Abstract
The usefulness of amplified fragment length polymorphism (AFLP) analysis was evaluated for the discrimination of Mycobacterium bovis (17 strains), M. tuberculosis (15 strains), and M. ulcerans (12 strains) at the inter- and intraspecific level. The AFLP technique is a whole-genome coverage genotypic fingerprinting method based on the selective PCR amplification of modified restriction fragments obtained through a double enzymatic digest and subsequent ligation of double-stranded restriction site-specific adapter oligonucleotides. Selective amplification of ApaI/TaqI templates with primer combination A02-T02 (both having an additional C at their 3′ end) generated autoradiographic AFLP fingerprints that were grouped by numerical analysis in two main AFLP clusters allowing clear separation of M. ulcerans (cluster I) from the M. tuberculosis complex members M. bovis and M. tuberculosis (cluster II). Calculation of similarities using the band-based Dice correlation coefficient instead of the Pearson product-moment correlation coefficient revealed a further subgrouping in cluster II. The two resulting subclusters corresponded with the phenotypic identity of M. bovis and M. tuberculosis, respectively, and could also be visually identified by two AFLP marker bands. Because of the relatively low degree of genotypic variation among the AFLP band patterns of the latter two taxa, no correlation could be found with previously reported molecular typing data or with geographical origin. The use of primer combination A02-T01 (the latter having an A as selective base) did not increase the resolving power within the M. tuberculosis complex but resulted in a visual subgrouping of the M. ulcerans strains that was not observed with primer combination A02-T02. Based on the presence or absence of a single AFLP marker band, the M. ulcerans isolates could be unambiguously classified in two continental types corresponding with the African and Australian origin of the strains, respectively. In conclusion, the radioactive AFLP method proved to be a reproducible and reliable taxonomic tool for the differentiation of the three mycobacterial species under study and also demonstrated its potential use for typing of M. ulcerans strains when employing multiple primer combinations.
Despite the constant evolution and innovation in the field of bacterial fingerprinting and DNA-based diagnostics, the molecular identification and typing of mycobacteria associated with human and animal diseases is still frequently hampered by a lack of resolution at the inter- or intraspecific level. A most striking example of these problems is found with the epidemiology of the Mycobacterium tuberculosis complex, including M. tuberculosis, M. bovis, M. bovis BCG, M. africanum, and M. microti. The first two species are widely recognized as the causal agents of human and bovine tuberculosis, respectively, but M. bovis infections have also been documented in humans due to the wide host range of this organism (25). The five members of the M. tuberculosis complex were originally grouped together on the basis of their phenotypic similarities (41), and the very high levels of DNA relatedness (85 to 100%) reported by Imaeda (13) even indicated that the four species should be placed in one single species using the general taxonomic criteria of Wayne and coauthors (40). Sequencing of the 16S rRNA gene (21) and the 16S-23S internal transcribed spacer (9) confirmed that the M. tuberculosis complex is a historical concept representing four taxa that should be separated at a subspecific or infrasubspecific level (41). Recently, the taxonomic situation within this complex became even more complicated with the addition of “M. canettii” (36) and the description of M. bovis subtypes bovis and caprae (24).
Del Portillo and coworkers (6) reported that PCR amplification of the 396-bp mtp40 fragment allowed differentiation between M. tuberculosis and M. bovis, but the universal applicability of this diagnostic marker was later questioned by Weil and coauthors (42). So far, spoligotyping (spacer oligotyping) based on DNA polymorphisms within the direct repeat locus of M. tuberculosis is one of the very few techniques that can distinguish among this species and M. bovis (3, 20). Initially, this technique was developed for strain typing of low-IS6110-copy-number isolates belonging to the M. tuberculosis complex. Restriction fragment length polymorphism (RFLP) analysis using the insertion element IS6110 as a probe is currently the most widely used method for strain differentiation within M. tuberculosis (32, 34). However, it has been shown that IS6110 RFLP can generate aberrant results with M. tuberculosis strains harboring fewer than five IS6110 copies (35) and is of limited value for typing of M. bovis as members of this taxon often only possess one IS6110 copy (5).
Next to members of the M. tuberculosis complex, the slowly growing mycobacterial species M. ulcerans is also becoming increasingly important as an emerging human pathogen, causing necrotizing skin ulceration, also referred to as Buruli ulcer (2). During the past decade, an increasing incidence of Buruli ulcer with a poorly understood epidemiology has been reported in West Africa (1, 18, 23) and Australia (8). So far, the number of molecular techniques available for typing of M. ulcerans strains is very limited (18). Jackson and coworkers (14) used an RFLP-based method using plasmid pTBN12 as a probe for typing of African and Australian M. ulcerans isolates. Similarly, the variability in the 3′ end of the 16S rRNA sequences has been used as a molecular marker for the geographical origin of M. ulcerans isolates from different continents (27).
In the majority of the above-mentioned typing methods, only a very limited part of the mycobacterial genome is covered through highly specific molecular targetting of one or more repetitive DNA elements. DNA fingerprinting techniques with whole-genome coverage, on the other hand, have not been widely used for typing within the M. tuberculosis complex or for strain differentiation of M. ulcerans. In this context, it has been assumed that the low degree of genetic polymorphism in these organisms may restrict the expected discriminative power of such generic techniques (22, 30). However, pulsed-field gel electrophoresis (PFGE) of macro-restriction fragments has been successfully applied in genomic and epidemiologic studies on M. tuberculosis and M. bovis (7, 29). Recently, a preliminary evaluation of restriction fragment length end labeling analysis demonstrated only limited use of this method for unraveling the genetic diversity within the M. tuberculosis complex (23).
Next to PFGE, whole-genome fingerprinting using the amplified fragment length polymorphism (AFLP) technique is one of the promising methods for typing of bacterial pathogens. Essentially, the AFLP method is based on (i) digestion of whole-genomic DNA with two endonucleases, (ii) ligation of double-stranded oligonucleotide adapters to the restriction halfsites, and (iii) selective amplification of the modified restriction fragments with adapter-specific primers that have an extension of one or more bases at their 3′ end (15). For the purpose of bacterial strain typing, major applications of AFLP have been reported within the genera Aeromonas, Acinetobacter, Campylobacter, Legionella, Staphylococcus, Streptococcus, Legionella (for a review, see reference 28), and recently, for Vibrio cholerae (17). In addition, AFLP proved to be a highly useful taxonomic tool for the differentiation of genomic groups in Aeromonas (11), Acinetobacter (16), and Burkholderia (4).
Recently, Goulding and coworkers (10) determined the value of fluorescent AFLP for genetic analysis of M. tuberculosis and concluded that their methodology can be used in conjunction with IS6110 RFLP typing to further unravel the epidemiology and evolution of this species. The present study describes the use of the conventional AFLP method using radioactive labeling and was undertaken to assess the discriminatory power of this technique (i) for the differentiation of M. bovis and M. tuberculosis at the species and the strain level, and (ii) for the geographical subgrouping and typing of M. ulcerans isolates.
MATERIALS AND METHODS
Strains used.
The present study comprised a total of 44 strains encompassing the species M. bovis (n = 17), M. tuberculosis (n = 15), and M. ulcerans (n = 12). All strains were part of the research collection of the Institute of Tropical Medicine. M. tuberculosis and M. ulcerans isolates were cultured on Löwenstein-Jensen medium for 3 to 5 weeks at 37 and 33°C, respectively. M. bovis isolates were cultured on Stonebrink medium (31) for 3 weeks at 37°C. Phenotypic identification was performed according to the minimal standards defined by Vincent Lévy-Frébault and Portaels (38).
DNA extraction.
Whole-genomic DNA was prepared using the standardized extraction method described by van Embden and coworkers (34). Briefly, bacteria were harvested into Tris-EDTA buffer (pH 8.0) and digested with lysozyme (1 mg/ml). After the addition of proteinase K (0.1 mg/ml) and sodium dodecyl sulfate (to 1%), tubes were incubated with NaCl (0.6 M) and N-acetyl-N,N,N-trimethyl ammonium bromide. Subsequent to extraction with chloroform-isoamyl alcohol, DNA was precipitated with isopropanol.
Preparation of AFLP templates.
AFLP templates were prepared from ∼1 μg of high-molecular-weight genomic DNA through double enzymatic digestion using the endonucleases ApaI and TaqI subsequently followed by restriction halfsite-specific ligation of double-stranded oligonucleotide adapters and selective precipitation according to the method of Janssen et al. (15). The adapters were prepared by mixing equimolar amounts of the partially complementary oligonucleotides 5′-TCGTAGACTGCGTACAGGCC-3′ and 5′-TGTACGCAGTCTAC-3′ (for ApaI) and 5′-GACGATGAGTCCTGAC-3′ and 5′-CGGTCAGGACTCAT-3′ (for TaqI).
AFLP reactions.
For AFLP fingerprinting, ApaI-TaqI restriction fragments tagged with specific adapters were used as template DNA for selective PCR amplification directed by the primers A02 (5′-GACTGCGTACAGGCCCC-3′) and T02 (5′-CGATGAGTCCTGACCGAC-3′) or T01 (5′-CGATGAGTCCTGACCGAA-3′) (selective bases at the 3′ end of the primers are underlined). Primer A02 was labeled at its 5′ end in a T4 kinase (Pharmacia Biotech, Uppsala, Sweden) assay using 32P-labeled ATP (Amersham International, Little Chalfont, Buckinghamshire, England) as described by Vos et al. (39). PCR amplifications were performed in a Perkin-Elmer 9600 thermal cycler according to the protocol of Vos et al. (39) with modifications of Janssen et al. (15) using the following cycle profile: (i) 13 cycles of denaturation at 94°C for 60 s, annealing using a decreasing stringency rate at [65 − (n − 0.7)]°C for 30 s, where n is the cycle number, and extension at 72°C for 60 s and (ii) 12 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 60 s.
Electrophoresis and visualization of PCR products.
Radioactive AFLP reactions were separated electrophoretically in a denaturing 5% polyacrylamide–8.3 M urea matrix (SequaGel; National Diagnostics) using TBE as electrophoresis buffer (15). A standard reference, i.e., AFLP template of M. tuberculosis strain H37Ra (ITM 8004), was included every fifth to sixth lane on each gel. Gels were vacuum dried on a gel dryer (model 583; Bio-Rad) for 55 min at 80°C. AFLP fingerprints were visualized autoradiographically by exposure of the dried gel to Hyperfilm-MP (Amersham International) for 5 to 24 h depending on the amount of radiation. Following exposure, films were developed manually in staining baths using Metinol and Acidofix solutions (Agfa Gevaert, Leverkusen, Germany).
Data processing.
Autoradiograms generated from radioactive AFLP fingerprinting were scanned using a high-resolution densitometric scanner (RayVen RSU1; X-Ray Scanner Cooperation). Transmission image data were stored in TIFF files and further processed by GelCompar software (version 4.2; Applied Maths, Kortrijk, Belgium). Following conversion, normalization using strain ITM 8004 as standard reference, and background subtraction, similarities between AFLP fingerprints were calculated using the Pearson product-moment correlation coefficient or the band-based Dice coefficient (37). Cluster analysis was performed by the unweighted pair-group method using arithmetic averages (37).
RESULTS AND DISCUSSION
The current investigation was initiated to evaluate the potential of the whole-genome coverage DNA fingerprinting technique AFLP to discriminate among strains of M. tuberculosis, M. bovis, and M. ulcerans at the inter- and intraspecific level. In the course of a taxonomic study on M. kansasii subspecies, Picardeau and coworkers (26) previously reported the use of a simplified version of the original AFLP protocol. For this purpose, the authors employed only one restriction enzyme instead of a double restriction digest and used an agarose gel instead of a highly resolving polyacrylamide matrix for electrophoresis. From a conceptual point of view, however, it can be argued that these two modifications may significantly reduce the resolving power of the AFLP technique. Typically, the AFLP patterns obtained in the current study with Mycobacterium genomes comprised 30 to 50 bands (Fig. 1 and 2), whereas the simplified method used by Picardeau et al. (26) generally yielded three to eight bands. In this regard, the latter authors concluded that AFLP was very suitable as a rapid prescreening technique, but recommended combined use with PFGE analysis for high-level strain characterization.
FIG. 1.
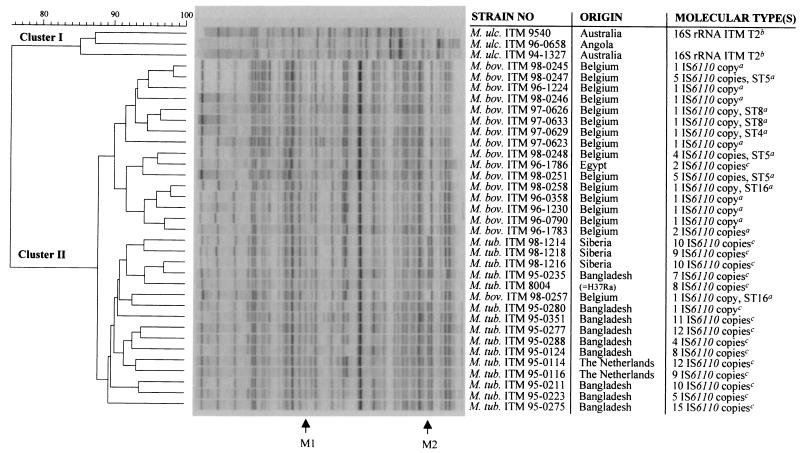
Numerical analysis of normalized AFLP band patterns generated from M. bovis (M. bov.) (n = 17), M. tuberculosis (M. tub.) (n = 15), and a subset of M. ulcerans (M. ulc.) strains (n = 3) using primer combination A02-T02. The dendrogram was constructed using the unweighted pair-group method using arithmetic averages with correlation levels expressed as percentage values of the band-based Dice coefficient. M1 and M2 denote species-specific AFLP marker bands differentiating M. tuberculosis from M. bovis. Footnotes: a, data from Rigouts et al. (L. Rigouts, M. Desmecht, J. Dufey, C. Saegerman, K. Kremer, H. Traore, D. van Soolingen, K. Walravens, J. Godfroid, and F. Portaels, submitted for publication); b, data from Portaels et al. (27); c, data not published (all these isolates showed different IS6110-RFLP profiles).
FIG. 2.
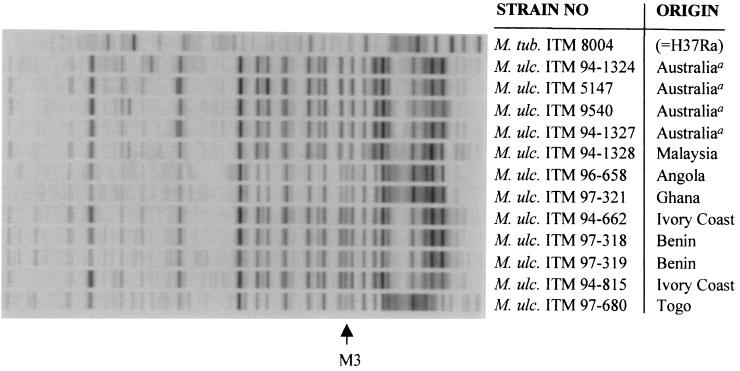
Normalized AFLP band patterns generated from 12 M. ulcerans strains and one M. tuberculosis reference strain using primer combination A02-T01. M3 denotes a continent-specific AFLP marker band.
In this study, the enzymes ApaI [GGGC(C/C)] and TaqI [(T/C)GA] were chosen for AFLP template preparation based on the theoretical assumption that both enzymes display relatively high cleavage frequencies in genomes of high G+C-content organisms such as Mycobacterium (G+C content, 68 to 71%). In our hands, the reproducibility of the AFLP method was good throughout the entire investigation and consistent with previously reported values (11, 15). Typically, normalized reference lanes displayed at least 93% relatedness between gels using the Pearson product-moment correlation coefficient. Using the primer combination A02-T02, both having one C extension at their respective 3′ ends, visual inspection as well as clustering analysis using the Pearson product-moment correlation coefficient revealed two main AFLP clusters in which M. ulcerans (AFLP cluster I) was maximally separated from M. tuberculosis and M. bovis both residing in AFLP cluster II (data not shown). A recent DNA-DNA hybridization study by Tønjum and coworkers (33) revealed that M. ulcerans and M. tuberculosis strains displayed up to 38% DNA homology. According to the authors, this relatively high relatedness was not known before and may offer new perspectives for the development of pathogenesis models (33). In the present study, numerical analysis of AFLP profiles showed that the M. ulcerans cluster was joined with the M. tuberculosis-M. bovis cluster at a Pearson correlation level of only 4%, indicating a relatively low genetic relationship between both taxa.
Our numerical data also showed that AFLP cluster II exhibited a further division in which M. tuberculosis and M. bovis were heterogeneously distributed among two subclusters (data not shown). Interestingly, visual inspection of the normalized band patterns revealed two species-specific AFLP marker bands (i.e., M1 and M2) allowing rapid classification of a given M. tuberculosis complex strain into M. tuberculosis or M. bovis. In fact, this visually observed taxonomic subgrouping was strongly supported when employing the band-based Dice coefficient instead of the Pearson coefficient for the calculation of interstrain correlations (Fig. 1). Because it compares entire densitometric curves, the Pearson coefficient has become well established as the standard coefficient of choice when comparing complex AFLP profiles for taxonomic purposes (11, 12, 28). However, this coefficient is sensitive to differences in background and, to a lesser extent, to variations in relative band intensities. The presence-absence algorithm of the Dice coefficient, on the other hand, is not influenced by fluctuations in intensity and assigned more statistical weight to the AFLP marker bands M1 and M2, thereby allowing a much better taxonomic differentiation compared to the Pearson coefficient. In contrast to AFLP marker band M1, which was clearly present in all M. tuberculosis strains but also gave a very weak signal in M. bovis, AFLP marker band M2 yielded an intense highly specific fragment for M. tuberculosis not found in M. bovis (Fig. 1). This resulted in two subclusters within AFLP cluster II with perfect grouping according to phenotypic species identity except for the position of strain ITM 98-0257. Nevertheless, visual inspection of the digitized AFLP fingerprints would have correctly classified this isolate into M. bovis based on the absence of marker band M2. The dendrogram shown in Fig. 1 comprises normalized AFLP patterns selected from three autoradiograms obtained after three separate AFLP experiments and includes M. tuberculosis isolates from various geographical origins, including Bangladesh, The Netherlands, and Siberia (Fig. 1). Considering these variations, it can be concluded that AFLP marker band M2 remains stable during independent AFLP analyses and is not subjected to strain-to-strain polymorphisms.
The high genomic relatedness between M. bovis and M. tuberculosis as previously observed by DNA-DNA hybridization studies (13) was also clearly reflected by numerical analysis of AFLP fingerprints, as both subclusters in AFLP cluster II were joined at a correlation level as high as 73% using the Pearson coefficient (data not shown) and 87.5% with the Dice coefficient (Fig. 1). In general, genomic species delineated by DNA-DNA hybridization studies display 40 to 60% similarity in AFLP clustering (28). Moreover, visual interpretation of the digitized band patterns shown in Fig. 1 clearly illustrates the high level of relatedness within both taxa. Occasionally, AFLP fingerprints exhibited strain-specific band differences. However, no correlation could be found between the observed AFLP polymorphisms and specific molecular types revealed by IS6110 RFLP or spoligotyping (Fig. 1). During the course of this study, a small subset of the strains included in Fig. 1 were also investigated using the primer combination A02-T01 (G.Huys, unpublished data). However, the change from primer T02 to T01 did not improve discrimination among molecular types in M. tuberculosis and M. bovis (G. Huys, unpublished data). A recent evaluation of fluorescent AFLP by Goulding and coworkers (10), on the other hand, indicated that this technique was in good agreement with IS6110 RFLP typing of M. tuberculosis and could also discriminate subtypes among strains harboring only one copy of IS6110. The latter investigation (10) and the present study essentially differed in various technical aspects, including the enzymes used for restriction (EcoRI/MseI instead of ApaI/TaqI) and the methodology used for labeling, electrophoresis, and data capture. However, the true explanation for the difference in discriminatory power between both studies probably lies with the fact that Goulding and colleagues used multiplex AFLP, i.e., the employment of four primer combinations only differing in the selective base (A, C, G, or T) of the EcoRI primer and labeled with four different fluorescent dyes. As a result, for each strain a composite AFLP fingerprint was generated from four AFLP patterns that were often much less discriminative when considered individually. It is believed that DNA polymorphisms in the M. tuberculosis complex are mainly concentrated in repetitive genomic sequences, resulting in unusually low structural gene variation (22), although it has been observed that West African strains are biochemically and genotypically more heterogenous than European isolates (19). It is thus possible that highly concentrated DNA polymorphisms can be overlooked when only a limited number of enzymes and/or PCR primers have been tested with AFLP, which explains the success of the multiplex AFLP approach (10). In our hands, the implementation of the multiplex AFLP concept with the radioactive methodology used in the present study was not feasible, as such a modification would result in an extremely labor-intensive and complicated protocol from the viewpoint of primer labeling and data analysis.
In contrast to M. tuberculosis and M. bovis, it was found that representatives of M. ulcerans displayed much more genetic heterogeneity. Each of the 12 M. ulcerans isolates under study generated a unique AFLP band pattern, and this degree of polymorphism strongly depended on the primer combination used. Whereas no correlation was found between AFLP polymorphisms and geographical origin with combination A02-T02, visual interpretation of normalized fingerprints could distinguish two subgroups in M. ulcerans when primers A02 and T01 were employed (Fig. 2). The observed subdivision into two subgroups corresponded with the Australian (n = 4) and African (n = 7) origin of the isolates and was based on the typical presence of AFLP marker band M3 in the African type. Similarly, RFLP analysis using plasmid pTBN12 (14) and partial 16S rRNA sequencing (27) could distinguish two subtypes linked to the Australian and African continent. The pTBN12 RFLP method could even distinguish between isolates originating from two Australian states and from two African countries (14). Portaels and coworkers (27) also found a third subgroup of two Mexican isolates representing the American M. ulcerans type, but these strains were not investigated in the present study. Interestingly, our AFLP data indicated that one Malaysian isolate (i.e., strain ITM 94-1328) also belonged to the Australian type, whereas recent IS2404 RFLP data suggested that isolates from Malaysia and Papua New Guinea may represent a fourth Southeast Asian M. ulcerans type (K. Chemlal, K. De Ridder, P. A. Fonteyne, W. M. Meyers, J. Swings, and F. Portaels, submitted for publication). In addition, it is also important to note that the subdivision of African and Australian AFLP types through visual comparison of band patterns was not supported by clustering analysis using Pearson or Dice correlation coefficients. This can be explained by the fact that the relative high number of strain-specific bands statistically outruled the one-band difference (i.e., AFLP marker band M3) significant for continental subgrouping and again highlights the relevance of profile comparison by eye.
In conclusion, the present investigation has demonstrated the usefulness of the AFLP technique as a reliable taxonomic tool for the differentiation of M. bovis, M. tuberculosis, and M. ulcerans. Clearly, further testing is needed to validate its potential for fingerprinting other members of the M. tuberculosis complex or other mycobacterial species. As a result of the low degree of genetic variation throughout their respective genomes, the radioactive AFLP methodology seems less promising for individual strain differentiation between M. bovis and M. tuberculosis. Within the more heterogeneous species M. ulcerans, on the other hand, AFLP proved to be a promising epidemiological method for differentiation of geographical types. Next to the further development of a nonradioactive AFLP methodology (10), future prospects may also include the recovery and molecular cloning of AFLP marker bands for the identification of species-specific or type-specific mycobacterial markers.
ACKNOWLEDGMENTS
This study was carried out in the framework of research project G.0368.98 of the Fund for Scientific Research, Flanders (Belgium) (F.W.O.-Vlaanderen). This work was also partially supported by the Directorate General for International Cooperation (DGIC), Belgium. K. Chemlal was supported by the Damien Foundation (Brussels, Belgium).
We thank Dirk Dewettinck for excellent technical assistance in the preparation of the figures.
REFERENCES
- 1.Aguiar J, Domingo M-C, Guédénon A, Meyers W, Steunou C, Portaels F. L'ulcère de Buruli, une maladie mycobactérienne importante et en recrudescence au Benin. Bull Seance Acad R Sci Outre-Mer. 1997;43:325–356. [Google Scholar]
- 2.Asiedu K, Portaels F. WHO/CDS/CPE/GBUI/2000.1 (ed.), Buruli ulcer: Mycobacterium ulcerans infection. Geneva, Switzerland: World Health Organization; 1999. Introduction; pp. 5–7. [Google Scholar]
- 3.Bauer J, Andersen A B, Kremer K, Miörner H. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J Clin Microbiol. 1999;37:2602–2606. doi: 10.1128/jcm.37.8.2602-2606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenye T, Schouls L M, Govan J R W, Kersters K, Vandamme P. Identification of Burkholderia species and genomovars from cystic fibrosis patients by AFLP fingerprinting. Int J Syst Microbiol. 1999;49:1657–1666. doi: 10.1099/00207713-49-4-1657. [DOI] [PubMed] [Google Scholar]
- 5.Collins D M, Erasmuson S K, Stephens D M, Yates G F, De Lisle G W. DNA fingerprinting of Mycobacterium bovis strains by restriction fragment analysis and hybridization with insertion elements IS1081 and IS6110. J Clin Microbiol. 1993;31:1143–1147. doi: 10.1128/jcm.31.5.1143-1147.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Portillo P, Murillo L A, Patarroyo M E. Amplification of a species-specific DNA fragment of Mycobacterium tuberculosis and its possible use in diagnosis. 1991. J Clin Microbiol. 1991;29:2163–2168. doi: 10.1128/jcm.29.10.2163-2168.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feizabadi M M, Robertson I D, Cousins D V, Hampson D J. Genomic analysis of Mycobacterium bovis and other members of the Mycobacterium tuberculosis complex by isoenzyme analysis and pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:1136–1142. doi: 10.1128/jcm.34.5.1136-1142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flood P, Street A, O'Brien P, Hayman J. Mycobacterium ulcerans infection on Philip Island, Victoria. Med J Aust. 1994;160:160. [PubMed] [Google Scholar]
- 9.Frothingham R, Hills H G, Wilson K H. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J Clin Microbiol. 1994;32:1639–1643. doi: 10.1128/jcm.32.7.1639-1643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goulding J N, Stanley J, Saunders N, Arnold C. Genome-sequence-based fluorescent amplified-fragment length polymorphism analysis of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:1121–1126. doi: 10.1128/jcm.38.3.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huys G, Coopman R, Janssen P, Kersters K. High-resolution genotypic analysis of the genus Aeromonas by AFLP fingerprinting. Int J Syst Bacteriol. 1996;46:572–580. doi: 10.1099/00207713-46-2-572. [DOI] [PubMed] [Google Scholar]
- 12.Huys G, Swings J. Evaluation of a fluorescent amplified fragment length polymorphism (FAFLP) methodology for the genotypic discrimination of Aeromonas taxa. FEMS Microbiol Lett. 1999;177:83–92. [Google Scholar]
- 13.Imaeda T. Deoxyribonucleic acid relatedness among selected strains of Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium bovis BCG, Mycobacterium microti, and Mycobacterium africanum. Int J Syst Microbiol. 1985;35:147–150. [Google Scholar]
- 14.Jackson K, Edwards R, Leslie D E, Hayman J. Molecular method for typing Mycobacterium ulcerans. J Clin Microbiol. 1995;33:2250–2253. doi: 10.1128/jcm.33.9.2250-2253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 16.Janssen P, Maquelin K, Coopman R, Tjernberg I, Bouvet P, Kersters K, Dijkshoorn L. Discrimination of Acinetobacter genomic species by AFLP fingerprinting. Int J Syst Bacteriol. 1997;47:1179–1187. doi: 10.1099/00207713-47-4-1179. [DOI] [PubMed] [Google Scholar]
- 17.Jiang S C, Matte M, Matte G, Huq A, Colwell R R. Genetic diversity of clinical and environmental isolates of Vibrio cholerae determined by amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 2000;66:148–153. doi: 10.1128/aem.66.1.148-153.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson P, Stinear T, Portaels F, Chemlal K, Dobos K, King H. WHO/CDS/COE/GBUI/2000.1 (ed.), Buruli ulcer: Mycobacterium ulcerans infection. Geneva, Switzerland: World Health Organization; 1999. Modern diagnostic techniques; pp. 23–30. [Google Scholar]
- 19.Källenius G, Koivula T, Ghebremichael S, Hoffner S E, Norberg R, Svensson E, Dias F, Marklund B-I, Svenson S B. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J Clin Microbiol. 1999;37:3872–3878. doi: 10.1128/jcm.37.12.3872-3878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F-C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W M, Martin C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, van Embden J D A. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marston B J, Diallo M O, Horsburgh C R, Jr, Diomande I, Saki M Z, Kanga J-M, Patrice G, Lipman H B, Ostroff S M, Good R. Emergence of Buruli ulcer disease in the Daloa region of Cote d'Ivoire. Am J Trop Med Hyg. 1995;52:219–224. doi: 10.4269/ajtmh.1995.52.219. [DOI] [PubMed] [Google Scholar]
- 24.Niemann S, Richter E, Rüsch-Gerdes S. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J Clin Microbiol. 2000;38:152–157. doi: 10.1128/jcm.38.1.152-157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Reilly L M, Daborn C J. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuberc Lung Dis. 1995;76S1:1–46. doi: 10.1016/0962-8479(95)90591-x. [DOI] [PubMed] [Google Scholar]
- 26.Picardeau M, Prod'hom G, Raskine L, LePennec M P, Vincent V. Genotypic characterization of five subspecies of Mycobacterium kansasii. J Clin Microbiol. 1997;35:25–32. doi: 10.1128/jcm.35.1.25-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portaels F, Fonteyne P-A, De Beenhouwer H, de Rijk P, Guédénon A, Hayman J, Meyers W M. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates of isolates. J Clin Microbiol. 1996;34:962–965. doi: 10.1128/jcm.34.4.962-965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savelkoul P H M, Aarts H J M, de Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker J L W, Schouls L, Lenstra J A. Amplified-fragment length polymorphism analysis: the state of the art. J Clin Microbiol. 1999;37:3083–3091. doi: 10.1128/jcm.37.10.3083-3091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh S P, Salamon H, Lahti C J, Farid-Moyer M, Small P M. Use of pulsed-field gel electrophoresis for molecular epidemiologic and population genetic studies of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:1927–1931. doi: 10.1128/jcm.37.6.1927-1931.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stonebrink B. The use of pyruvate containing egg medium in the culture of isoniazid resistant strains of Mycobacterium tuberculosis var. hominis. Acta Tuberc Scand. 1958;35:67–80. [Google Scholar]
- 32.Suffys, P. N., M. E. Ivens de Araujo, and W. M. Degrave. 1997. The changing face of the epidemiology of tuberculosis due to molecular strain typing - a review. Mem. Inst. Oswaldo Cruz Rio de Janeiro 92:297–316. [DOI] [PubMed]
- 33.Tønjum T, Welty D B, Jantzen E, Small P L. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J Clin Microbiol. 1998;36:918–925. doi: 10.1128/jcm.36.4.918-925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Soolingen D, de Haas P E W, Hermans P W, Groenen P M, van Embden J D. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Soolingen D, Hoogenboezem T, de Haas P E W, Hermans P W M, Koedam M A, Teppema K S, Brennan P J, Besra G S, Portaels F, Top J, Schouls L M, van Embden J D A. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol. 1997;47:1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- 37.Vauterin L, Vauterin P. Computer-aided objective comparison of electrophoresis patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–42. [Google Scholar]
- 38.Vincent Lévy-Frébault V, Portaels F. Proposed minimal standards for the genus Mycobacterium and for the description of new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992;42:315–323. doi: 10.1099/00207713-42-2-315. [DOI] [PubMed] [Google Scholar]
- 39.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLPTM: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wayne L G, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky M I, Moore L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Trüper H G. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 41.Wayne L G, Kubica G P. Genus Mycobacterium Lehmann and Neumann 1896. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Baltimore, Md: Williams and Wilkins; 1986. pp. 1436–1457. [Google Scholar]
- 42.Weil A, Plikaytis B B, Butler W R, Woodley C L, Shinnick T M. The mtp40 gene is not present in all strains of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:2309–2311. doi: 10.1128/jcm.34.9.2309-2311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]