Abstract
Dysregulation of autophagy may contribute to the progression of various muscle diseases, including Duchenne muscular dystrophy (DMD). Heme oxygenase-1 (HO-1, encoded by Hmox1), a heme-degrading enzyme, may alleviate symptoms of DMD, inter alia, through anti-inflammatory properties. In the present study, we determined the role of HO-1 in the regulation of autophagy and mitophagy in mdx animals, a commonly used mouse model of the disease. In the gastrocnemius of 6-week-old DMD mice, the mRNA level of mitophagy markers: Bnip3 and Pink1, as well as autophagy regulators, e.g., Becn1, Map1lc3b, Sqstm1, and Atg7, was decreased. In the dystrophic diaphragm, changes in the latter were less prominent. In older, 12-week-old dystrophic mice, diminished expressions of Pink1 and Sqstm1 with upregulation of Atg5, Atg7, and Lamp1 was depicted. Interestingly, we demonstrated higher protein levels of autophagy regulator, LC3, in dystrophic muscles. Although the lack of Hmox1 in mdx mice influenced blood cell count and the abundance of profibrotic proteins, no striking differences in mRNA and protein levels of autophagy and mitophagy markers were found. In conclusion, we demonstrated complex, tissue, and age-dependent dysregulation of mitophagic and autophagic markers in DMD mice, which are not affected by the additional lack of Hmox1.
Keywords: DMD, Duchenne muscular dystrophy, mitophagy, autophagy, mdx, heme oxygenase-1, HO-1
1. Introduction
Skeletal muscles, characterized by high metabolic activity [1] and constantly exposed to mechanical and oxidative stress, are particularly susceptible to the formation of dysfunctional organelles and protein aggregates [2]. Whether in response to the stressor cells decide to undergo apoptosis or autophagy, is strictly dependent on the nature of the stimulus and the balance between inhibitory and activating signals [3,4]. The sustained excessive level of autophagy flux degrades a large number of vital molecules and organelles, while not sufficient autophagic machinery causes protein inclusions and accumulation of damaged organelles [1,5]. Therefore, autophagy plays a pivotal role in the proper functioning and repair of the muscle [6,7].
A growing number of studies indicate dysregulation of autophagic flux in myoblasts as a hallmark of numerous muscular diseases, including muscular dystrophies [8]. In Duchenne muscular dystrophy (DMD), the disease caused by a lack of dystrophin, a protein essential for myocyte integrity, dysfunctional autophagy was demonstrated to contribute to muscle weakness and wasting [9,10,11,12]. Furthermore, systemic metabolic impairment, including disruption in Ca2+ homeostasis, augmented reactive oxygen species (ROS) production, and mitochondrial dysfunction, greatly contribute to the progression of this incurable disorder (reviewed in: [13]). Therefore, autophagy and organelle-specific autophagy, namely mitophagy, may be of particular importance in DMD [11,12].
Accordingly, normalization of defective autophagy/mitophagy was suggested as an experimental therapy for DMD. Resveratrol, a polyphenol found in grapes and red wine, led to mitophagy stabilization, reduction of ROS levels, and improvement of muscle condition in mdx animals, a mouse model of DMD [11]. Kuno et al. demonstrated that treatment of dystrophic mice with this polyphenol for 56 weeks ameliorates mitophagy and decreases cardiomyopathy, the life-threatening heart complication during DMD progression [8]. Interestingly, Whitehead et al. noted that the use of simvastatin (an HMG-CoA reductase inhibitor, generally used to normalize cholesterol levels) has a positive effect on autophagy in the DMD mouse model [14,15]. Recently, restoration of functional autophagy in dystrophic skeletal muscle was also observed after treatment with a hydrogen sulfide donor [16]. Of note, it was also underlined that expression of mitophagy- and autophagy-related proteins may vary during mice lifetime [17].
An important aspect of maintaining homeostasis in skeletal muscles is counteracting oxidative stress. In DMD, sustained oxidative stress aggravates muscle damage and contributes to disease progression, hence the use of antioxidants may be of therapeutic importance [18]. One of the candidates might be heme oxygenase-1 (HO-1, encoded by Hmox1), a cytoprotective enzyme that through heme degradation generates anti-inflammatory end products (mostly CO) with protective roles in physiological and pathological conditions. In our previous study, chemical inhibition of HO-1 activity as well as global, genetic knockout of Hmox1 in dystrophic animals, increased the inflammatory reaction, deteriorated the condition of the muscles, and decreased mice endurance to exercise [19]. HO-1 was also demonstrated to control the activation of autophagy to counteract stress conditions and inflammation in various diseases, including sepsis, diabetes, or reperfusion after hypoxia in the liver (reviewed in: [20]). Moreover, the appearance of abnormal mitochondria together with impaired Pink1/Parkin-dependent mitophagy and mitochondrial biogenesis were noticed in Hmox1-lacking animals (reviewed in: [21]). Of note, to the best of our knowledge, no data regarding the possible role of HO-1 in the control of the autophagy or mitophagy pathways in DMD has been published.
Therefore, we aimed to investigate the role of HO-1 in dysregulated autophagy and mitophagy in mdx mice. The mitochondrial dysfunction in dystrophic animals was evident at the mRNA and protein level in the diaphragm and gastrocnemius muscles. Although a decrease in the expression of genes regulating mostly mitophagy was observed in 6-week-old mdx mice, in double knockout animals–lacking dystrophin and Hmox1 (mdx/Hmox1−/−), no changes were observed, questioning our hypothesis about the beneficial effect of this cytoprotective enzyme in this process. However, at the protein level, the expression of autophagy markers was increased by dystrophin deficiency, indicating additional regulation at the translational level. As we observed muscle and age-dependent effects, more research is still required to better understand molecular mechanisms of “self-eating” as well as HO-1 role in the autophagy dysregulation seen in DMD (especially in experimental settings with HO-1 overproduction/overexpression), where it could be a potential therapeutic agent.
2. Results
2.1. Verification of the Animal Models Used in the Study
Although mdx mice, with a nonsense point mutation (C-to-T transition) in exon 23 that aborts full-length dystrophin expression [22], are not severely affected (as compared to humans), and the differences in the disease outcomes between skeletal muscles and the diaphragm are known [23], they are commonly employed model animals in DMD research. To investigate the role of HO-1 in DMD, we generated mdx mice lacking the Hmox1 gene, encoding HO-1, (named as mdx/Hmox1−/−), as described in our previous study [19]. In such double knockouts, expression of Hmox1 was not detectable, and it did not differ between 6-week-old wild-type (WT) and mdx animals (Figure 1A).
Figure 1.
Validation of the animal model. Hmox1 expression in 6-week-old animals (A), qRT-PCR; n = 6/group. The protein level of HO-1 in gastrocnemius muscle (B), ELISA, n = 6. Dystrophic mice exhibit decreased muscular strength (C), grip test, n = 6–7/group; and no difference in body weight (D), n = 5–7/group. The expression of DMD markers: Spp1 in the diaphragm (E), qRT-PCR; n = 10–11/group and its protein level (osteopontin, OPN) in the serum of 6- and 12-week-old mice (F), ELISA; n = 5–6/group. The expression of OPN, matrix metalloproteinase-9 (MMP-9), and plasminogen activator inhibitor-1 (PAI-1) in the diaphragm of 6-week-old mice (G), Proteome Profiler™ array, representative results. Serum level of lactate dehydrogenase, LDH (H) and creatine kinase, CK (I) activity measured in 12-week-old mice; n = 6–8/group. The number of white blood cells (WBC) (J) and the percentage of lymphocytes (K), granulocytes (L), and monocytes (M); blood cell count, n = 12–14/group; Results presented as mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.005, **** p < 0.0001 by one-way ANOVA with Tukey’s post hoc test.
On the protein level, the lack of HO-1 in dystrophic animals was also confirmed, together with potent upregulation of its level in mdx vs. WT mice (Figure 1B). In dystrophic mice, we detected decreased muscle strength (Figure 1C) without any differences in body weight (Figure 1D) in comparison to WT counterparts. Both parameters were not affected by the lack of HO-1 (Figure 1C,D).
Further analysis confirmed the DMD phenotype, as the upregulated level of Spp1 encoding osteopontin (OPN), the fibrotic and inflammatory factor [24,25], was detected in the diaphragm of 6-week-old mdx mice and it was persistently elevated in 12-week-old animals (Figure 1E). The protein level of this DMD marker was upregulated in the serum of older (Figure 1F) dystrophic mice.
However, no differences were found in Spp1 expression in the diaphragm of 6- and 12-week-old mdx/Hmox1−/− animals in comparison to mdx mice (Figure 1E). In serum, its protein level was higher in mice lacking Hmox1 only in older animals (Figure 1F). A similar pattern was found in proteome profiler array analysis as OPN was one of the highly upregulated proteins in dystrophic animals (Figure 1G). Moreover, the level of factors regulating extracellular matrix remodeling, affecting muscle regeneration like plasminogen activator inhibitor-1 (PAI-1) and matrix metalloproteinase-9 (MMP-9) [26], was potently increased in the diaphragm of 6-week-old mdx mice (Figure 1G). Lack of Hmox1 accelerated the expression of the abovementioned factors on the protein level (Figure 1G) suggesting aggravated phenotype of the disease. In older animals, as in our previous work [19], HO-1 deficiency resulted in the elevated activity of lactate dehydrogenase (LDH) (Figure 1H) and creatine kinase (CK) (Figure 1I), the indicators of muscle damage, already increased in dystrophic animals. Furthermore, although the mice of the three genotypes did not exert any differences in the number of white blood cells (WBC) (Figure 1J), the percentage of lymphocytes among WBC was decreased in dystrophic animals and was further diminished in mdx/Hmox1−/− mice (Figure 1K). Oppositely, a higher percentage of granulocytes and monocytes was found in mdx mice and was even further increased in double knockouts (Figure 1L,M).
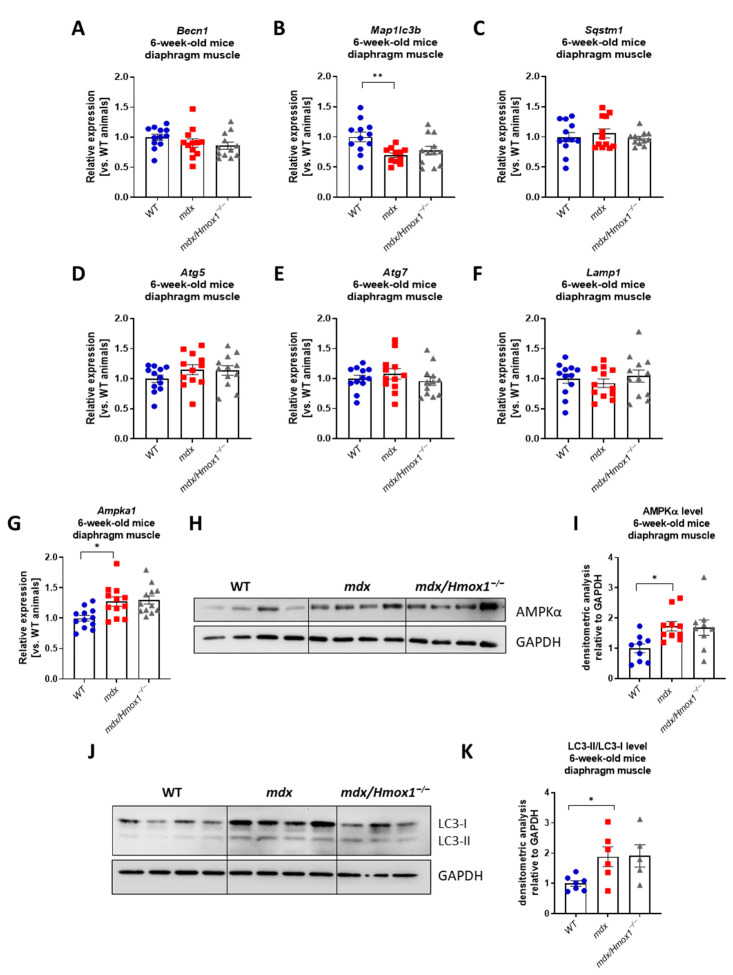
2.2. The Expression of Genes Modulating Autophagy Is Selectively Regulated in Gastrocnemius and the Diaphragm of 6-Week-Old Dystrophic Mice
To analyze the role of autophagy in DMD progression, the panel of six markers, namely beclin-1 (Becn1), microtubule-associated proteins 1A/1B light chain 3 (Map1lc3b), sequestosome 1 (Sqstm1), autophagy-related gene 5 (Atg5), autophagy-related gene 7 (Atg7), and lysosomal-associated membrane protein 1 (Lamp1) has been evaluated in two muscles. In gastrocnemius, the mRNA level of those factors was decreased in dystrophic animals (the tendency was observed for Atg5 and Lamp1) (Figure 2A–F). In the diaphragm, which is also potently affected by disease progression, no differences in the expression of genes regulating autophagy, except for the diminished Map1lc3b detected in dystrophic animals, were observed (Figure 3A–F). Interestingly, we did not see any differences between mdx and mdx/Hmox1−/− mice in both analyzed muscles (Figure 2A–F and Figure 3A–F). In the gastrocnemius muscle of dystrophic animals, the protein level of AMP-activated protein kinase (AMPKα), a regulator of the autophagy-mitophagy pathway [27], was not significantly affected (Figure 2G,H). On the other hand, the upregulation of the AMPKα mRNA and protein was evident in the diaphragm (Figure 3G–I).
Figure 2.
The expression of several genes regulating autophagy is decreased in gastrocnemius of 6-week-old mdx mice without the further influence of Hmox1 deficiency. Analysis of mRNA levels of factors regulating autophagy in gastrocnemius muscle of WT, mdx, and mdx/Hmox1−/− mice. The expression of Becn1 (A), Map1lc3b (B), Sqstm1 (C), Atg5 (D), Atg7 (E), and Lamp1 (F) was analyzed by qRT-PCR; n = 9–12/group; Results presented as mean ± SEM. The protein level of AMPKα presented by Western blot (G) and its densitometric analysis (H), n = 7–8/group. GAPDH was used as a loading control. Results presented as mean ± SEM. * p < 0.05 by one-way ANOVA with Tukey’s post hoc test.
Figure 3.
The expression of genes regulating autophagy is not changed by the additional lack of Hmox1 in the diaphragm of 6-week-old dystrophic mice. Analysis of mRNA levels of genes regulating autophagy in diaphragm muscle of WT, mdx, and mdx/Hmox1−/− animals. The expression of Becn1 (A), Map1lc3b (B), Sqstm1 (C), Atg5 (D), Atg7 (E), Lamp1 (F), and Ampka1 (G) was analyzed by qRT-PCR; n = 11–12/group. The protein level of AMPKα presented as a representative Western blot (H) and its densitometric analysis (I), n = 9–10/group. GAPDH was used as a loading control. Representative Western blot analysis of LC3-I/LC3-II protein (J) and densitometric analysis of LC3-II/LC3-I ratio (K), n = 5–7/group. GAPDH was used as a loading control. Results presented as mean ± SEM. * p < 0.05, ** p < 0.01 by one-way ANOVA with Tukey’s post hoc test.
However, the effect of HO-1 deficiency was observed neither in gastrocnemius nor in the diaphragm. Finally, although Map1lc3b expression was not changed, the protein levels of LC3s (encoded by this gene) were upregulated in the dystrophic diaphragm (Figure 3J). The ratio of LC3-II/LC3-I proteins was significantly increased in mdx mice in comparison to WT (Figure 3K) but the lack of HO-1 did not affect the level of the aforementioned proteins (Figure 3J–K).
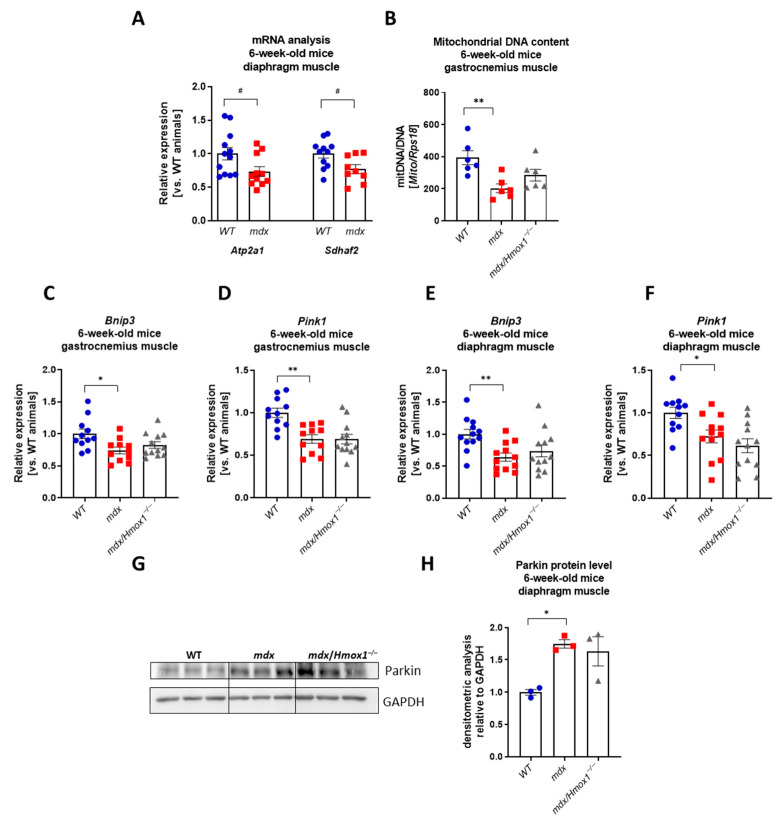
2.3. 6-Week-Old Mdx Mice Exhibit Mitochondrial Dysfunction and Decreased Expression of the Genes Regulating Mitophagy
Mitochondrial dysfunction is a pathological feature of DMD [28]. Knowing that mitochondria are a major source of cellular ATP involved in Ca2+ regulation and apoptotic signaling, we analyzed the expression of ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 (ATP2a1) regulating calcium metabolism and succinate dehydrogenase complex assembly factor 2 (Sdhaf2), encoding protein building mitochondrial complex II. A statistically significant decrease in their expression (Figure 4A) in the dystrophic diaphragm might indicate changes in mitochondrial signaling and homeostasis. Concomitantly, mitochondrial DNA content was decreased in mdx mice (Figure 4B). Therefore, we aimed to characterize mitophagy, the process responsible for the degradation of damaged or dysfunctional mitochondria [6]. The expression level of the Bcl2/adenovirus E1B 19 kDa protein-interacting protein 3 (Bnip3) was decreased in mdx mice both in the gastrocnemius and diaphragm (Figure 4C,E). However, no differences in the level of the Bnip3 transcript between mdx/Hmox1−/− and mdx individuals were found. Moreover, when PTEN-induced putative kinase 1 (Pink1) mRNA level was evaluated, we have found a very similar pattern of the expression. Lack of dystrophin resulted in a decreased level of Pink1 transcript in 6-week-old mdx mice, in both analyzed muscles. Nevertheless, HO-1 deficiency did not affect Pink1 expression in the gastrocnemius and diaphragm of 6-week-old mice (Figure 4D,F). Concomitantly with the previous findings for autophagy-related markers, the protein level of Parkin was induced in diaphragm (Figure 4G,H) of 6-week-old dystrophic animals, without any effect of HO-1 depletion. The opposite regulation of those mitophagy-related factors on mRNA and protein level together with decreased mitochondrial DNA content in dystrophic animals indicates the high complexity of this process and emphasizes the necessity of further experiments.
Figure 4.
Genes and proteins regulating mitochondrial homeostasis in 6-week-old mice. mRNA level of Atp2a1 and Sdhaf2 in 6-week-old WT and mdx mice analyzed by qRT-PCR (A); n = 9–12/group. Decreased mitochondrial DNA content in mdx mice (B), qRT-PCR, n = 6/group. Analysis of the mRNA level of Bnip3 and Pink1 genes regulating mitophagy in gastrocnemius (C,D) and diaphragm (E,F) muscles from WT, mdx, and mdx/Hmox1−/− genotypes, qRT-PCR; n = 11–12/group. Western blot analysis of Parkin protein (G) and densitometric analysis of Parkin level in diaphragm muscle (H), n = 3/group. GAPDH was used as a loading control. Results presented as mean ± SEM. # p < 0.05 with Student’s t-test * p < 0.05, ** p < 0.01 by one-way ANOVA with Tukey’s post hoc test.
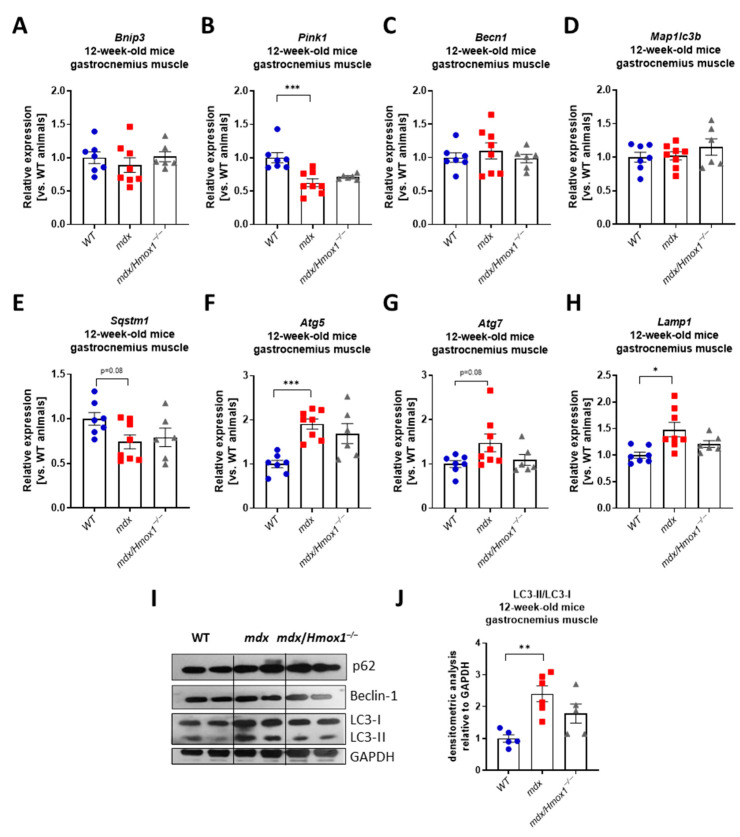
2.4. The Lack of HO-1 Does Not Affect Mitophagic and Autophagic Genes Expression in Older mdx Mice
To check if the lack of the effect of Hmox1 deficiency observed in our analysis might be the result of the relatively young age of the mice used in this study, we decided to perform a similar analysis in older, 12-week-old animals. We have concentrated on gastrocnemius, as we observed prominent changes in this muscle in 6-week-old mdx mice. Expression of Bnip3, Becn1, and Map1lc3b did not differ between WT and mdx animals whereas the expression of Pink1 and Sqstm1 was diminished in dystrophic muscle, similarly to 6-week-old mice (Figure 5A–E). Interestingly, an increase in the expression of Atg5, Atg7, and Lamp1 was found (Figure 5F–H). Although changes in p62 and Beclin-1 protein levels were rather moderate, a prominent rise in LC3-II/LC3-I proteins ratio was noticed when comparing WT and mdx mice (Figure 5I,J). Again, the lack of Hmox1 expression did not affect greatly the mRNA and protein level of analyzed mitophagy and autophagy markers (Figure 5A–J).
Figure 5.
Expression of genes regulating mitophagy and autophagy in 12-week-old WT, mdx, and mdx/Hmox1−/− mice. Analysis of mRNA level of genes regulating autophagy in gastrocnemius muscle. The expression of Bnip3 (A), Pink1 (B), Becn1 (C), Map1lc3b (D), Sqstm1 (E), Atg5 (F), Atg7 (G), and Lamp1 (H) was analyzed by qRT-PCR; n = 6–8/group. Western blot analysis of p62, Beclin-1, LC3-I, and LC3-II level (I) and densitometric analysis of LC3-II/LC3-I ratio (J), n = 5–6/group. GAPDH was used as a loading control. Results presented as mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.005 by one-way ANOVA with Tukey’s post hoc test.
3. Discussion
DMD, a disease resulting in progressive muscle weakness is caused by a mutation in the Dmd gene leading to the loss of dystrophin protein [23]. Although the cause of the disease has been known for a long time, no effective drug has been found yet. Therefore, numerous studies are still being conducted, mostly using the animal model of DMD—mdx mice. [29], which were also utilized in the present study. In our hands, mdx mice already at 6 weeks of age demonstrated changes in the expression of disease markers, like increased levels of OPN, MMP-9, and PAI-1, but more importantly, they exerted decreased muscle functionality as assessed by grip strength assay.
Literature data on autophagy disturbances in the mdx mice and DMD patients are inconclusive. Some of them suggest autophagic flux to be insufficient, and that reactivation of autophagy improves muscle function and reduces muscle damage [10]. Pauly et al. demonstrated that pharmacological activation of AMPK triggered autophagy in dystrophic mice, leading to the removal of defective mitochondria and improvement of diaphragm structure and function [9]. On the other hand, several studies have shown activation of the autophagy in dystrophic muscles of mdx animals as well as in individuals with DMD, but this process may be greatly affected by age and disease progression [17,30]. Nevertheless, to check if the disturbed expression of genes regulating mitophagy and autophagy may be further affected by the factors known to modulate DMD pathology, we concentrated on assessing the possible role of cytoprotective, anti-oxidant, and anti-inflammatory HO-1 in this regulation.
In our previous study, we observed that around 3-month-old double knockout animals lacking dystrophin and Hmox1 demonstrate impaired exercise capacity and aggravated DMD pathology in comparison to mdx animals [19]. Our results showed an increased level of both CK and LDH activity in 6- and 12-week-old mdx/Hmox1−/− animals, together with prominently alleviated inflammatory cell infiltration. However, when more in-depth analysis of M1-like and M2-like subpopulations of macrophages was performed, the lack of Hmox1 did not result in any significant changes. In the present study, we found an increase in the expression of some profibrotic markers and, similarly to our previous work, the level of tissue damage markers in serum of 12-week-old mice, such as LDH and CK activity. Moreover, a higher percentage of granulocytes and monocytes at the expense of lymphocytes within WBC was noticed already in 6-week-old mdx/Hmox1−/− in comparison to mdx animals. Interestingly, we have previously shown that similar changes could be observed in the 8-week-old dystrophic animals treated with the pharmacological inhibitor of HO-1 activity, SnPPIX [19]. Of note, in such relatively young animals, the diminished expression of both mitophagy and autophagy-regulating genes in the gastrocnemius muscle of mdx mice was evident. Interestingly, we did not find prominent changes in the expression of those genes in the diaphragm, except the downregulation of Map1lc3b, a ubiquitin-like modifier involved in the formation of autophagosomal vacuoles (autophagosomes), suggesting the muscle-specific effects. Moreover, Hmox1 deficiency did not affect the expression of the analyzed factors. Even in the older animals, no significant exacerbation of this phenomenon was observed in dystrophic mice deficient in Hmox1. Interestingly, when the autophagy regulators were assessed on the protein level, upregulation of AMPKα in the dystrophic diaphragm and LC3 proteins both in diaphragm and gastrocnemius muscle was evident
Several different molecular mechanisms of mitophagy have been described [31]. The most frequently depicted is based on the Pink1/Parkin interaction [32]. Kang et al. demonstrated the accumulation of damaged mitochondria in the myocardium of mdx mice caused by disturbances in mitophagy through decreased expression of Pink1 and Parkin [12], which stays in line with the results obtained in the present study in the gastrocnemius and diaphragm muscles of 6-week-old animals. However, the protein level of Parkin was higher in dystrophic tissues than in WT mice suggesting the posttranscriptional regulation. Moreover, in our study, we confirmed decreased mitochondrial DNA content in dystrophic animals [17], which could be correlated with higher Parkin protein level, as its overexpression was shown to induce mitochondrial removal from the cells [33]. Nevertheless, no effect of Hmox1 deletion on mitochondrial DNA content was noticed when compared to mdx mice.
Another component of mitophagy machinery is BNIP3, a member of a subfamily of death-inducing mitochondrial proteins, present on the outer mitochondrial membrane (OMM) and interacting with the autophagosome-localized LC3 protein [34]. Similar to our findings, De Palma et al. reported lower Bnip3 expression in the tibialis anterior muscle and diaphragm isolated from 4-month-old mdx mice [10] and Sebori et al. found reduced expression of this gene in the quadriceps of even older, 22-week-old dystrophic mice [11]. According to disturbances in mitophagy markers, we also confirmed, previously described in the literature [35], decreased levels of Atp2a1, encoding the SERCA1 pump responsible for Ca2+ removal from the cytosol into the lumen of the sarcoplasmic reticulum following muscular contraction. Dystrophic muscle with impaired Ca2+ handling and increased sarcoplasmic Ca2+ levels are prone to myofiber death through Ca2+-dependent protease activation and mitochondrial Ca2+ overload in dystrophic muscle. Moreover, a reduced level of Sdhaf2 indicating impaired tissue energetics and mitochondrial function was also found in our analysis of mdx mice.
We noted a decrease in the expression of Atg5, Atg7, Becn1, Map1lc3b, Lamp1, and Sqstm1 genes in the gastrocnemius muscle, but in the diaphragm of 6-week-old mice, only Map1lc3b was reduced. Similarly to our study, Panza et al. reported diminished expression of among others, Lamp1, Becn1, and Atg7, in quadriceps of 7-week-old animals [16] and the expression of Becn1, Map1lc3b, Lamp1, and Sqstm1 was decreased in the same muscle of 22-week-old mice [11]. No difference in the mRNA levels of the Map1lc3a, Map1lc3b, and Sqstm1 genes was found between WT and mdx mice hearts [8]. In our hands, in older, 12-week-old animals, the pattern of analyzed gene expression was complex with down-regulation of Pink1 and Sqstm1 and up-regulation of Atg5, Atg7, and Lamp1. Although the protein level of p62 and Beclin-1 (encoded by Sqstm1 and Becn1, respectively) was not greatly affected by the lack of the dystrophin, the upregulation in the LC3s (encoded by Map1lc3b) protein level and the ratio of LC3-II/LC3-I was noticed. Such discrepancies in mRNA and protein levels could be explained by the multi-stage regulation of this process, and they may indicate the complexity of autophagy regulation in DMD. For example, despite the upregulation of several autophagy-related proteins that could suggest induction of the self-degradation machinery, increased p62 protein is often correlated with disturbed autophagy [36,37]. The inconsistency of the published results regarding the status of autophagy/mitophagy in DMD may result also from additional factors, including the age of animals or type of analyzed muscles (which may greatly differ in the metabolic state). Some discrepancies may be also the cause of various genetic backgrounds of animals used in different studies (e.g., in this study, animals with a mixed genetic background C57BL/10ScSn/J × C57BL/6xFVB were used, whereas e.g., Sebori et al. [11] analyzed C57BL/10 (WT) and C57BL/10ScSn-Dmdmdx/J (mdx) strains. Moreover, it has been shown that mRNA levels may be increased when autophagy is impaired, and mRNA transcripts may be stable when the autophagic flux is active, proving that transcriptional regulation of autophagy does not always closely correlate with functional autophagic flux [38]. Furthermore, such discrepancies could be related to the used methodology, as a measurement of autophagic flux in tissue samples is challenging [38]. For example, a higher level of LC3-II might indicate enhanced autophagosome synthesis or, among others, reduced autophagosome turnover [39]. Caution during interpretation should be advised, especially as western blots show only snapshots in time. For proper interpretation of the data, usage of autophagy inductors or inhibitors should be applied to measure autophagy flux; however, its utilization in the animal models remains to be difficult and prone to misinterpretation. In conclusion, whether autophagy/mitophagy is beneficial or harmful to dystrophic skeletal muscle may depend on its size and the specific context in which it occurs.
The main aim of the performed study was to check if the disturbed expression of genes regulating mitophagy and autophagy may be affected by cytoprotective, anti-oxidant, and anti-inflammatory HO-1. Although we have previously found HO-1 deficiency as the condition accelerating inflammation, fibrosis, and muscle damage in mdx mice [19], in the present work we did not show any effect of Hmox1 absence on the mRNA and protein level of mitophagy/autophagy factors. Until now, no data regarding the possible role in the control of these pathways by HO-1 in DMD has been published; however, such effects in other disease models were published. Unuma et al. showed that induction of autophagy by HO-1 in rats treated with lipopolysaccharide (LPS) promotes the removal of damaged mitochondria in hepatocytes, thereby reducing oxidative stress and preventing sepsis-induced liver damage [40]. Moreover, Zhao et al. found that HO-1 counteracts oxidative stress and inflammation, and enhances autophagy in the myocardium to prevent its damage in diabetic cardiomyopathy [41]. In turn, mice lacking the Hmox1 gene in cardiomyocytes showed abnormalities related to the dysregulation of the Pink1/Parkin pathway [42]. Nevertheless, we cannot exclude the possibility, that such a strong effect of dystrophic phenotype on the autophagy and mitophagy homeostasis, observed by us and others [10], masks any influence of Hmox1 deletion in the mdx mouse model. It would be of interest to investigate, whether chemical upregulation of HO-1 and/or genetic Hmox1 overexpression, could affect dystrophy-related alterations in these processes.
Even though we did not confirm our hypothesis that “self-eating machinery” is impaired in Hmox1-lacking dystrophic animals, it cannot be excluded that such a situation might be present in DMD patients. HO-1 level and its activity vary in the human population, as a consequence of Hmox1 promoter polymorphism. Although not studied in DMD patients, it is well known that variations in the number of (GT)n repeats affect the cytoprotective, anti-inflammatory, and pro-angiogenic functions of HO-1 and may influence the progression of various diseases [43]. More studies in DMD individuals or human induced pluripotent stem cells, obtained from dystrophic boys with different HO-1 levels, and differentiated to, e.g., skeletal muscle cells or cardiomyocytes, are required to fully answer this question.
In summary, our experiments did not confirm the role of HO-1 in the regulation of the autophagy and mitophagy markers in DMD as postulated in other diseases. It might be suggested that lack of dystrophin disturbs autophagy to such an extent, which is not affected more by the additional lack of HO-1, which, nevertheless, aggravates other conditions in mdx animals. Although more detailed experiments are required, especially analyzing the autophagic flux in vivo and a more detailed examination of the mitochondrial structure and number, our study suggests disturbances in mitochondrial metabolism and mitophagy already in 6-week-old mdx mice. Those results may help to better understand the pathology of DMD and enable the development of effective therapy improving the length and quality of life of patients with this incurable disease.
4. Materials and Methods
4.1. Animal Models
The study was approved by the 2nd Institutional Animal Care and Use Committee (IACUC) in Kraków, Poland (approval number: 323/2018). Both mdx (C57BL/10ScSn-Dmdmdx/J) and control (C57BL/10ScSnJ) mice were purchased from the Jackson Laboratory whereas HO-1 knockout animals were generated by breeding of Hmox1−/+ mice at C57BL/6×FVB background originally gifted by Dr. Anupam Agarwal, Birmingham, USA. Crossing animals leading to the delivery of double knockouts on C57BL/10ScSn and C57BL/6xFVB mixed background (mice lacking dystrophin and HO-1, mdx/Hmox1−/−) was done as described previously [19]. Mice genotyping was performed by PCR method using DNA isolated by Genomic mini kit (A&A Biotechnology, Gdańsk, Poland) from mouse tails. Animals were housed in specific pathogen-free conditions with water and food available ad libitum. 6-week-old and 12-week-old male littermates or age-matched wild type (WT), mdx, and mdx/Hmox1−/− (all on C57BL/10ScSn and C57BL/6xFVB mixed background) mice were used for experiments.
4.2. Grip Strength Assay
Muscular strength was assessed in 6-week-old mice as previously described [44] using a triangular pull bar (Ugo Basile SRL, Gemonio (VA) Italy). Briefly, mice were allowed to grasp the grid using forelimbs and after moving toward the bar, they were pulled back horizontally until the grip was released. Results were calculated as the mean of five measurements (carried out with a 1-min break between measurements), normalized to body weight, and expressed as gf/g body weight (gf/g BW).
4.3. Total Blood Cell Count
Blood collected directly from vena cava to the EDTA-coated tubes was analyzed using scil Vet ABC™ Hematology Analyzer (HORIBA ABX, Warszawa, Poland). The number of white blood cells (WBC, 103/µL), as well as the percentage of lymphocytes, granulocytes, and monocytes, among WBC, was determined.
4.4. Analysis of Gene Expression by Real-Time Quantitative PCR (qRT-PCR)
Total RNA was isolated from the gastrocnemius and diaphragm muscles by using the modified Chomczynski method [45] with QIAzol (Qiagen, Hilden, Germany) as the lysis reagent. Tissues were homogenized using TissueLyser (Qiagen, Hilden, Germany) and then subjected to chloroform extraction and isopropanol precipitation. The quality and concentration of RNA were determined spectrophotometrically (NanoDrop, Thermo Fisher Scientific, Waltham, MA, USA). Moreover, 1 µg of RNA was reverse-transcribed into cDNA using RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA), and qRT-PCR reactions were performed using SYBR Green JumpStart Taq Ready Mix (Sigma-Aldrich, St. Louis, MO, USA) and specific primers (listed in Table 1) in StepOnePlus real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). Eef2 served as a housekeeping gene. For each sample, the mRNA levels of the gene of interest were normalized to Eef2 based on the comparative Ct method using the equation 2−ΔCt (ΔCt = Ct gene of interest − Ct Eef2) and presented as the relative expression in comparison to WT animals.
Table 1.
The sequences of primers used for the determination of gene expression on mRNA level by qRT-PCR.
| Gene | Full Gene Name | Primer Sequence |
|---|---|---|
| Ampka1 | AMP-activated protein kinase | F: 5′-TGTGACAAGCACATTTTCCAA-3′ |
| R: 5′-CCGATCTCTGTGGAGTAGCA-3′ | ||
| Atg5 | Autophagy-related gene 5 | F: 5′-CTGAAGATGGAGAGAAGAGG-3′ |
| R: 5′-GGGGACAATGCTAATATGAAG-3′ | ||
| Atg7 | Autophagy-related gene 7 | F: 5′-CTGTTCACCCAAAGTTCTTG-3′ |
| R: 5′-TCTAAGAAGGAATGTGAGGAG-3′ | ||
| Becn1 | Beclin-1 | F: 5′-CAATAATTTCAGACTGGGTCG-3′ |
| R: 5′-ATTTGTCTGTCAGAGACTCC-3′ | ||
| Bnip3 | Bcl2/adenovirus E1B-19 kDa interacting protein 3 |
F: 5′-ACCACAAGATACCAACAGAG-3′ |
| R: 5′-AATCTTCCTCAGACAGAGTG-3′ | ||
| Eef2 | Elongation factor-2 | F: 5′-AGAACATATTATTGCTGGCG-3′ |
| R: 5′-CAACAGGGTCAGATTTCTTG-3′ | ||
| Hmox1 | Heme oxygenase-1 | F: 5′-CCTCACTGGCAGGAAATCATC-3′ |
| R: 5′-CCTCGTGGAGACGCTTTACATA-3′ | ||
| Lamp1 | Lysosomal-associated membrane protein 1 | F: 5′-ATTGCAGTTTGGGATGAATG-3′ |
| R: 5′-TTGCACTTGTATGAGTTTCC-3′ | ||
| Map1lc3b | Microtubule-associated proteins 1A/1B light chain 3 |
F: 5′-GCTCATCAAGATAATCAGACG-3′ |
| R: 5′-GCATAAACCATGTACAGGAAG-3′ | ||
| Pink1 | PTEN-induced kinase 1 | F: 5′-ACTTACAGAAGATCCAGAGATG-3′ |
| R: 5′-CTTCATAACGAGGAACAGTG-3′ | ||
| Spp1 | Secreted phosphoprotein 1 | F: 5′-CCATCTCAGAAGCAGAATCTCCTT-3′ |
| R: 5′-GGTCATGGCTTTCATTGGAATT-3′ | ||
| Sqstm1 | Sequestosome 1 | F: 5′-AATGTGATCTGTGATGGTTG-3′ |
| R: 5′-GAGAGAAGCTATCAGAGAGG-3′ |
4.5. Mitochondrial DNA Content
DNA content measurement was performed by real-time quantitative PCR using SYBR Green JumpStart Taq Ready Mix (Sigma-Aldrich, St. Louis, MO, USA) and specific primers (listed in Table 2, based on [46]). Ribosomal protein S18 (Rps18) was used for normalization. DNA isolation from gastrocnemius muscle was performed using a Genomic mini kit (A&A Biotechnology, Gdańsk, Poland). Results are presented as the relative expression in comparison to WT animals.
Table 2.
The sequences of primers used for the determination of mitochondrial DNA content by qRT-PCR.
| Primer Sequence | |
|---|---|
|
Mito (mitochondrial) |
F: 5′-CTAGAAACCCCGAAACCAAA-3′ |
| R: 5′-CCAGCTATCACCAAGCTCGT-3′ | |
|
Rps18 (genomic) |
F: 5′-TGTGTTAGGGGACTGGTGGACA-3′ |
| R: 5′-CATCACCCACTTACCCCCAAAA-3′ |
4.6. Proteome Profiler Array
Diaphragm tissues were disrupted using a tissue homogenizer (TissueLyser, Qiagen, Hilden, Germany). The protein content was determined by bicinchoninic acid (BCA, Sigma-Aldrich, St. Louis, MO, USA) assay, and 100 µg of total protein was used for proteome profiling using Proteome Profiler™ array according to the manufacture’s protocol (R&D Systems, Minneapolis, MN, USA).
4.7. CK and LDH Activity Assessment
For the assays, blood was collected from the vena cava. Clear, non-hemolyzed serum was prepared by allowing the blood to clot at room temperature for 30 min and then centrifuged at 2000× g for 10 min at 4 °C. The activity of CK and LDH was measured in serum using the diagnostic Liquick Cor-CK and Liquick Cor-LDH kits, respectively, according to the vendor’s instruction (Cormay, Warsaw, Poland). The absorbance values obtained in 10 times diluted serum were then converted to LDH or CK activity (U/l) using the formula supplied with the kit.
4.8. ELISA Assay
OPN concentration in 750 times diluted serum of 6- and 12-week-old animals was determined by ELISA following the vendor’s protocol (R&D Systems, Minneapolis, MN, USA, cat no. MOST00). HO-1 content in gastrocnemius muscle of 6-week-old mice was analyzed using Mouse Heme Oxygenase 1 ELISA Kit (Abcam, Cambridge, UK, cat no. ab204524). The results measured as ng/mL are shown as the percentage of WT animals.
4.9. Western Blot
Fragments of muscles (gastrocnemius/diaphragm) were homogenized using an automatic TissueLyser (Qiagen, Hilden, Germany) in ice-cold PBS containing 1% Triton X-100 and protease inhibitors (1 μg/mL phenylmethylsulfonyl fluoride, 1 μg/mL leupeptin, and 1 μg/mL aprotinin). Total protein concentration was determined by the bicinchoninic acid protein assay kit (Sigma-Aldrich, St. Louis, MO, USA), according to the vendor’s protocol. Protein lysates were subjected to western blot as described previously [47]. The primary antibodies used were rabbit anti-LC3-I/LC3-II, rabbit anti-p62, rabbit anti-Beclin-1, mouse anti-Parkin, rabbit anti-AMPKα, and mouse anti-GAPDH (listed in Table 3). Appropriate peroxidase-conjugated secondary antibodies (goat anti-mouse 1:20000; BD Pharmingen, San Diego, CA, USA and goat anti-rabbit, 1:5000, Cell Signaling Technology, Danvers, MA, USA) were used to detect the proteins of interest by chemiluminescence (Immobilon Western Chemiluminescent HRP Substrate, Millipore, Burlington, MA, USA).
Table 3.
List of primary antibodies used for western blot.
| Antibody | Company | Catalog Number | Dilution |
|---|---|---|---|
| AMPKα | Cell Signaling Technology Danvers, MA, USA |
2603 | 1:500 |
| Beclin-1 | Cell Signaling Technology Danvers, MA, USA |
3495 | 1:500 |
| GAPDH | Santa Cruz Biotechnology Dallas TX, USA |
sc-59540 | 1:1000 |
| LC3-I/II | Cell Signaling Technology Danvers, MA, USA |
12741 | 1:500 |
| Parkin | Cell Signaling Technology Danvers, MA, USA |
4211 | 1:500 |
| SQSTM1/p62 | Cell Signaling Technology Danvers, MA, USA |
39749 | 1:500 |
4.10. Statistical Analyses
Data are presented as mean ± SEM. Differences between groups were tested for statistical significance using the one-way ANOVA followed by Tukey’s post hoc test when more than two groups were analyzed or the unpaired 2-tailed Student’s t-test for the comparison of two groups (Figure 4A) p < 0.05 was considered significant. The outliers were identified based on Grubb’s test.
Acknowledgments
We are grateful to Ewa Werner, Karolina Hajduk, Łukasz Szczepanik, and Janusz Drebot from the Animal Facility of the Faculty of Biochemistry, Biophysics, and Biotechnology for the mice care and breeding. We would like to thank the administrative staff of the Department of Medical Biotechnology (Agnieszka Andrychowicz-Róg, Joanna Uchto-Bajołek, and Joanna Strzęp-Knapiak) for their assistance.
Author Contributions
Conceptualization, J.D. and A.Ł.; formal analysis, A.Ł.; funding acquisition, A.Ł.; investigation, O.M., K.K., P.P., K.R. and A.Ł.; methodology, O.M., K.K., P.P., K.R. and A.Ł.; project administration, A.Ł.; supervision, A.Ł.; writing—original draft, O.M. and A.Ł.; writing—review and editing, O.M., K.K., P.P., K.R, J.D. and A.Ł. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants 2016/21/B/NZ1/00293 and 2019/35/B/NZ3/02817 (to A.Ł.) from the National Science Centre.
Institutional Review Board Statement
All animal experiments were performed in accordance with national and European legislation, after approval by the 2nd Institutional Animal Care and Use Committee (IACUC) in Kraków, Poland.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. Autophagy Is Required to Maintain Muscle Mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Neel B.A., Lin Y., Pessin J.E. Skeletal muscle autophagy: A new metabolic regulator. Trends Endocrinol. Metab. 2013;24:635–643. doi: 10.1016/j.tem.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maiuri M.C., Zalckvar E., Kimchi A., Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 4.Katayama M., Kawaguchi T., Berger M.S., Pieper R.O. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–558. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 5.Sandri M., Coletto L., Grumati P., Bonaldo P. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J. Cell Sci. 2013;126:5325–5333. doi: 10.1242/jcs.114041. [DOI] [PubMed] [Google Scholar]
- 6.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandri M. New findings of lysosomal proteolysis in skeletal muscle. Curr. Opin. Clin. Nutr. Metab. Care. 2011;14:223–229. doi: 10.1097/MCO.0b013e3283457a75. [DOI] [PubMed] [Google Scholar]
- 8.Kuno A., Hosoda R., Sebori R., Hayashi T., Sakuragi H., Tanabe M., Horio Y. Resveratrol Ameliorates Mitophagy Disturbance and Improves Cardiac Pathophysiology of Dystrophin-deficient mdx Mice. Sci. Rep. 2018;8:15555. doi: 10.1038/s41598-018-33930-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pauly M., Daussin F., Burelle Y., Li T., Godin R., Fauconnier J., Koechlin-Ramonatxo C., Hugon G., Lacampagne A., Coisy-Quivy M., et al. AMPK Activation Stimulates Autophagy and Ameliorates Muscular Dystrophy in the mdx Mouse Diaphragm. Am. J. Pathol. 2012;181:583–592. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 10.De Palma C., Morisi F., Cheli S., Pambianco S., Cappello V., Vezzoli M., Rovere-Querini P., Moggio M., Ripolone M., Francolini M., et al. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis. 2012;3:e418. doi: 10.1038/cddis.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sebori R., Kuno A., Hosoda R., Hayashi T., Horio Y. Resveratrol Decreases Oxidative Stress by Restoring Mitophagy and Improves the Pathophysiology of Dystrophin-Deficient mdx Mice. Oxidative Med. Cell. Longev. 2018;2018:9179270. doi: 10.1155/2018/9179270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang C., Badr M.A., Kyrychenko V., Eskelinen E.-L., Shirokova N. Deficit in PINK1/PARKIN-mediated mitochondrial autophagy at late stages of dystrophic cardiomyopathy. Cardiovasc. Res. 2018;114:90–102. doi: 10.1093/cvr/cvx201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Łoboda A., Dulak J. Muscle and cardiac therapeutic strategies for Duchenne muscular dystrophy: Past, present, and future. Pharm. Rep. 2020;72:1227–1263. doi: 10.1007/s43440-020-00134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehead N.P., Kim M.J., Bible K.L., Adams M.E., Froehner S.C. A new therapeutic effect of simvastatin revealed by functional improvement in muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2015;112:12864–12869. doi: 10.1073/pnas.1509536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitehead N.P. Enhanced autophagy as a potential mechanism for the improved physiological function by simvastatin in muscular dystrophy. Autophagy. 2016;12:705–706. doi: 10.1080/15548627.2016.1144005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panza E., Vellecco V., Iannotti F.A., Paris D., Manzo O.L., Smimmo M., Mitilini N., Boscaino A., de Dominicis G., Bucci M., et al. Duchenne’s muscular dystrophy involves a defective transsulfuration pathway activity. Redox Biol. 2021;45:102040. doi: 10.1016/j.redox.2021.102040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore T.M., Lin A.J., Strumwasser A.R., Cory K., Whitney K., Ho T., Ho T., Lee J.L., Rucker D.H., Nguyen C.Q., et al. Mitochondrial Dysfunction Is an Early Consequence of Partial or Complete Dystrophin Loss in mdx Mice. Front. Physiol. 2020;11:690. doi: 10.3389/fphys.2020.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozakowska M., Pietraszek-Gremplewicz K., Jozkowicz A., Dulak J. The role of oxidative stress in skeletal muscle injury and regeneration: Focus on antioxidant enzymes. J. Muscle Res. Cell Motil. 2015;36:377–393. doi: 10.1007/s10974-015-9438-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietraszek-Gremplewicz K., Kozakowska M., Bronisz-Budzynska I., Ciesla M., Mucha O., Podkalicka P., Madej M., Glowniak U., Szade K., Stępniewski J., et al. Heme Oxygenase-1 Influences Satellite Cells and Progression of Duchenne Muscular Dystrophy in Mice. Antioxid. Redox Signal. 2018;29:128–148. doi: 10.1089/ars.2017.7435. [DOI] [PubMed] [Google Scholar]
- 20.Vasconcellos L.R., Siqueira M.S., Moraes R., Carneiro L.A., Bozza M.T., Travassos L.H. Heme Oxygenase-1 and Autophagy Linked for Cytoprotection. CPD. 2018;24:2311–2316. doi: 10.2174/1381612824666180727100909. [DOI] [PubMed] [Google Scholar]
- 21.Ryter S.W. Heme Oxgenase-1, a Cardinal Modulator of Regulated Cell Death and Inflammation. Cells. 2021;10:515. doi: 10.3390/cells10030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sicinski P., Geng Y., Ryder-Cook A.S., Barnard E.A., Darlison M.G., Barnard P.J. The molecular basis of muscular dystrophy in the mdx mouse: A point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 23.McGreevy J.W., Hakim C.H., McIntosh M.A., Duan D. Animal models of Duchenne muscular dystrophy: From basic mechanisms to gene therapy. Dis. Models Mech. 2015;8:195–213. doi: 10.1242/dmm.018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vetrone S.A., Montecino-Rodriguez E., Kudryashova E., Kramerova I., Hoffman E.P., Liu S.D., Miceli M.C., Spencer M.J. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J. Clin. Investig. 2009;119:1583–1594. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagel C.N., Wasgewatte Wijesinghe D.K., Taghavi Esfandouni N., Mackie E.J. Osteopontin, inflammation and myogenesis: Influencing regeneration, fibrosis and size of skeletal muscle. J. Cell Commun. Signal. 2014;8:95–103. doi: 10.1007/s12079-013-0217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman F.A., Krause M.P. PAI-1, the Plasminogen System, and Skeletal Muscle. Int. J. Mol. Sci. 2020;21:7066. doi: 10.3390/ijms21197066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia J., Bissa B., Brecht L., Allers L., Choi S.W., Gu Y., Zbinden M., Burge M.R., Timmins G., Hallows K., et al. AMPK, a key regulator of metabolism and autophagy, is activated by lysosomal damage via a novel galectin-directed ubiquitin signal transduction system. Mol. Cell. 2020;77:951–969.e9. doi: 10.1016/j.molcel.2019.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong A.V., Sanson M., Richard I., Israeli D. A revised model for mitochondrial dysfunction in Duchenne muscular dystrophy. Eur. J. Transl. Myol. 2021;31:10012. doi: 10.4081/ejtm.2021.10012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manning J., O’Malley D. What has the mdx mouse model of duchenne muscular dystrophy contributed to our understanding of this disease? J. Muscle Res. Cell Motil. 2015;36:155–167. doi: 10.1007/s10974-015-9406-4. [DOI] [PubMed] [Google Scholar]
- 30.Fiacco E., Castagnetti F., Bianconi V., Madaro L., De Bardi M., Nazio F., D’Amico A., Bertini E., Cecconi F., Puri P.L., et al. Autophagy regulates satellite cell ability to regenerate normal and dystrophic muscles. Cell Death Differ. 2016;23:1839–1849. doi: 10.1038/cdd.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen G., Kroemer G., Kepp O. Mitophagy: An Emerging Role in Aging and Age-Associated Diseases. Front. Cell Dev. Biol. 2020;8:200. doi: 10.3389/fcell.2020.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin S.M., Youle R.J. PINK1- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 2012;125:795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narendra D., Tanaka A., Suen D.-F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anding A.L., Baehrecke E.H. Cleaning House: Selective Autophagy of Organelles. Dev. Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider J.S., Shanmugam M., Gonzalez J.P., Lopez H., Gordan R., Fraidenraich D., Babu G.J. Increased sarcolipin expression and decreased sarco(endo)plasmic reticulum Ca2+ uptake in skeletal muscles of mouse models of Duchenne muscular dystrophy. J. Muscle Res. Cell Motil. 2013;34:349–356. doi: 10.1007/s10974-013-9350-0. [DOI] [PubMed] [Google Scholar]
- 36.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottlieb R.A., Andres A.M., Sin J., Taylor D.P.J. Untangling Autophagy Measurements. Circ. Res. 2015;116:504–514. doi: 10.1161/CIRCRESAHA.116.303787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murugan S., Amaravadi R.K. Methods for Studying Autophagy Within the Tumor Microenvironment. Adv. Exp. Med. Biol. 2016;899:145–166. doi: 10.1007/978-3-319-26666-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unuma K., Aki T., Matsuda S., Funakoshi T., Yoshida K., Uemura K. Inducer of heme oxygenase-1 cobalt protoporphyrin accelerates autophagy and suppresses oxidative damages during lipopolysaccharide treatment in rat liver: Autophagy and suppression of liver damage. Hepatol. Res. 2013;43:91–96. doi: 10.1111/j.1872-034X.2012.01049.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y., Zhang L., Qiao Y., Zhou X., Wu G., Wang L., Peng Y., Dong X., Huang H., Si L., et al. Heme Oxygenase-1 Prevents Cardiac Dysfunction in Streptozotocin-Diabetic Mice by Reducing Inflammation, Oxidative Stress, Apoptosis and Enhancing Autophagy. PLoS ONE. 2013;8:e75927. doi: 10.1371/journal.pone.0075927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suliman H.B., Keenan J.E., Piantadosi C.A. Mitochondrial quality-control dysregulation in conditional HO-1–/– mice. JCI Insight. 2017;2 doi: 10.1172/jci.insight.89676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loboda A., Jazwa A., Grochot-Przeczek A., Rutkowski A.J., Cisowski J., Agarwal A., Jozkowicz A., Dulak J. Heme oxygenase-1 and the vascular bed: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2008;10:1767–1812. doi: 10.1089/ars.2008.2043. [DOI] [PubMed] [Google Scholar]
- 44.Bronisz-Budzyńska I., Kozakowska M., Podkalicka P., Kachamakova-Trojanowska N., Łoboda A., Dulak J. The role of Nrf2 in acute and chronic muscle injury. Skelet. Muscle. 2020;10:35. doi: 10.1186/s13395-020-00255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 46.Malik A.N., Czajka A., Cunningham P. Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion. 2016;29:59–64. doi: 10.1016/j.mito.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Podkalicka P., Mucha O., Bronisz-Budzyńska I., Kozakowska M., Pietraszek-Gremplewicz K., Cetnarowska A., Głowniak-Kwitek U., Bukowska-Strakova K., Cieśla M., Kulecka M., et al. Lack of miR-378 attenuates muscular dystrophy in mdx mice. JCI Insight. 2020;5 doi: 10.1172/jci.insight.135576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.