and relocalization of proteostasis modulators in cells lacking the ataxia protein sacsin. Hum. Mol. Genet. 2017;26:3130–3143. doi: 10.1093/hmg/ddx197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard M., Larivière R., Parfitt D.A., Deane E.C., Gaudet R., Nossova N., Blondeau F., Prenosil G., Vermeulen E.G.M., Duchen M., et al. Mitochondrial dysfunction and Purkinje cell loss in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) Proc. Natl. Acad. Sci. USA. 2012;109:1661–1666. doi: 10.1073/pnas.1113166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mesdom P., Colle R., Lebigot E., Trabado S., Deflesselle E., Fève B., Becquemont L., Corruble E., Verstuyft C. Human Dermal Fibroblast: A Promising Cellular Model to Study Biological Mechanisms of Major Depression and Antidepressant Drug Response. Curr. Neuropharmacol. 2020;18:301–318. doi: 10.2174/1570159X17666191021141057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kálmán S., Garbett K., Janka Z., Mirnics K. Human dermal fibroblasts in psychiatry research. Neuroscience. 2016;320:105–121. doi: 10.1016/j.neuroscience.2016.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nejaddehbashi F., Bayati V., Mashali L., Hashemitabar M., Abbaspour M., Moghimipour E., Orazizadeh M. Isolating human dermal fibroblasts using serial explant culture. Stem Cell Investig. 2019;6:23. doi: 10.21037/sci.2019.08.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arellano C.M., Vilches A., Clemente E., Pascual S.I.P., Bolinches-Amorós A., Castro A.A., Espinós C., Rodriguez M.L., Jendelova P., Erceg S. Generation of a human iPSC line from a patient with autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) caused by mutation in SACSIN gene. Stem Cell Res. 2018;31:249–252. doi: 10.1016/j.scr.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Li X., Gehring K. Structural studies of parkin and sacsin: Mitochondrial dynamics in neurodegenerative diseases. Mov. Disord. 2015;30:1610–1619. doi: 10.1002/mds.26357. [DOI] [PubMed] [Google Scholar]
- 27.Kozlov G., Denisov A.Y., Girard M., Dicaire M.-J., Hamlin J., McPherson P.S., Brais B., Gehring K. Structural Basis of Defects in the Sacsin HEPN Domain Responsible for Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay (ARSACS) J. Biol. Chem. 2011;286:20407–20412. doi: 10.1074/jbc.M111.232884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson J.F., Siller E., Barral J.M. The Sacsin Repeating Region (SRR): A Novel Hsp90-Related Supra-Domain Associated with Neurodegeneration. J. Mol. Biol. 2010;400:665–674. doi: 10.1016/j.jmb.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Ménade M., Kozlov G., Trempe J.-F., Pande H., Shenker S., Wickremasinghe S., Li X., Hojjat H., Dicaire M.-J., Brais B., et al. Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations. J. Biol. Chem. 2018;293:12832–12842. doi: 10.1074/jbc.RA118.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greer P.L., Hanayama R., Bloodgood B., Mardinly A., Lipton D.M., Flavell S., Kim T.-K., Griffith E., Waldon Z., Maehr R., et al. The Angelman Syndrome Protein Ube3A Regulates Synapse Development by Ubiquitinating Arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dabbaghizadeh A., Paré A., Cheng-Boivin Z., Dagher R., Minotti S., Dicaire M.J., Bernard B., Young J.C., Durham H.D., Gentil B.J. The J domain of sacsin disrupts intermediate filament assembly. bioRxiv. 2021 doi: 10.1101/2021.07.14.452331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Ménade M., Kozlov G., Hu Z., Dai Z., McPherson P.S., Brais B., Gehring K. High-Throughput Screening for Ligands of the HEPN Domain of Sacsin. PLoS ONE. 2015;10:e0137298. doi: 10.1371/journal.pone.0137298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grynberg M., Erlandsen H., Godzik A. HEPN: A common domain in bacterial drug resistance and human neurodegenerative proteins. Trends Biochem. Sci. 2003;28:224–226. doi: 10.1016/S0968-0004(03)00060-4. [DOI] [PubMed] [Google Scholar]
- 34.Bradshaw T.Y., Romano L.E.L., Duncan E.J., Nethisinghe S., Abeti R., Michael G.J., Giunti P., Vermeer S., Chapple J.P. A reduction in Drp1-mediated fission compromises mitochondrial health in autosomal recessive spastic ataxia of Charlevoix Saguenay. Hum. Mol. Genet. 2016;25:3232–3244. doi: 10.1093/hmg/ddw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pilliod J., Moutton S., Lavie J., Maurat E., Hubert C., Bellance N., Anheim M., Forlani S., Mochel F., N’Guyen K., et al. New practical definitions for the diagnosis of autosomal recessive spastic ataxia of Charlevoix-Saguenay. Ann. Neurol. 2015;78:871–886. doi: 10.1002/ana.24509. [DOI] [PubMed] [Google Scholar]
- 36.Morani F., Doccini S., Chiorino G., Fattori F., Galatolo D., Sciarrillo E., Gemignani F., Züchner S., Bertini E.S., Santorelli F.M. Functional Network Profiles in ARSACS Disclosed by Aptamer-Based Proteomic Technology. Front. Neurol. 2020;11:603774. doi: 10.3389/fneur.2020.603774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bchetnia M., Bouchard L., Mathieu J., Campeau P.M., Morin C., Brisson D., Laberge A.M., Vézina H., Gaudet D., Laprise C. Genetic burden linked to founder effects in Saguenay-Lac-Saint-Jean illustrates the importance of genetic screening test availability. J. Med. Genet. 2021;58:653–665. doi: 10.1136/jmedgenet-2021-107809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longo F., De Ritis D., Miluzio A., Fraticelli D., Baets J., Scarlato M., Santorelli F.M., Biffo S., Maltecca F. Assessment of Sacsin Turnover in Patients With ARSACS: Implications for Molecular Diagnosis and Pathogenesis. Neurology. 2021;97:e2315–e2327. doi: 10.1212/WNL.0000000000012962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiffault I., Dicaire M., Tetreault M., Huang K., Demers-Lamarche J., Bernard G., Duquette A., Larivière R., Gehring K., Montpetit A., et al. Diversity of ARSACS Mutations in French-Canadians. Can. J. Neurol. Sci./J. Can. Sci. Neurol. 2013;40:61–66. doi: 10.1017/S0317167100012968. [DOI] [PubMed] [Google Scholar]
- 40.Bouchard J.P., Barbeau A., Bouchard R.W. Autosomal recessive spastic ataxia of Charlevoix-Saguenay. Can. J. Neurol. Sci./J. Can. des Sci. Neurol. 1978;5:61–69. doi: 10.1017/S0317167100024793. [DOI] [PubMed] [Google Scholar]
- 41.McKenzie E., Sharma P., Parboosingh J., Suchowersky O. Forge Canada Consortium Novel SACS Mutation Deviates from the French Canadian ARSACS Phenotype. Can. J. Neurol. Sci. J. Can. Sci. Neurol. 2014;41:88–89. doi: 10.1017/S0317167100016334. [DOI] [PubMed] [Google Scholar]
- 42.Richter A., Rioux J.D., Bouchard J.-P., Mercier J., Mathieu J., Ge B., Poirier J., Julien D., Gyapay G., Weissenbach J., et al. Location Score and Haplotype Analyses of the Locus for Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay, in Chromosome Region 13q11. Am. J. Hum. Genet. 1999;64:768–775. doi: 10.1086/302274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El Euch-Fayache G., Lalani I., Amouri R., Turki I., Ouahchi K., Hung W.-Y., Belal S., Siddique T., Hentati F. Phenotypic Features and Genetic Findings in Sacsin-Related Autosomal Recessive Ataxia in Tunisia. Arch. Neurol. 2003;60:982–988. doi: 10.1001/archneur.60.7.982. [DOI] [PubMed] [Google Scholar]
- 44.Bouhlal Y., El Euch-Fayeche G., Hentati F., Amouri R. A Novel SACS Gene Mutation in a Tunisian Family. J. Mol. Neurosci. 2009;39:333–336. doi: 10.1007/s12031-009-9212-9. [DOI] [PubMed] [Google Scholar]
- 45.Hammer M.B., Eleuch-Fayache G., Gibbs J.R., Arepalli S.K., Chong S.B., Sassi C., Bouhlal Y., Hentati F., Amouri R., Singleton A.B. Exome sequencing: An efficient diagnostic tool for complex neurodegenerative disorders. Eur. J. Neurol. 2013;20:486–492. doi: 10.1111/j.1468-1331.2012.03883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez C.M. ARSACS goes global. Neurology. 2004;62:10–11. doi: 10.1212/WNL.62.1.10. [DOI] [PubMed] [Google Scholar]
- 47.Grieco G.S., Malandrini A., Comanducci G., Leuzzi V., Valoppi M., Tessa A., Palmeri S., Benedetti L., Pierallini A., Gambelli S., et al. Novel SACS mutations in autosomal recessive spastic ataxia of Charlevoix-Saguenay type. Neurology. 2004;62:103–106. doi: 10.1212/01.WNL.0000104491.66816.77. [DOI] [PubMed] [Google Scholar]
- 48.Criscuolo C., Banfi S., Orio M., Gasparini P., Monticelli A., Scarano V., Santorelli F.M., Perretti A., Santoro L., De Michele G., et al. A novel mutation in SACS gene in a family from southern Italy. Neurology. 2004;62:100–102. doi: 10.1212/WNL.62.1.100. [DOI] [PubMed] [Google Scholar]
- 49.Ricca I., Morani F., Bacci G.M., Nesti C., Caputo R., Tessa A., Santorelli F.M. Clinical and molecular studies in two new cases of ARSACS. Neurogenetics. 2019;20:45–49. doi: 10.1007/s10048-019-00564-7. [DOI] [PubMed] [Google Scholar]
- 50.Prodi E., Grisoli M., Panzeri M., Minati L., Fattori F., Erbetta A., Uziel G., D’Arrigo S., Tessa A., Ciano C., et al. Supratentorial and pontine MRI abnormalities characterize recessive spastic ataxia of Charlevoix-Saguenay. A comprehensive study of an Italian series. Eur. J. Neurol. 2013;20:138–146. doi: 10.1111/j.1468-1331.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 51.Pensabene M.C., Melis M., De Corato L., Di Stefano C., Pizzicannella G., Mondillo M., Amico A., Tatulli D., Floris R. Autosomal recessive spastic ataxia of charlevoix-saguenay: Findings from MRI in two adult Italian siblings. Radiol. Case Rep. 2020;15:507–510. doi: 10.1016/j.radcr.2019.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anesi L., De Gemmis P., Pandolfo M., Hladnik U. Two Novel Homozygous SACS Mutations in Unrelated Patients Including the First Reported Case of Paternal UPD as an Etiologic Cause of ARSACS. J. Mol. Neurosci. 2011;43:346–349. doi: 10.1007/s12031-010-9448-4. [DOI] [PubMed] [Google Scholar]
- 53.Terracciano A., Casali C., Grieco G.S., Orteschi D., Di Giandomenico S., Seminara L., Di Fabio R., Carrozzo R., Simonati A., Stevanin G., et al. An inherited large-scale rearrangement in SACS associated with spastic ataxia and hearing loss. Neurogenetics. 2009;10:151–155. doi: 10.1007/s10048-008-0159-8. [DOI] [PubMed] [Google Scholar]
- 54.Masciullo M., Modoni A., Fattori F., Santoro M., Denora P.S., Tonali P., Santorelli F.M., Silvestri G. A novel mutation in the SACS gene associated with a complicated form of spastic ataxia. J. Neurol. 2008;255:1429–1431. doi: 10.1007/s00415-008-0936-1. [DOI] [PubMed] [Google Scholar]
- 55.Gücüyener K., Özgül K., Paternotte C., Erdem H., Prud’Homme J.F., Özgüç M., Topaloğlu H. Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay in Two Unrelated Turkish Families. Neuropediatrics. 2001;32:142–146. doi: 10.1055/s-2001-16616. [DOI] [PubMed] [Google Scholar]
- 56.Richter A.M., Ozgul R.K., Poisson V.C., Topaloglu H. Private SACS mutations in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) families from Turkey. Neurogenetics. 2004;5:165–170. doi: 10.1007/s10048-004-0179-y. [DOI] [PubMed] [Google Scholar]
- 57.Oguz K., Haliloglu G., Temucin C., Gocmen R., Has A., Doerschner K., Dolgun A.B., Alikasifoglu M. Assessment of Whole-Brain White Matter by DTI in Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. Am. J. Neuroradiol. 2013;34:1952–1957. doi: 10.3174/ajnr.A3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurt S., Kartal E., Aksoy D., Cevik B., Eken A.G., Sahbaz I., Basak A.N. Coexistence of autosomal recessive spastic ataxia of Charlevoix Saguenay and spondyloepiphyseal dysplasia in a Turkish patient. J. Neurol. Sci. 2015;357:290–291. doi: 10.1016/j.jns.2015.06.050. [DOI] [PubMed] [Google Scholar]
- 59.Incecik F., Hergüner O.M., Bisgin A. Autosomal-Recessive Spastic Ataxia of Charlevoix-Saguenay: A Turkish Child. J. Pediatr. Neurosci. 2018;13:355–357. doi: 10.4103/JPN.JPN_8_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samanci B., Gokalp E.E., Bilgic B., Gurvit H., Artan S., Hanagasi H.A. A novel SACS p.Pro4154GlnfsTer20 mutation in a family with autosomal recessive spastic ataxia of Charlevoix-Saguenay. Neurol. Sci. 2021;42:1–5. doi: 10.1007/s10072-021-05117-1. [DOI] [PubMed] [Google Scholar]
- 61.Vermeer S., Meijer R.P.P., Pijl B.J., Timmermans J., Cruysberg J.R.M., Bos M.M., Schelhaas H.J., Van De Warrenburg B.P.C., Knoers N.V.A.M., Scheffer H., et al. ARSACS in the Dutch population: A frequent cause of early-onset cerebellar ataxia. Neurogenetics. 2008;9:207–214. doi: 10.1007/s10048-008-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ouyang Y., Segers K., Bouquiaux O., Wang F., Janin N., Andris C., Shimazaki H., Sakoe K., Nakano I., Takiyama Y. Novel SACS mutation in a Belgian family with sacsin-related ataxia. J. Neurol. Sci. 2008;264:73–76. doi: 10.1016/j.jns.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 63.Baets J., Deconinck T., Smets K., Goossens D., Bergh P.V.D., Dahan K., Schmedding E., Santens P., Rasic V.M., Van Damme P., et al. Mutations in SACS cause atypical and late-onset forms of ARSACS. Neurology. 2010;75:1181–1188. doi: 10.1212/WNL.0b013e3181f4d86c. [DOI] [PubMed] [Google Scholar]
- 64.Criscuolo C., Saccà F., De Michele G., Mancini P., Combarros O., Infante J., Garcia A., Banfi S., Filla A., Berciano J. Novel mutation of SACS gene in a Spanish family with autosomal recessive spastic ataxia. Mov. Disord. 2005;20:1358–1361. doi: 10.1002/mds.20579. [DOI] [PubMed] [Google Scholar]
- 65.Sánchez M.G., Pérez J.E., Pérez M.R., Redondo A.G. Novel SACS mutation in autosomal recessive spastic ataxia of Charlevoix-Saguenay. J. Neurol. Sci. 2015;358:475–476. doi: 10.1016/j.jns.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 66.Xiromerisiou G., Dadouli K., Marogianni C., Provatas A., Ntellas P., Rikos D., Stathis P., Georgouli D., Loules G., Zamanakou M., et al. A novel homozygous SACS mutation identified by whole exome sequencing-genotype phenotype correlations of all published cases. J. Mol. Neurosci. 2020;70:131–141. doi: 10.1007/s12031-019-01410-z. [DOI] [PubMed] [Google Scholar]
- 67.Krygier M., Konkel A., Schinwelski M., Rydzanicz M., Walczak A., Sildatke-Bauer M., Płoski R., Slawek J. Autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS)—A Polish family with novel SACS mutations. Neurol. Neurochir. Polska. 2017;51:481–485. doi: 10.1016/j.pjnns.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Rudenskaya G.E., Kadnikova V.A., Ryzhkova O.P. Spastic ataxia of Charlevoix-Saguenay: The first Russian case report and literature review. Zhurnal Nevrol. Psikhiatrii Im. SS Korsakova. 2020;120:85–91. doi: 10.17116/jnevro202012002185. [DOI] [PubMed] [Google Scholar]
- 69.Tzoulis C., Johansson S., Haukanes B.I., Boman H., Knappskog P.M., Bindoff L.A. Novel SACS Mutations Identified by Whole Exome Sequencing in a Norwegian Family with Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. PLoS ONE. 2013;8:e66145. doi: 10.1371/journal.pone.0066145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vill K., Müller-Felber W., Gläser D., Kuhn M., Teusch V., Schreiber H., Weis J., Klepper J., Schirmacher A., Blaschek A., et al. SACS variants are a relevant cause of autosomal recessive hereditary motor and sensory neuropathy. Qual. Life Res. 2018;137:911–919. doi: 10.1007/s00439-018-1952-6. [DOI] [PubMed] [Google Scholar]
- 71.Pyle A., Griffin H.R., Yu-Wai-Man P., Duff J., Eglon G., Pickering-Brown S., Santibanez-Korev M., Horvath R., Chinnery P.F. Prominent Sensorimotor Neuropathy Due to SACS Mutations Revealed by Whole-Exome Sequencing. Arch. Neurol. 2012;69:1351–1354. doi: 10.1001/archneurol.2012.1472. [DOI] [PubMed] [Google Scholar]
- 72.Parkinson M.H., Bartmann A.P., Clayton L.M.S., Nethisinghe S., Pfundt R., Chapple J.P., Reilly M.M., Manji H., Wood N., Bremner F., et al. Optical coherence tomography in autosomal recessive spastic ataxia of Charlevoix-Saguenay. Brain. 2018;141:989–999. doi: 10.1093/brain/awy028. [DOI] [PubMed] [Google Scholar]
- 73.Miressi F., Faye P.-A., Pyromali I., Bourthoumieux S., Derouault P., Husson M., Favreau F., Sturtz F., Magdelaine C., Lia A.-S. A mutation can hide another one: Think Structural Variants! Comput. Struct. Biotechnol. J. 2020;18:2095–2099. doi: 10.1016/j.csbj.2020.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palmio J., Kärppä M., Baumann P., Penttilä S., Moilanen J.S., Udd B. Novel compound heterozygous mutation in SACS gene leads to a milder autosomal recessive spastic ataxia of Charlevoix-Saguenay, ARSACS, in a Finnish family. Clin. Case Rep. 2016;4:1151–1156. doi: 10.1002/ccr3.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petrov I. Novel Mutation in SACS Gene in a Patient with Autosomal Recessive Spastic Ataxia Charlevoix-Saguenay. Mov. Disord. Clin. Pract. 2021;8:963–965. doi: 10.1002/mdc3.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Breckpot J., Takiyama Y., Thienpont B., Van Vooren S., Vermeesch J.R., Ortibus E., Devriendt K. A novel genomic disorder: A deletion of the SACS gene leading to Spastic Ataxia of Charlevoix–Saguenay. Eur. J. Hum. Genet. 2008;16:1050–1054. doi: 10.1038/ejhg.2008.58. [DOI] [PubMed] [Google Scholar]
- 77.Pascual-Castroviejo I., I Pascual-Pascual S., Viaño J., Martínez V. Carlevoix-Saguenay type recessive spastic ataxia. A report of a Spanish case. Rev. Neurol. 2000;31:36–38. [PubMed] [Google Scholar]
- 78.Okawa S., Sugawara M., Watanabe S., Toyoshima I., Imota T. A novel sacsin mutation in a Japanese woman showing clinical uniformity of autosomal recessive spastic ataxia of Charlevoix-Saguenay. J. Neurol. Neurosurg. Psychiatry. 2006;77:280–282. doi: 10.1136/jnnp.2005.077297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamamoto Y., Hiraoka K., Araki M., Nagano S., Shimazaki H., Takiyama Y., Sakoda S. Novel compound heterozygous mutations in sacsin-related ataxia. J. Neurol. Sci. 2005;239:101–104. doi: 10.1016/j.jns.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 80.Ouyang Y., Takiyama Y., Sakoe K., Shimazaki H., Ogawa T., Nagano S., Yamamoto Y., Nakano I. Sacsin-related ataxia (ARSACS): Expanding the genotype upstream from the gigantic exon. Neurology. 2006;66:1103–1104. doi: 10.1212/01.wnl.0000204300.94261.ea. [DOI] [PubMed] [Google Scholar]
- 81.Hara K., Onodera O., Endo M., Kondo H., Shiota H., Miki K., Tanimoto N., Kimura T., Nishizawa M. Sacsin-related autosomal recessive ataxia without prominent retinal myelinated fibers in Japan. Mov. Disord. 2005;20:380–382. doi: 10.1002/mds.20315. [DOI] [PubMed] [Google Scholar]
- 82.Shimazaki H., Sakoe K., Niijima K., Nakano I., Takiyama Y. An unusual case of a spasticity-lacking phenotype with a novel SACS mutation. J. Neurol. Sci. 2007;255:87–89. doi: 10.1016/j.jns.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 83.Shimazaki H., Takiyama Y., Sakoe K., Ando Y., Nakano I. A phenotype without spasticity in sacsin-related ataxia. Neurology. 2005;64:2129–2131. doi: 10.1212/01.WNL.0000166031.91514.B3. [DOI] [PubMed] [Google Scholar]
- 84.Hara K., Shimbo J., Nozaki H., Kikugawa K., Onodera O., Nishizawa M. Sacsin-related ataxia with neither retinal hypermyelination nor spasticity. Mov. Disord. 2007;22:1362–1363. doi: 10.1002/mds.21557. [DOI] [PubMed] [Google Scholar]
- 85.Kamada S., Okawa S., Imota T., Sugawara M., Toyoshima I. Autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS): Novel compound heterozygous mutations in the SACS gene. J. Neurol. 2008;255:803–806. doi: 10.1007/s00415-008-0672-6. [DOI] [PubMed] [Google Scholar]
- 86.Shimazaki H., Takiyama Y., Honda J., Sakoe K., Namekawa M., Tsugawa J., Tsuboi Y., Suzuki C., Baba M., Nakano I. Middle Cerebellar Peduncles and Pontine T2 Hypointensities in ARSACS. J. Neuroimaging. 2013;23:82–85. doi: 10.1111/j.1552-6569.2011.00647.x. [DOI] [PubMed] [Google Scholar]
- 87.Aida I., Ozawa T., Fujinaka H., Goto K., Ohta K., Nakajima T. Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay without Spasticity. Intern. Med. 2021:7401–7421. doi: 10.2169/internalmedicine.7401-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haga R., Miki Y., Funamizu Y., Kon T., Suzuki C., Ueno T., Nishijima H., Arai A., Tomiyama M., Shimazaki H., et al. Novel compound heterozygous mutations of the SACS gene in autosomal recessive spastic ataxia of Charlevoix-Saguenay. Clin. Neurol. Neurosurg. 2012;114:746–747. doi: 10.1016/j.clineuro.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 89.Kwon K.-Y., Huh K., Eun B.-L., Yoo H.-W., Kamsteeg E.-J., Scheffer H., Koh S.-B. A Probable Korean Case of Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. Can. J. Neurol. Sci./J. Can. Sci. Neurol. 2015;42:271–273. doi: 10.1017/cjn.2015.38. [DOI] [PubMed] [Google Scholar]
- 90.Bong J.B., Kim S.W., Lee S.T., Choi J.R., Shin H.Y. Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. J. Korean Neurol. Assoc. 2019;37:69–72. doi: 10.17340/jkna.2019.1.13. [DOI] [Google Scholar]
- 91.Cho H., Lyoo C.H., Park S.E., Seo Y., Han S.-H., Han J. Optical Coherence Tomography Findings Facilitate the Diagnosis of Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. Korean J. Ophthalmol. 2021;35:330–331. doi: 10.3341/kjo.2021.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu L., Li X., Zi X., Shen L., Hu Z., Huang S., Yu D., Li H., Xia K., Tang B., et al. A novel hemizygous SACS mutation identified by whole exome sequencing and SNP array analysis in a Chinese ARSACS patient. J. Neurol. Sci. 2016;362:111–114. doi: 10.1016/j.jns.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 93.Sun W., Meng Y.U., Zhuo Y., Wang Z., Yuan Y. Novel spastic ataxia of Charlevoix-Saguenay gene compound heterozygous mutations in late onset autosomal recessive spastic ataxia of Charlevoix-Saguenay. Chin. J. Neurol. 2017;50:831–836. [Google Scholar]
- 94.Luo W., Chen Y., Cen Z., Zheng X., Chen S., Xie F. Novel Compound Heterozygous SACS Mutations in a Case with a Spasticity-Lacking Phenotype of Sacsin-Related Ataxia. Neurol. India. 2021;69:219. doi: 10.4103/0028-3886.310115. [DOI] [PubMed] [Google Scholar]
- 95.Lu Q., Shang L., Tian W.T., Cao L., Zhang X., Liu Q. Complicated paroxysmal kinesigenic dyskinesia associated with SACS mutations. Ann. Transl. Med. 2020;8:8. doi: 10.21037/atm.2019.11.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li S., Chen Y., Yuan X., Wei Q., Ou R., Gu X., Shang H. Identification of compound heterozygous mutations of SACS gene in two patients from a pedigree with spastic ataxia of Charlevoix-Saguenay. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2018;35:507–510. doi: 10.3760/cma.j.issn.1003-9406.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 97.Wang Z., Song Y., Wang X., Li X., Xu F., Si L., Dong Y., Yao T., Zhu J., Lai H., et al. Autosomal recessive spastic ataxia of Charlevoix-Saguenay caused by novel mutations in SACS gene: A report of two Chinese families. Neurosci. Lett. 2021;752:135831. doi: 10.1016/j.neulet.2021.135831. [DOI] [PubMed] [Google Scholar]
- 98.Srikajon J., Pitakpatapee Y., Limwongse C., Chirapapaisan N., Srivanitchapoom P. Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay (ARSACS) in a Thai Patient: The Classic Clinical Manifestations, Funduscopic Feature, and Brain Imaging Findings with a Novel Mutation in the SACS Gene. Tremor Other Hyperkinetic Mov. 2020;10:1. doi: 10.5334/tohm.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Al-Ajmi A., Shamsah S., Janicijevic A., Williams M., Al-Mulla F. Novel frameshift mutation in the SACS gene causing spastic ataxia of charlevoix-saguenay in a consanguineous family from the Arabian Peninsula: A case report and review of literature. World J. Clin. Cases. 2020;8:1477–1488. doi: 10.12998/wjcc.v8.i8.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Habibzadeh P., Tabatabaei Z., Inaloo S., Nashatizadeh M.M., Synofzik M., Ostovan V.R., Faghihi M.A. Case Report: Expanding the Genetic and Phenotypic Spectrum of Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. Front. Genet. 2020;11:585136. doi: 10.3389/fgene.2020.585136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sharifi Z., Taheri M., Fallah M.-S., Abiri M., Golnabi F., Bagherian H., Zeinali R., Farahzadi H., Alborji M., Tehrani P.G., et al. Comprehensive Mutation Analysis and Report of 12 Novel Mutations in a Cohort of Patients with Spinal Muscular Atrophy in Iran. J. Mol. Neurosci. 2021;71:2281–2298. doi: 10.1007/s12031-020-01789-0. [DOI] [PubMed] [Google Scholar]
- 102.Faruq M., Narang A., Kumari R., Pandey R., Garg A., Behari M., Dash D., Srivastava A., Mukerji M. Novel mutations in typical and atypical genetic loci through exome sequencing in autosomal recessive cerebellar ataxia families. Clin. Genet. 2014;86:335–341. doi: 10.1111/cge.12279. [DOI] [PubMed] [Google Scholar]
- 103.Agarwal P.A., Ate-Upasani P., Ramprasad V.L. Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay (ARSACS)-First Report of Clinical and Imaging Features from India, and a Novel SACS Gene Duplication. Mov. Disord. Clin. Pract. 2017;4:775–777. doi: 10.1002/mdc3.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuchay R.A.H., Mir Y.R., Zeng X., Hassan A., Musarrat J., Parwez I., Kernstock C., Traschütz A., Synofzik M. ARSACS as a Worldwide Disease: Novel SACS Mutations Identified in a Consanguineous Family from the Remote Tribal Jammu and Kashmir Region in India. Cerebellum. 2019;18:807–812. doi: 10.1007/s12311-019-01028-2. [DOI] [PubMed] [Google Scholar]
- 105.Sheetal S., Kumar S.A., Byju P. SACS Mutation-Positive Autosomal Recessive Spastic Ataxia of Charlevoix Saguenay (ARSACS) from Kerala. Ann. Indian Acad. Neurol. 2020;23:374–376. doi: 10.4103/aian.AIAN_16_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qavi A., Agarwal A., Garg D., Kharat A. Autosomal recessive spastic ataxia of charlevoix-saguenay (ARSACS): Case report of a novel nonsense mutation in the SACS gene. Ann. Indian Acad. Neurol. 2020;23:395–397. doi: 10.4103/aian.AIAN_670_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ali Z., Klar J., Jameel M., Khan K., Fatima A., Raininko R., Baig S., Dahl N. Novel SACS mutations associated with intellectual disability, epilepsy and widespread supratentorial abnormalities. J. Neurol. Sci. 2016;371:105–111. doi: 10.1016/j.jns.2016.10.032. [DOI] [PubMed] [Google Scholar]
- 108.Manzoor H., Brüggemann N., Hussain H.M.J., Bäumer T., Hinrichs F., Wajid M., Münchau A., Naz S., Lohmann K. Novel homozygous variants in ATCAY, MCOLN1, and SACS in complex neurological disorders. Park. Relat. Disord. 2018;51:91–95. doi: 10.1016/j.parkreldis.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 109.Blumkin L., Bradshaw T., Michelson M., Kopler T., Dahari D., Lerman-Sagie T., Lev D., Chapple J.P., Leshinsky-Silver E. Molecular and functional studies of retinal degeneration as a clinical presentation of SACS-related disorder. Eur. J. Paediatr. Neurol. 2015;19:472–476. doi: 10.1016/j.ejpn.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 110.Narayanan V., Rice S.G., Olfers S.S., Sivakumar K. Autosomal recessive spastic ataxia of Charlevoix-Saguenay: Compound heterozygotes for nonsense mutations of the SACS gene. J. Child. Neurol. 2011;26:1585–1589. doi: 10.1177/0883073811412825. [DOI] [PubMed] [Google Scholar]
- 111.Liew W.K., Ben-Omran T., Darras B.T., Prabhu S.P., Darryl C., Vatta M., Yang Y., Eng C.M., Chung W.K. Clinical application of whole-exome sequencing: A novel autosomal recessive spastic ataxia of Charlevoix-Saguenay sequence variation in a child with ataxia. JAMA Neurol. 2013;70:788–791. doi: 10.1001/jamaneurol.2013.247. [DOI] [PubMed] [Google Scholar]
- 112.McElroy J.P., Krupp L., Johnson B., McCauley J.L., Qi Z., Caillier S.J., Gourraud P.-A., Yu J., Nathanson L., Belman A.L., et al. Copy number variation in pediatric multiple sclerosis. Mult. Scler. J. 2013;19:1014–1021. doi: 10.1177/1352458512469696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wagner F., Titelbaum D.S., Engisch R., Coskun E.K., Waugh J.L. Subtle Imaging Findings Aid the Diagnosis of Adolescent Hereditary Spastic Paraplegia and Ataxia. Clin. Neuroradiol. 2019;29:215–221. doi: 10.1007/s00062-018-0665-5. [DOI] [PubMed] [Google Scholar]
- 114.Dougherty S.C., Harper A., Al Saif H., Vorona G., Haines S.R. A Chromosomal Deletion and New Frameshift Mutation Cause ARSACS in an African-American. Front. Neurol. 2018;9:956. doi: 10.3389/fneur.2018.00956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fogel B.L., Lee J.Y., Lane J., Bs A.W., Chan S., Huang A., Bs G.E.O., Klein E., Mamah C., Perlman S., et al. Mutations in rare ataxia genes are uncommon causes of sporadic cerebellar ataxia. Mov. Disord. 2012;27:442–446. doi: 10.1002/mds.24064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pedroso J.L., Braga-Neto P., Abrahao A., Rivero R.L.M., Abdalla C., Abdala N., Barsottini O.G.P. Autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS): Typical clinical and neuroimaging features in a Brazilian family. Arq. Neuro-Psiquiatr. 2011;69:288–291. doi: 10.1590/S0004-282X2011000300004. [DOI] [PubMed] [Google Scholar]
- 117.Burguêz D., De Oliveira C.M., Rockenbach M.A.B.C., Fussiger H., Vedolin L.M., Winckler P.B., Maestri M.K., Finkelsztejn A., Santorelli F.M., Jardim L.B., et al. Autosomal recessive spastic ataxia of Charlevoix-Saguenay: A family report from South Brazil. Arq. Neuro-Psiquiatr. 2017;75:339–344. doi: 10.1590/0004-282x20170044. [DOI] [PubMed] [Google Scholar]
- 118.Filho F.M.R., Parkinson M.H., Pedroso J.L., Poh R., Faber I., Lourenço C.M., Júnior W.M., Junior M.C.F., Kok F., Sallum J., et al. Clinical, ophthalmological, imaging and genetic features in Brazilian patients with ARSACS. Park. Relat. Disord. 2019;62:148–155. doi: 10.1016/j.parkreldis.2018.12.024. [DOI] [PubMed] [Google Scholar]
- 119.Souza P.V.S., Bortholin T., Naylor F.G.M., Pinto W.B.V.D.R., Oliveira A.S.B. Early-onset axonal Charcot-Marie-Tooth disease due to SACS mutation. Neuromuscul. Disord. 2018;28:169–172. doi: 10.1016/j.nmd.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 120.Nickerson S.L., Marquis-Nicholson R., Claxton K., Ashton F., Leong I.U.S., Prosser D.O., Love J.M., George A.M., Taylor G., Wilson C., et al. SNP Analysis and Whole Exome Sequencing: Their Application in the Analysis of a Consanguineous Pedigree Segregating Ataxia. Microarrays. 2015;4:490–502. doi: 10.3390/microarrays4040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Burnett B., Li F., Pittman R.N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- 122.Feng S.-T., Wang Z.-Z., Yuan Y.-H., Wang X.-L., Sun H.-M., Chen N.-H., Zhang Y. Dynamin-related protein 1: A protein critical for mitochondrial fission, mitophagy, and neuronal death in Parkinson’s disease. Pharmacol. Res. 2020;151:104553. doi: 10.1016/j.phrs.2019.104553. [DOI] [PubMed] [Google Scholar]
- 123.Oliver D., Reddy P.H. Dynamics of Dynamin-Related Protein 1 in Alzheimer’s Disease and Other Neurodegenerative Diseases. Cells. 2019;8:961. doi: 10.3390/cells8090961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Itoh K., Nakamura K., Iijima M., Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anderson J.F., Siller E., Barral J.M. The Neurodegenerative-Disease-Related Protein Sacsin Is a Molecular Chaperone. J. Mol. Biol. 2011;411:870–880. doi: 10.1016/j.jmb.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 126.Campanella C., Pace A., Caruso Bavisotto C., Marzullo P., Marino Gammazza A., Buscemi S., Palumbo Piccionello A. Heat Shock Proteins in Alzheimer’s Disease: Role and Targeting. Int. J. Mol. Sci. 2018;19:2603. doi: 10.3390/ijms19092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mays C.E., Armijo E., Morales R., Kramm C., Flores A., Tiwari A., Bian J., Telling G.C., Pandita T.K., Hunt C.R., et al. Prion disease is accelerated in mice lacking stress-induced heat shock protein 70 (HSP70) J. Biol. Chem. 2019;294:13619–13628. doi: 10.1074/jbc.RA118.006186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tao J., Berthet A., Citron Y.R., Tsiolaki P.L., Stanley R., Gestwicki J.E., Agard D.A., McConlogue L. Hsp70 chaperone blocks α-synuclein oligomer formation via a novel engagement mechanism. J. Biol. Chem. 2021;296:100613. doi: 10.1016/j.jbc.2021.100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zeng H., Tang J.-G., Yang Y.-F., Tan Z.-P., Tan J.-Q. A Novel Homozygous SACS Mutation Identified by Whole-Exome Sequencing in a Consanguineous Family with Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. Cytogenet. Genome Res. 2017;152:16–21. doi: 10.1159/000477428. [DOI] [PubMed] [Google Scholar]
- 130.Rezende Filho F.M., Bremner F., Pedroso J.L., de Andrade J.B.C., Marianelli B.F., Lourenço C.M., Marques-Júnior W., França M.C., Kok F., Sallum J.M., et al. Retinal architecture in autosomal recessive spastic ataxia of charlevoix-saguenay (ARSACS): Insights into disease pathogenesis and biomarkers. Mov. Disord. 2021;36:2027–2035. doi: 10.1002/mds.28612. [DOI] [PubMed] [Google Scholar]
- 131.Lessard I., Lavoie C., Côté I., Mathieu J., Brais B., Gagnon C. Validity and reliability of the LEMOCOT in the adult ARSACS population: A measure of lower limb coordination. J. Neurol. Sci. 2017;377:193–1966. doi: 10.1016/j.jns.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 132.Bui H.T., Gagnon C., Audet O., Mathieu J., Leone M. Measurement properties of a new wireless electrogoniometer for quantifying spasticity during the pendulum test in ARSACS patients. J. Neurol. Sci. 2017;375:181–185. doi: 10.1016/j.jns.2017.01.065. [DOI] [PubMed] [Google Scholar]
- 133.Audet O., Bui H.T., Allisse M., Comtois A.-S., Leone M. Assessment of the impact of an exercise program on the physical and functional capacity in patients with autosomal recessive spastic ataxia of Charlevoix-Saguenay: An exploratory study. Intractable Rare Dis. Res. 2018;7:164–171. doi: 10.5582/irdr.2018.01060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vogel A.P., Stoll L.H., Oettinger A., Rommel N., Kraus E.-M., Timmann D., Scott D., Atay C., Storey E., Schöls L., et al. Speech treatment improves dysarthria in multisystemic ataxia: A rater-blinded, controlled pilot-study in ARSACS. J. Neurol. 2019;266:1260–1266. doi: 10.1007/s00415-019-09258-4. [DOI] [PubMed] [Google Scholar]
- 135.Martinelli C., Battaglini M., Pucci C., Gioi S., Caracci C., Macaluso G., Doccini S., Santorelli F.M., Ciofani G. Development of Nanostructured Lipid Carriers for the Delivery of Idebenone in Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. ACS Omega. 2020;5:12451–12466. doi: 10.1021/acsomega.0c01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ricca I., Tessa A., Trovato R., Bacci G.M., Santorelli F.M. Docosahexaenoic acid in ARSACS: Observations in two patients. BMC Neurol. 2020;20:215. doi: 10.1186/s12883-020-01803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nethisinghe S., Abeti R., Kesavan M., Wigley W.C., Giunti P. Hsp90 Inhibition: A Promising Therapeutic Approach for ARSACS. Int. J. Mol. Sci. 2021;22:11722. doi: 10.3390/ijms222111722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zuehlke A.D., Moses M.A., Neckers L. Heat shock protein 90: Its inhibition and function. Philos. Trans. R. Soc. B Biol. Sci. 2017;373:20160527. doi: 10.1098/rstb.2016.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.