Abstract
In a recent genotypic survey of β-lactam-resistant pneumococci recovered in different areas of United States during 1997, eight clonal types that each represented 3 to 40 isolates accounted for 134 of 144 isolates (G. Gherardi, C. Whitney, R. Facklam, and B. Beall, J. Infect. Dis. 181:216–229, 2000). We determined the degree of pspA gene diversity among these 134 isolates and for 11 previously characterized internationally disseminated multiresistant strains. Thirty-four different pspA restriction profiles were determined for an amplicon encompassing the variable portion of the structural gene that encodes the surface-exposed domain of PspA and a variable-length proline-rich putative cell wall-associated domain. These restriction profiles closely correlated with those of 33 different pspA sequence types of an approximately 230-residue region corresponding to residues 182 to 410 of the strain Rx1 PspA. These residues encompass a 100-residue clade-defining region known to contain cross-protective epitopes for which 17 sequence types were found. Distinct, conserved pspA sequence types were found for the majority of strains within seven of the eight U.S. clonal types assessed, while one pulsed-field gel electrophoresis type was represented by isolates of three distinct PspA clades. Sequence typing of pspA provides an added level of specificity in the subtyping of isolates and is a necessary first step in determining the components needed in a PspA vaccine which could elicit effective cross-protective coverage.
Streptococcus pneumoniae is a major cause of morbidity and mortality worldwide and is the leading cause of bacterial pneumonia, meningitis, bacteremia, and otitis media. Pneumococcal polyvalent purified capsular polysaccharide vaccines are serotype specific in their protection, contain only 23 of the 90 known capsular types, and are unable to elicit effective immunologic responses in children younger than 2 years of age and many elderly individuals (6, 13). Although conjugate vaccines should be much more effective in these individuals, they are still limited to offering protection against specific capsular types. For these reasons, there is much interest in developing pneumococcal protein vaccines. One protein vaccine candidate, PspA, is an antigenically variable surface virulence factor (16) that interferes with complement-mediated clearance of pneumococci during bacteremia (1, 2). Antibodies to PspA protect mice against lethal systemic infection, and this protection is often cross-protective for strains of different capsular serotypes expressing distinct PspA molecule types (1, 2, 17, 19). Immunization of humans with a single recombinant PspA stimulated antibodies broadly cross-reactive to heterologous PspA molecules (23). Although it is known that PspA is broadly cross-reactive, cross-reactivity does not necessarily correspond to cross-protection, and the total array of common circulating PspA types should be determined before the degree of cross-protection afforded by specific sequence types can be assessed. It is particularly important to assess PspA variability among strains with serotypes not targeted by impending conjugate vaccines and circulating strains that have become highly resistant to antibiotics used for treatment. Highlighting the importance of the latter group is the recent report that high-level penicillin resistance was an independent parameter of mortality in pneumococcal bacteremia in a population with high human immunodeficiency virus seroprevalence (27). For the study presented here, we examined the pspA sequences of multiresistant, genetically characterized U.S. isolates recovered in 1997 and 11 previously characterized internationally disseminated multiresistant strains.
MATERIALS AND METHODS
Isolates.
Eight genetically related sets comprised 134 of 144 multiresistant isolates that were recovered in 1997 and that were described previously (9). These isolates were obtained from the Emerging Infections Program/Active Bacterial Core Surveillance (available at http://www.cdc.gov/ncidod/dbmd/abcs) conducted in seven states within the United States.
Previously characterized clones.
The following previously genetically characterized strains were provided by L. McDougal and F. Tenover and were designated according to the Pneumococcal Molecular Epidemiology Network (15). These included clones Spain23F-1 (strain SP193, ATCC 700669) (4, 21), Spain6B-2 (GM17, SP194, ATCC 700670) (22), France9V-3 (strain SP195, ATCC 700671; highly related to strain 665) (4, 24), Tennessee23F-4 (strain SP196, ATCC 51916) (20, 24), England14-9 (strain SP200, ATCC 700676) (10), Spain14-5 (strain VH14, SP197, ATCC 700672) (5), Hungary19A-6 (SP220, ATCC 700673) (21), South Africa19A-7 (SP198, strain 17619, ATCC 700674) (25), South Africa6B-8 (strain SP199, ATCC 700675) (24), Slovakia14-10 (strain 91-006571, SP221, ATCC 700677) (8), and Slovakia19A-11 (strain 91-0006571, SP222, ATCC 700678) (8).
PCR, restriction profiling, and sequencing.
Primers F (GCCAGCGTCGCTATCTTAGGGGCTGG) and R (GAACCATCAGTATTGTAGAAGTACCA) were used for PCR and were derived by comparisons of conserved regions between the two sequences corresponding to GenBank accession numbers U89711 (28) and A41971 (9). Primer R was used to obtain the sequences of 550 to 780 bases used for analysis. Restriction analysis of the amplicon generated with primers F and R was accomplished as described previously for penicillin-binding protein gene amplicons (9) following digestion of 10 μl of unpurified PCR products with 2 U each of the enzymes RsaI, HinfI, and HaeIII. Amplicons showing identical triple-digest patterns were additionally compared by DdeI digestion in the same manner to ensure that there was true representation of highly similar amplicons. Amplicons from each of the 11 internationally dispersed clones were also profiled by restriction analysis and sequenced. Independent amplicons from at least two 1997 isolates from each identical restriction profile set were sequenced. When possible, pspA amplicons from isolates that differed in serotype, geographic source, and pulsed-field gel electrophoresis (PFGE) profile from within each identical pspA restriction profile set were used for sequence analysis.
DNA sequence analysis.
The translation of the primer R-generated sequence complement was used for amino acid sequence comparisons by using the Wisconsin Package, version 10.0, software (Genetics Computer Group, Madison, Wis.).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the partial pspA sequences encompassing roughly codons 180 to 410 of the RxA pspA gene (19) are listed in Table 1.
TABLE 1.
pspA RFLP types and sequence accession numbers for 1997 multiresistant isolates and internationally disseminated clones that were genotyped
| Reference strain (PFGE type) | No. of isolates | PFGE subtypesa | Serotype (no. of isolates) | pspA GenBank accession no.b | pspA RFLP typec | pspA clade (closest match/ % identity)d |
|---|---|---|---|---|---|---|
| Spain23F-1(A1) | 16 | A2–A3, A5, A6 | 23F (11), 19F (5) | AF254259 | a1 | 3 (BG8090/97.1) |
| 2 | A2 | 23F (2) | AF255542 | a2 | 3 (BG8090/97.1) | |
| 3 | A2 | 23F (2), 19F(1) | AF255543 | a3 | 3 (BG8090/92.2) | |
| 1 | A2 | 19F (1) | AF254256 | a4 | 3 (BG8090/96.1) | |
| 1 | A4 | 23F (1) | AF288751 | a5 | 5 (ATCC6303/98.3) | |
| France9V-3 (B1) | 33 | B2–B5, B8–B15, B21, B22 | 9V (16), 9A (10), 14 (9) | AF252286 | b1 | 3 (AC122/100) |
| 1 | B6 | 9V (1) | AF252286 | b4 | 3 (AC122/100) | |
| 2 | B2 | 9V (2) | AF253404 | b2 | 3 (AC122/100) | |
| 1 | B7 | 9V (1) | AF253405 | b3 | 3 (AC122/99) | |
| 1 | B13 | 14 (1) | AF253406 | c1 | 1 (AC94/97) | |
| England14-9 (C1) | 31 | C1–C11 | 14 (30) | AF253406 | c1 | 1 (AC94/97) |
| Strain 121 (D1) | 1 | D1 | 6B (1) | AF255548 | d3 | 3 (BG8090/99) |
| 2 | D4 | 23F (1), 6B (1) | AF255547 | d1 | 3 (BG8090/99) | |
| 1 | D7 | 6A (1) | AF255547 | d2 | 3 (BG8090/99) | |
| 2 | D3 (1), D5 (1) | 6B (2) | AF255549 | d4 | 1 (AC94/93) | |
| 1 | D2 | 6B (1) | AF255550 | d5 | 1 (BG6692/100) | |
| 1 | D8 | 6B (1) | AF255551 | d6 | 1 (BG6692/100) | |
| 2 | D6, D9 | 23F (1), 6B (1) | AF255552 | d7 | 2 (DBL5/96) | |
| Tennessee23F-4 (E1) | E1 | 23F | AF253407 | e1 | 1 (BG6843/92) | |
| 6 | E2, E5 | 23F (6) | AF255900 | e2 | 1 (BG6843/92) | |
| 3 | E2, E7, E8 | 23F (3) | AF255901 | e3 | 1 (BG6843/92) | |
| 3 | E2 | 23F (3) | AF255902 | e4 | 1 (BG6843/92) | |
| 3 | E2 | 23F (3) | AF255903 | e5 | 1 (BG6843/92) | |
| 1 | E3 | 23F (1) | AF255904 | e6 | 1 (BG6843/92) | |
| 1 | E7 | 23F (1) | AF255905 | e7 | 1 (BG6843/92) | |
| 1 | E4 | 23F (1) | AF255906 | e8 | 1 (BG6843/92) | |
| 1 | E6 | 23F (1) | AF255907 | e9 | 1 (BG6843/92) | |
| Strain 127 (G1) | 2 | G1 | 19F (3) | AF25554 | g1 | 3 (AC122/100) |
| 1 | G2 | 19F (1) | AF255545 | g2 | 3 (AC122/100) | |
| Strain 69 (H1) | 4 | H1 | 14 (4) | AF255908 | h1 | 3 (AC122/96.1) |
| Spain6B-2 (K1) | 4 | K1–K5 | 6B | AF254257 | k1 | 1 (DBL1/100) |
| South Africa6B-8 (M) | M1 | 6B | AF254258 | m1 | 1 (BG6692/98) | |
| Slovakia19A-11 (N) | N1 | 19A | AF254255 | n1 | 5 (ATCC6303/94.9) | |
| Hungary19A-6 (O) | O1 | 19A | AF254254 | o1 | 5 (ATCC6303/99.2) | |
| South Africa19A-7 (P) | P1 | 19A | AF253408 | p1 | 4 (BG7817/99) | |
| Spain14-5 (α) | α1 | 14 | AF253406 | c1 | 1 (AC94/97) | |
| Slovakia14-10 (β) | β1 | 14 | AF255546 | c1 | 1 (AC94/97) |
Isolates were genotyped previously (9). For each group those isolates that differed by only one to six bands from subtype 1 were assigned a common type (A, B, C, etc.). For each type, isolates with a difference from subtype 1 of more than six bands of were considered unrelated isolates and were assigned a different PFGE type. Reference strains 121, 127, and 69 represent new pneumococcal PFGE types (D, G, and H, respectively) (9) and are included in the number of 1997 U.S. isolates given in the second column.
Each different accession number represents a unique pspA sequence of 550 to 775 bases encompassing the clade-defining region and variable-length proline-rich repeat region.
The following pspA RFLP types shared identical clade-defining regions: a1 and a2; b1, b2, b4, g1, and g2; b3 and c1; d1 to d3; d5 and d6; and e1 to e9.
Percent identity to the clade-defining region of closest match from reference 11.
RESULTS AND DISCUSSION
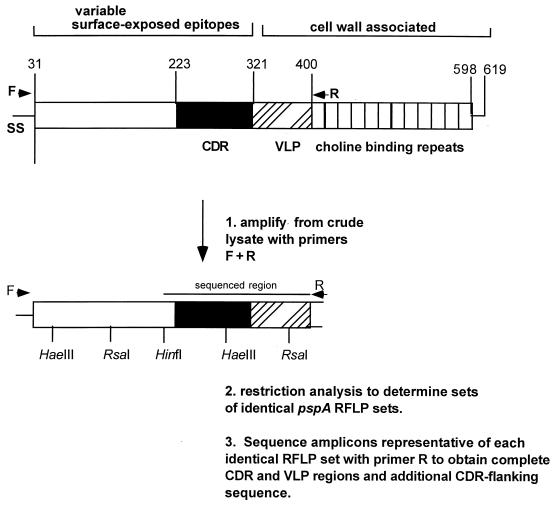
Primers were selected based upon sequences conserved between the pspA DNA sequence of strain EF5668 (19) and the PspA amino acid sequence of JY2008 (28), which were the only two PspA sequences accessible from GenBank at the time that this study was initiated. Two primers were selected for amplification based on their ability to generate a single 1,000- to 1,500-bp amplicon from all pneumococcal strains tested. Primer F annealed to the conserved signal sequence codons 12 to 20, while primer R annealed to codons 411 to 419 and 465 to 473 of the strain EF5668 and strain JY2008 pspA genes, respectively (Fig. 1). Thus, this amplicon carries the sequence that encodes the α-helical surface-exposed region with most antigenic epitopes and the closely situated variable-length proline-alanine repeat region (VLP in Fig. 1) (3, 11, 18, 28). The surface-exposed region encompasses the 100-residue pspA clade-defining region which includes protection-eliciting epitopes (3, 11).
FIG. 1.
General features of PspA and method used for sequence typing of pspA amplicons. The pspA codon positions are taken from references 3 and 11 and are based upon the Rx1 pspA sequence (27). Abbreviations: SS, signal sequence; CDR, clade-defining region containing cross-protective epitopes; VLP, variable-length proline-rich repeat region.
Previously, sequence-based divisions among diverse PspA proteins classified these proteins into three families that were further subdivided into six clades (3, 11). The clade-defining regions of PspA proteins in different clades vary at >20% of amino acid positions (3). Family 1 is composed of clades 1 and 2, and family 2 is composed of clades 3 to 5. Family 3 has only one highly divergent member. Although both clades and families are easily recognized sequence-based divisions of PspA proteins, it is the family-level division that appears to be the most important serologically. Rabbit antisera made to recombinant PspA proteins could reliably distinguish PspA families but could not always distinguish PspA clades within the same family (23; S. Hollingshead and D. E. Briles, unpublished data).
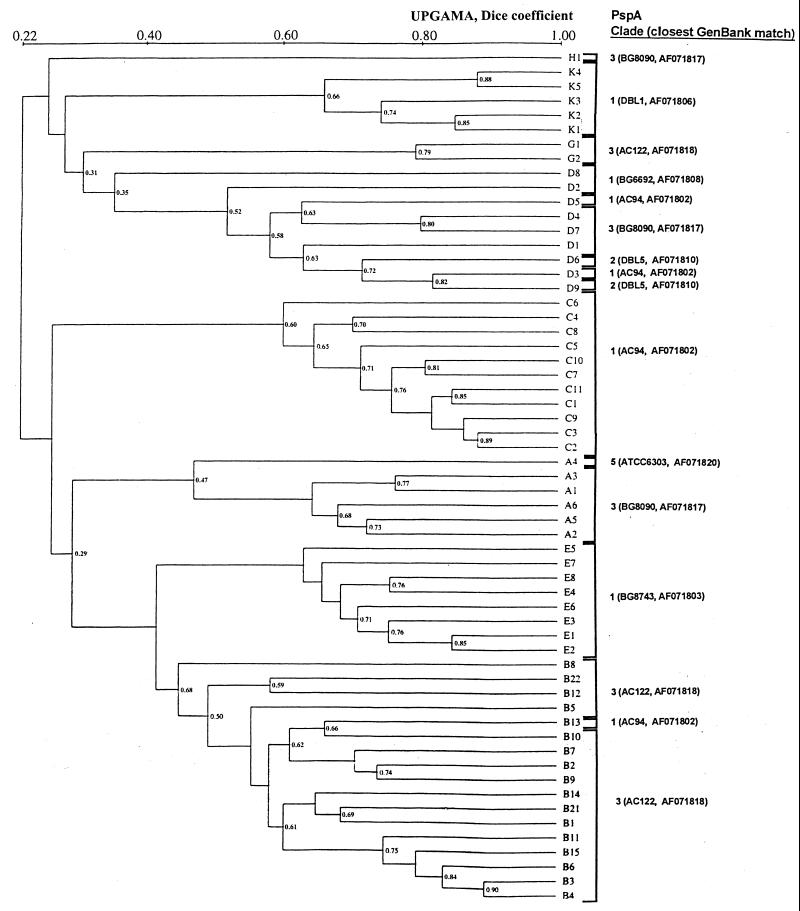
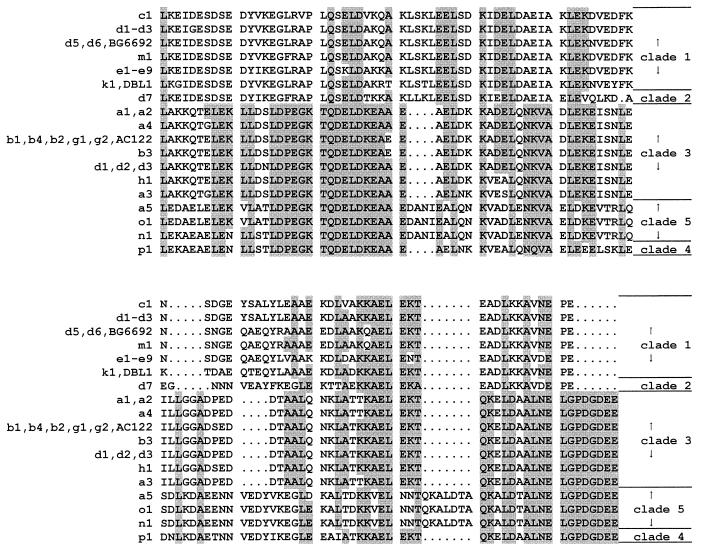
Figure 1 summarizes the method of pspA amplicon-based subtyping used. Table 1 summarizes the pspA amplicon sequencing and restriction pattern results. The PspA sequences of only families 1 and 2, consisting of clades 1 to 5, were found among the multiresistant pneumococcal strains used for this study. Seventy-three of the 134 1997 U.S. isolates (54%), representing five distinct PFGE types, were found to be of PspA clade 3, and the sequences of their clade 3-defining regions diverged from those of previously sequenced clade 3-defining regions by no more than 7.8%. Fifty-eight isolates (43%), representing three PFGE types, were found to contain clade 1 PspA sequences, with their clade-defining regions having 92 to 100% identity to previously sequenced clade 1-defining regions. Only one clade 2 PspA sequence was found among these 1997 U.S. isolates, from two independent PFGE type D isolates comprising two different serotypes (subtypes D2 and D9 in Fig. 2). Only one clade 5 isolate was found among the 1997 U.S. isolates and consisted of a PFGE type A isolate (subtype A4 in Fig. 2). The sequences of the clade 5-defining regions of two internationally disseminated clones (PFGE types N1 and O1 in Table 1) were found to have 95 to 99% amino sequence identity to the clade-defining region of clade 5 strain ATCC 6303 (11). Finally, a single clade 4 PspA sequence was found from the internationally disseminated strain South Africa19A-7 (PFGE type P1 in Table 1). The relation of clades 3 to 5 (family 2) compared to clades 1 and 2 (family 1) is evident in Fig. 3, in which the clade-defining regions representing all of the sequences examined in this study are aligned.
FIG. 2.
PspA clades of previously determined PFGE types (9) of multiresistant, invasive pneumococcal isolates. The dendrogram gives underestimated Dice coefficients because closely migrating bands could not be properly resolved with the program used. Visually identical PFGE profiles gave average Dice coefficients of ∼0.86 to 0.93. UPGMA, unweighted pair group method with arithmetic averages. The strains with the closest matches in their PspA clade-defining regions (11) are listed along with their pspA GenBank accession numbers.
FIG. 3.
Alignment of the PspA clade-defining regions that contain cross-protective epitopes from different pspA amplicon RFLP types. BG6692 and DBL1 refer to PspA protein sequences from reference 11. The PileUp program was used as described previously (11). Positions shared by nine or more PspA sequences are shaded.
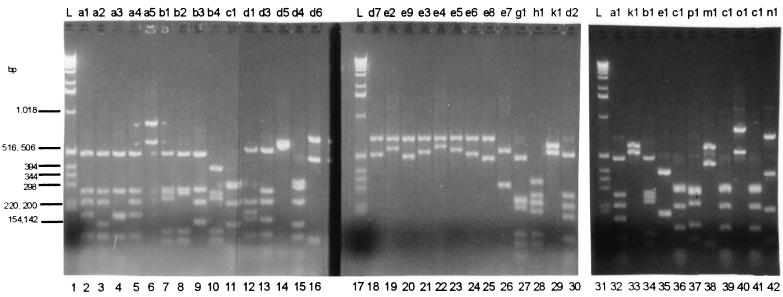
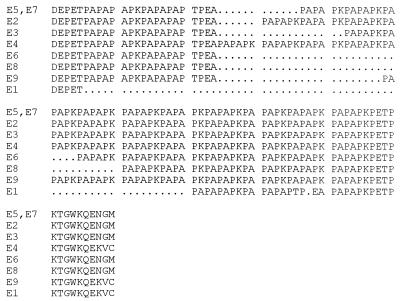
As reported previously (3, 11), the 650- to 775-bp region of pspA sequenced was remarkably variable, with the more distantly related sequences (e.g., clade 1 versus clade 3) sharing only about 40% sequence identity at either the DNA or the protein level. For isolates of PFGE types A, B, C, H, and K, one specific pspA amplicon restriction fragment length polymorphism (RFLP) pattern represented the majority of isolates (89 of 101 [88.1%]) obtained within a PFGE type (Table 1). For isolates of PFGE types A, B, C, and K, the major pspA sequence and RFLP type was additionally shared with those of previously characterized internationally disseminated antibiotic-resistant strains of the same PFGE types. With only three exceptions, each specific RFLP type was associated with a specific pspA sequence type. Closely related RFLP patterns b1 and b4 both corresponded to the GenBank accession number AF252286 sequence, and similarly, RFLP patterns d1 and d2 both corresponded to the GenBank accession number AF255547 sequence. One strain (Slovakia14-10) contained only one conservative substitution in PspA relative to the other isolates of pspA RFLP type c1. The sequence differences between sets of highly related amplicons sharing the same clade-defining region were most often the result of differences in the number of repetitive sequences in the variable-length proline-rich domain. For example, the accession number AF25429 and AF255542 sequences (corresponding to RFLP types a1 and a2, respectively) differ only by the deletion of a PAPAPKPEQ direct repeat (data not shown), and this 27-bp deletion appears to account for the difference in the amplicon restriction profile (Fig. 4; compare lanes 2 and 3). Similarly, the nine sequences corresponding to RFLP types e1 to e9, respectively, shared identical clade-defining regions, and the principal difference between them was in the number of proline-rich repeats in the variable-length proline-rich region (Fig. 5).
FIG. 4.
RFLP patterns of pspA amplicons. Amplicons generated with primers F and R were subjected to triple digestion with HaeIII, HinfI, and RsaI prior to agarose gel electrophoresis. The RFLP type designations at the top are from Table 1. Lanes 1, 17, and 31, size ladder (L); lanes 2 to 16 and 18 to 30, 1997 multiresistant U.S. isolates; lanes 32 to 42, internationally disseminated multiresistant strains: lane 32, Spain23F-1; lane 33, Spain6B-2; lane 34, France9V-3; lane 35, Tennessee23F-4; lane 36, Spain14-5; lane 37, S. Africa19A-7; lane 38, S. Africa6B-8; lane 39, England14-9; lane 40, Hungary19A-6; lane 41, Slovakia14-10; lane 42, Slovakia19A-11.
FIG. 5.
Alignment of PFGE type E (subtypes E1 to E9) PspA variable-length proline-rich regions.
Only two pspA sequence types were shared among strains that did not appear to be highly related according to the results of PFGE. The PspA sequence with GenBank accession number AF253406 (768 bases) was shared exclusively among PFGE type C clinical isolates: PFGE type C isolate England14-9, PFGE type α isolate Spain14-5, and a PFGE type B isolate. The GenBank accession number AF255546 sequence, which differed from the GenBank accession number AF253406 sequence by only one conservative substitution (data not shown), was also found in the distinct PFGE type β strain Slovakia14-10. It is interesting that all of these strains with the pspA sequence corresponding to GenBank accession number AF253406 (and the single strain with the GenBank accession number AF255546 sequence), although they appeared to be nonrelated according to the results of PFGE and penicillin-binding protein gene-dhf restriction profiling (data not shown), were all serotype 14 (Table 1). Further analysis of the degree of relatedness between these strains should be facilitated by multilocus sequence typing (7).
With the exceptions of PFGE type D isolates, the majority of isolates within a given PFGE type exhibited identity or a high degree of homology within their pspA sequences, which is evident in the alignment of the approximately 100-residue clade-defining region (Fig. 3). The PFGE type D isolates displayed the widest range of variability in the pspA clade-defining region, with isolates found to be of clade 3 (four isolates), clade 1 (four isolates), and clade 2 (two isolates). The clade 5 PspA protein from the isolate of PFGE subtype A4 is quite divergent from the clade 3 PspA proteins shared by the other 23 PFGE type A isolates (including Spain23F-1). This is consistent with the fact that PFGE subtype A4 is the most divergent PFGE subtype within type A (Fig. 2). This sequence (GenBank accession number AF25426 [RFLP type a5]) differed from the sequence found for Hungary19A-6 (RFLP type o1) by only 2 codon substitutions over a 227-codon overlap and by only 1 conservative substitution in the clade-defining region (Fig. 3). Additionally, 1 of the 40 PFGE type B isolates had the major PFGE type C pspA sequence (GenBank accession number AF253406) and RFLP type c1 (Table 1).
Isolates of PFGE type D are a more broadly divergent group on the basis of the deeper branching points in the dendrogram (Fig. 2), which is consistent with PFGE type D having three different PspA clades. The deeper branching points could possibly indicate an older age of that clonal type, which would allow more time for horizontal pspA transfer events. However, isolates of PFGE type B have deeper branch points than isolates of the PFGE types other than D, and yet 39 of the 40 PFGE type B isolates (except for subtype B13) have very conserved pspA sequences.
Comparisons of approximately 160- to 230-codon regions of pspA consisting of the clade-defining region plus the flanking sequence that includes 20 to 50 codons on the N-terminal side and the variable-length proline-rich domain on the C-terminal side, in combination with RFLP analysis of the pspA amplicon, provides a potential tool for distinguishing between highly related isolates (Fig. 3 and 4; Table 1). A total of 145 multiresistant pneumococci were found to exhibit 34 pspA amplicon restriction profiles that were predictive of specific sequence types. In a similar manner, restriction profiling of pspA amplicons was previously found to be useful in revealing related subsets of pspA (A. Brooks-Walter, L. S. McDaniel, S. K. Hollingshead, M. J. Crain, and D. E. Briles, Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother., abstr. B12, p. 28, 1995) and was used to assist in demonstrating the clonal origin of serotype 9L isolates from western Canada (26). All of the isolates in the present study were found to contain family 1 or family 2 pspA alleles that encoded PspA proteins primarily of clade 1 and clade 3, respectively. The most prevalent PFGE types of resistant pneumococci from within the United States were found to have pspA amplicons and sequence types identical to those for highly related internationally dispersed multiresistant strains. PspA has recently been shown to have a mosaic structure that presumably results from extensive immunologic selection for intraspecies recombination events (11, 12). Although different serotypes and clonal backgrounds appear to vary in their degrees of virulence (14), it is unknown if or how different PspA types affect pneumococcal virulence. For epidemiologic studies and for PspA-based vaccine considerations, it may possibly prove to be important to track pneumococcal strains with regard to the sequence types of this variable protein.
ACKNOWLEDGMENTS
We thank the Georgia, California, Connecticut, Minnesota, Oregon, Maryland, New York, and Tennessee Emerging Infections Program Network/Active Bacterial Core Surveillance for isolates. We are grateful to Linda McDougal and Fred Tenover for sharing characterized pneumococcal clones and information. Zhongya Li provided excellent technical assistance. We thank Ruth Franklin and Deloise Jackson for the bulk of the serotyping results.
G.G. was a recipient of a career award from the Italian Fondazione Cariverona Progetto Sanità.
REFERENCES
- 1.Briles D E, King J D, Gray M L, McDaniel L S, Swialto E, Benton K A. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine. 1996;14:858–867. doi: 10.1016/0264-410x(96)82948-3. [DOI] [PubMed] [Google Scholar]
- 2.Briles D E, Tart R C, Wu H-Y, Ralph B A, Russell M W, McDaniel L S. Systemic and mucosal protective immunity to pneumococcal surface protein A. Ann N Y Acad Sci. 1996;797:118–126. doi: 10.1111/j.1749-6632.1996.tb52954.x. [DOI] [PubMed] [Google Scholar]
- 3.Briles D E, Hollingshead S, Swiatlo E, Dillard J P, Smith P, Benton K A, Ralph B A, Brooks-Walter A, Crain M J, Hollingshead S K, McDaniel L S. PspA and PspC: their potential for use as pneumococcal vaccines. Microb Drug Resist. 1997;3:401–408. doi: 10.1089/mdr.1997.3.401. [DOI] [PubMed] [Google Scholar]
- 4.Coffey T J, Dowson C G, Daniels C G, Zhou M, Martin J, Spratt C, G. B, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 5.Coffey T J, Berron S, Daniels M, Garcia-Leoni M E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 6.Cowan M J, Ammann A J, Wara D W, Howie V M, Schultz L, Doyle N, Kaplan M. Pneumococcal polysaccharide immunization in infants and children. Pediatrics. 1978;62:721–727. [PubMed] [Google Scholar]
- 7.Enright M C, Spratt B G. A multilocus typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 8.Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and in Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 9.Gherardi G, Whitney C, Facklam R, Beall B. Major sets of genetically related β-lactam resistant pneumococci in the United States based upon PFGE and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J Infect Dis. 2000;181:216–229. doi: 10.1086/315194. [DOI] [PubMed] [Google Scholar]
- 10.Hall L M, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollingshead S K, Becker R, Briles D E. University of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect Immun. 2000;68:5889–5900. doi: 10.1128/iai.68.10.5889-5900.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollingshead S K, Bessen D, Briles D E. Archeological footprints of horizontal gene transfer: mosaic cell surface proteins of Streptococcus pyogenes and Streptococcus pneumoniae. In C. Kado and M. Syvanen (ed.), Horizontal gene transfer: implications and consequences. New York, N.Y: Chapman & Hall; 1998. [Google Scholar]
- 13.Jernigan D B, Cetron M S, Breiman R F. Defining the public health impact of drug resistant Streptococcus pneumoniae: report of a working group. Morb Mortal Wkly Rep. 1996;45:1–20. [PubMed] [Google Scholar]
- 14.Kelly T, Dillard J P, Yother J. Effect of genetic switching of capsular type on virulence of Streptococcus pneumoniae. Infect Immun. 1994;62:1813–1819. doi: 10.1128/iai.62.5.1813-1819.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klugman K. Pneumococcal molecular epidemiology network. ASM News. 1998;64:371. [Google Scholar]
- 16.McDaniel L S, Yother J, Vijayakumar M, McGarry L, Guild W R, Briles D E. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA) J Exp Med. 1987;165:381–394. doi: 10.1084/jem.165.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDaniel L S, Sheffield J S, Delucchi P, Briles D E. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59:222–228. doi: 10.1128/iai.59.1.222-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniel L S, Ralph B A, McDaniel D O, Briles D E. Location of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb Pathog. 1994;17:323–337. doi: 10.1006/mpat.1994.1078. [DOI] [PubMed] [Google Scholar]
- 19.McDaniel L S, McDaniel D O, Hollingshead S K, Briles D E. Comparison of the PspA sequence from Streptococcus pneumoniae EF5668 to the previously identified PspA sequence from strain Rx1 and ability of PspA from EF5668 to elicit protection against pneumococci of different capsular types. Infect Immun. 1998;66:4748–4754. doi: 10.1128/iai.66.10.4748-4754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDougal L K, Rasheed J K, Biddle J W, Tenover F C. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother. 1995;39:2282–2288. doi: 10.1128/aac.39.10.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 22.Munoz R, Musser J M, Crain M, Briles D E, Morton A, Parkinson A J, Sorenson U, Tomasz A. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 23.Nabors G S, Braun P A, Herrmann D J, Heise M L, Pyle D J, Gravenstein S, Schilling M, Ferguson L M, Hollingshead S K, Briles E E, et al. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) stimulated cross-reactive antibodies to heterologous PspA molecules. Vaccine. 2000;18:1743–1754. doi: 10.1016/s0264-410x(99)00530-7. [DOI] [PubMed] [Google Scholar]
- 24.Sibold C, Wang J, Henrichsen J, Hakenbeck R. Genetic relationships of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect Immun. 1992;60:4119–4126. doi: 10.1128/iai.60.10.4119-4126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 26.Swiatlo E, Crain M J, McDaniel L S, Brooks-Walter A, Coffey T J, Spratt B G, Morrison D A, Briles D E. DNA polymorphisms and variant penicillin-binding proteins as evidence that relatively penicillin-resistant pneumococci in western Canada are clonally related. J Infect Dis. 1996;174:884–888. doi: 10.1093/infdis/174.4.884. [DOI] [PubMed] [Google Scholar]
- 27.Turett G S, Blum S, Fazal B A, Justman J E, Telzak E E. Penicillin resistance and other predictors of mortality in pneumococcal bacteremia in a population with high human immunodeficiency virus seroprevalence. Clin Infect Dis. 1999;29:321–327. doi: 10.1086/520209. [DOI] [PubMed] [Google Scholar]
- 28.Yother J, Briles D E. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol. 1992;174:601–609. doi: 10.1128/jb.174.2.601-609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]