Abstract
Congenital cataracts (CC) are responsible for approximately one-tenth of childhood blindness cases globally. Here, we report an African American family with a recessively inherited form of CC. The proband demonstrated decreased visual acuity and bilateral cataracts, with nuclear and cortical cataracts in the right and left eye, respectively. Exome sequencing revealed a novel homozygous variant (c.563A > G; p.(Asn188Ser)) in GJA3, which was predicted to be pathogenic by structural analysis. Dominantly inherited variants in GJA3 are known to cause numerous types of cataracts in various populations. Our study represents the second case of recessive GJA3 allele, and the first report in African Americans. These results validate GJA3 as a bona fide gene for recessively inherited CC in humans.
Keywords: GJA3, congenital cataract, African American, exome sequencing, nuclear cataract, cortical cataract
1. Introduction
Congenital cataract (CC) is a clinically heterogenous condition that leads to imperviousness of the crystalline lens [1]. Cataracts accounts for approximately 10% of childhood vision impairment and blindness across the world, and can be subdivided according to the anatomical location within the lens and appearance, but are also broadly categorized by etiology [2]. Among genetic, traumatic, metabolic and infectious aspects, hereditary cataracts account for 22.3% of childhood cases worldwide [3]. The prevalence of CC is estimated to be 1 to 15 per 10,000 children globally [4]. Cataracts in infancy and early childhood can also impact visual development due to amblyopia [5]. Though pathogenesis of CC can be caused by multiple factors, genetic factors are the most common, with a predominance of autosomal dominant inheritance [6].
Cataract could manifest as an isolated ocular disease and/or as a component of complex syndromic disorders [7] such as Lowe oculocerebrorenal syndrome, a rare X-linked disorder, with characteristic features of intellectual disability, CC and later onset, renal dysfunction [8]. Cataracts as part of other syndromes, e.g., Hereditary hyperferritinemia-cataract syndrome (HHCS), display variation in onset and severity. The morphological features of cataracts in HHCS include numerous white breadcrumb-like opacities, abundant in the lens cortex [9]. In CC, the lens microarchitecture is disrupted and vacuoles form, which cause light scattering due to variability in the lens’s opaque density [10]. Over 100 genes have been reported to be involved in CC [7].
Approximately 25% of genetic mutations causative of CCs involve mutations in connexin genes [11]. The lens expresses three distinct connexins, connexin 43 (Cx43), Cx46, and Cx50, which oligomerize to form gap junctions. The avascular lens requires an extensive intercellular communication system using gap junctions to maintain tissue homeostasis, namely crystallin transparency and solubility [12]. Of three connexins, variants in Cx46 (GJA3) and Cx50 (GJA8) cause disruptions in hemichannel formation, the most common cause of autosomal dominant CC. Structurally impaired gap junctions due to genetic mutations disrupt hemichannel function and affect lens homeostasis, which ultimately alters the orderly arrangement of crystallin and causes cataract formation [13]. Recently, Cx46 is reported to play a key role in H2O2-induced apoptosis in human lens epithelial cells [14]. Predominantly heterozygous variants of GJA3, apart from one allele, are reported in individuals with congenital cataracts [15,16]. Here, we report a novel recessively inherited variant of GJA3 in an African American family.
2. Results
2.1. Clinical Findings
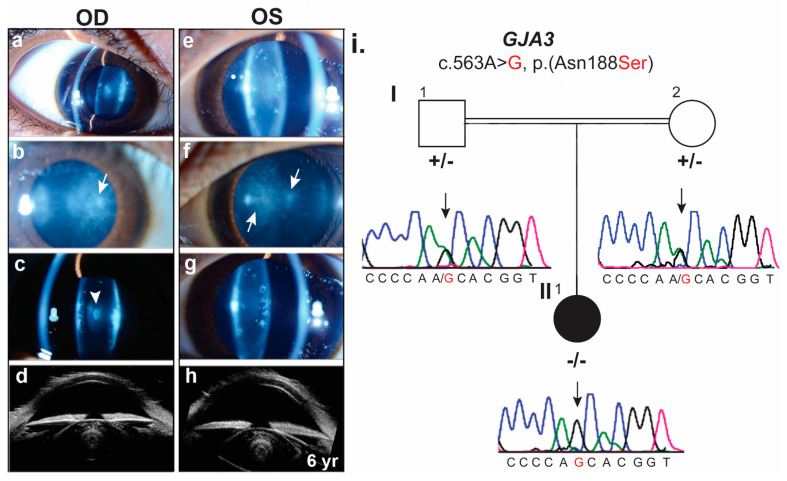
The proband (II: 1, Figure 1) was an African American female diagnosed with juvenile onset bilateral cataracts at age 6 after failed vision screening. Parents of the proband were siblings. Siblings of the proband include three full and additional half siblings, none of whom have cataracts. The proband’s medical history was significant due to depressive disorder, attention deficit hyperactivity disorder, and eczema. Her physical growth was normal for age in height but well below average for weight with weight Z-scores of −3 to −3.5, below the first percentile based on Centers for Disease Control and Prevention (Girls, 2–20 years) data. Prior ocular history was significant only for high myopia diagnosed at age 5. Her presenting best corrected visual acuity was 20/100 in the right and 20/150 in the left eye. Her cycloplegic refraction was −7.50 + 1.00 × 180 and −9.00 + 3.00 × 180, in each eye, respectively. Her slit lamp examination demonstrated nuclear and cortical cataracts in the right and left eye, respectively, with dense fleck-like deposits in the anterior and posterior subcapsular regions, and diffuse “dust-like” pulverulent opacities (Figure 1a–h). At the time of cataract surgery, the proband was found to have small corneal diameters of 10 mm in each eye and increased axial lengths of 24.5 and 24.6 mm, in each eye, respectively. Ultrasound biomicroscopy was performed at the time of cataract surgery (Figure 1a–h).
Figure 1.
Clinical phenotype of proband affected by juvenile bilateral cataracts. (a–h) Slit lamp photographs of the OD (right) and OS (left) eye (a–g), respectively, demonstrate nuclear and cortical cataracts with dense fleck-like deposits in the anterior and posterior subcapsular regions, and diffuse “dust-like” pulverulent opacities (arrows). OD shows a small white central nuclear opacity (arrowhead; photographs courtesy of William Buie). Ultrasound images of OD and OS are shown in panels d and h, respectively. (i). Family of proband with CC showing segregation of GJA3 missense variant. The affected individual is shown by a filled symbol. Sanger sequencing DNA chromatograms of GJA3 for the heterozygous normal parents (I:1, I:2), and affected individual (II:1) are shown on the right side.
2.2. Mutation Detection in GJA3
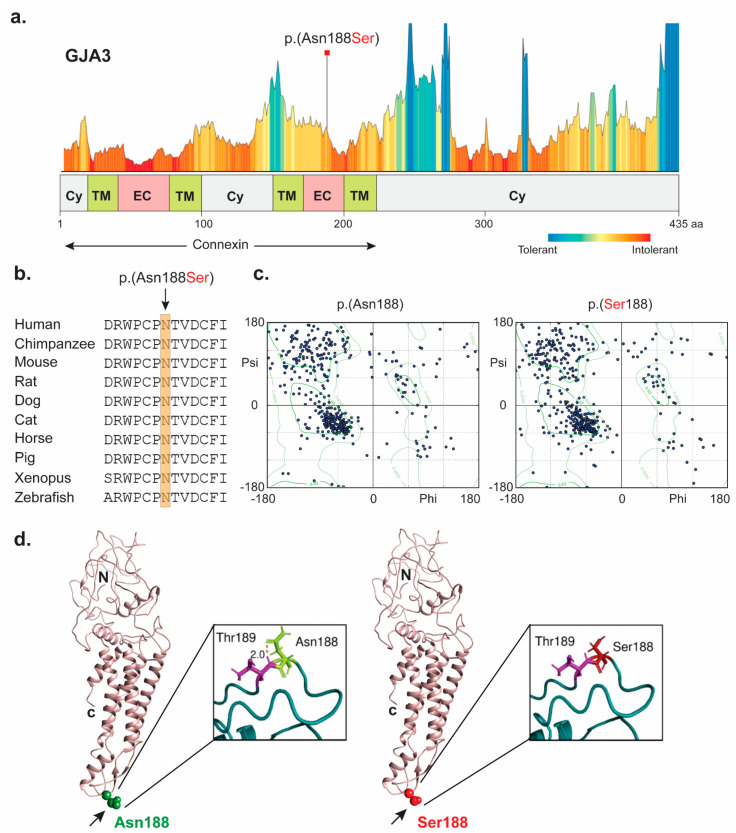
Bioinformatics analysis of WES data generated using the DNA sample of proband revealed a homozygous variant (c.563A > G, p.(Asn188Ser)) in GJA3, which segregated with the phenotype in a recessive pattern (Figure 1i). The identified missense variant (p.(Asn188Ser)) is predicted to be the part of intolerant region (Figure 2a), deleterious (Supplementary Table S1), and replaces evolutionary conserved residue (Figure 2b). The p.Asn188 is located in the second extracellular loop (E2) of the protein (Figure 2d), a domain critical for the activity of the protein [12,13]. The mutant residue (serine) is predicted to be small in size with loss of interactions, due to the lack of carbonyl group (as in asparagine), which interacts with the adjacent residues. The phi angle with the residue of interest shifted further outside of the permitted region of the Ramachandran plot (Figure 2c), further enforcing the fact that the loss of this stabilizing interaction affects the protein structure.
Figure 2.
Computational analysis of human GJA3 missense variant identified in the proband. (a) Tolerance and metadome landscape representation of GJA3 with protein domains underneath. Connexin domain is marked with arrows. Missense variant (p.Asn188Ser) identified in the study is highlighted in red and found to be present in intolerant region. p.Asn188 is predicted to be present in extracellular region of connexin domain. (b) Amino acids conservation in orthologous species for the p.(Asn188Ser) variant. The wild type residue (p.Asn188) is conserved across a wide variety of species. (c). Shown are the Ramachandran plots of the wild type (left) and mutant (right) proteins PDB structures generated by Phyre2 and visualized by Chimera. Favored and allowed regions are shown in light color with two respective boundaries. The favored regions (inner boundary line) represent conformations with no steric clashes. (d) 3D structural modeling in Chimera of the GJA3 protein, with the wild type and mutant residues shown, in ribbon and electrostatic potential representation, respectively. Asparagine and serine both are polar, non-charged (neutral) residues shown in white. However, blue and red colors represent amino acids with positive and negative charge, respectively. Abbreviations: Cy: cytoplasm; TM: transmembrane domain; EC: extracellular; C: c terminus; and N: n terminus.
3. Discussion
The major CC-associated proteins include transcription factors, crystallin, structural proteins, and membrane proteins (connexins) [14]. The lens expresses three distinct connexins—Cx43, Cx46, and Cx50—all of which appear to have different functions in maintaining lens homeostasis [8]. Extracellular domains of connexins play a key role in both mediating hemichannel docking [15,16] and regulating voltage gating of the channel [17]. Gap junctions within the lens help maintain an environment that favors crystallin solubility and fiber transparency by coupling the metabolically active epithelium and the organelle lacking lens fibers into a syncytium.
In human, GJA3 (Cx43) has been associated with a variety of inherited forms of CC (Table 1), though the most common remains zonular cataracts [12]. In our study, parents of the proband were siblings and carriers of the same variant of GJA3. Her clinical presentation showed bilateral cataracts upon slit lamp examination, with decreased corneal diameter and increased axial length of both eyes. There is a high proportion of genetically inherited forms of cataract with a substantial heterogeneity in both genetics and phenotype. Therefore, the arising phenotype could be the consequence of pleiotropic effect of lens proteins, which would thus show significant inter and intra-familial inconsistencies.
Table 1.
GJA3 variants, inheritance, cataract type, ethnicity or origin, and ACMG classification.
| DNA Change | Amino Acid Change | Zygosity | Cataract Type | Ethnicity/Location |
|---|---|---|---|---|
| c.-17-22C > G or c.-39C > G | - | Heterozygous | Nuclear | Chinese |
| c.1A > G | p.(Met1Val) | Heterozygous | Congenital | Indian |
| c.5G > A | p.(Gly2Asp) | Heterozygous | Nuclear pulverulent and posterior polar | Chinese |
| c.7G > C | p.(Asp3His) | Heterozygous | Congenital | Caucasian, Middle Eastern, Asian |
| c.7G > T | p.(Asp3Tyr) | Heterozygous | Zonular pulverulent | Hispanic |
| c.32T > C | p.(Leu11Ser) | Heterozygous | Ant-egg cataract | Danish |
| c.56C > T | p.(Thr19Met) | Heterozygous | Posterior-polar, nuclear-lamellar | Indian, Southeastern Australia |
| c.64G > A | p.(Gly22Ser) | Heterozygous | Pulverized cataract | Chinese |
| c.82G > T | p.(Val28Leu) | Heterozygous | Congenital | British |
| c.82G > A | p.(Val28Met) | Heterozygous | Posterior cortical and anterior capsular | Indian |
| c.84G > A | p.(Val28Val) | Heterozygous | Congenital, posterior subcapsular cataract, nystagmus | Indian |
| c.92T > A | p.(Ile31Asn) | Heterozygous | Bilateral microphthalmia, microcornea, and membranous cataract | Indian |
| c.96C > A | p.(Phe32Leu) | Heterozygous | Congenital nuclear pulverulent | Chinese |
| c.98G > T | p.(Arg33Leu) | Heterozygous | Congenital | Indian |
| c.125A > C | p.(Glu42Ala) | Heterozygous | pulverulent | Chinese |
| c.130G > A | p.(Val44Met) | Heterozygous | Central nuclear cataract with punctate cortical opacities | Chinese, Caucasian, Middle Eastern and Asian |
| c.134G > C | p.(Trp45Ser) | Heterozygous | Congenital nuclear cataract | Chinese |
| c.139G > A | p.(Asp47Asn) | Heterozygous | Congenital nuclear cataract | Chinese |
| c.143A > G | p.(Glu48Gly) | Heterozygous | Congenital | Chinese |
| c.148T > C | p.(Ser50Pro) | Heterozygous | Congenital | European, Chinese |
| c.163A > G | p.(Asn55Asp) | Heterozygous | Congenital | Chinese |
| c.176C > T | p.(Pro59Leu) | Heterozygous | Nuclear punctate, Congenital | Caucasian, Middle Eastern and Asian, Chinese |
| c.184G > A | p.(Glu62Lys) | Heterozygous | Congenital | Caucasian |
| c.188A > G | p.(Asn63Ser) | Heterozygous | Congenital Zonular Pulverulent | Caucasian |
| c.199G > C | p.(Asp67His) | Heterozygous | Cataract | Chinese |
| c.226C > G | p.(Arg76Gly) | Heterozygous | Cataract | Indian |
| c.227G > A | p.(Arg76His) | Heterozygous | Congenital with incomplete penetrance | Australian |
| c.260C > T | p.(Thr87Met) | Heterozygous | Peal box cataract | Indian |
| c.268C > T | p.(Leu90Phe) | Heterozygous | Nuclear | Chinese |
| c.415G > A | p.(Val139Met) | Heterozygous | Nuclear | Chinese |
| c.427G > A | p.(Gly143Arg) | Heterozygous | Congenital Coppock-like cataract | Chinese |
| c.428G > A | p.(Gly143Glu) | Heterozygous | Congenital nuclear cataract | Chinese |
| c.443C > T | p.(Thr148Ile) | Heterozygous | Bilateral pulverulent nuclear | Chinese |
| c.466A > C | p.(Lys156Gln) | Heterozygous | - | Southeastern Australia |
| c.559C > T | p.(Pro187Ser) | Heterozygous | Congenital, central nuclear opacity | Chinese |
| c.560C > T | p.(Pro187Leu) | Heterozygous | Congenital zonular pulverulent cataract | Caucasian |
| c.563A > T | p.(Asn188Ile) | Heterozygous | Congenital nuclear coralliform cataracts | Chinese |
| c.563A > C | p.(Asn188Thr) | Heterozygous | congenital nuclear pulverulent cataract | Chinese |
| c.563A > G | p.(Asn188Ser) | Homozygous | Juvenile onset, nuclear | African American |
| c.578T > C | p.(Phe193Ser) | Heterozygous | Syndromic | Caucasian |
| c.589C > T | p.(Pro197Ser) | Heterozygous | Congenital | Indian |
| c.596A > C | p.(Glu199Ala) | Heterozygous | Congenital | European |
| c.616T > A | p.(Phe206Ile) | Heterozygous | Congenital, nuclear | Chinese |
| c.771dupC | p.(Ser258Glnfs*68) | Heterozygous | Isolated lamellar cataract | British |
| c.950dupG | p.(His318Profs*8) | Homozygous | Nuclear, secondary glaucoma after cataract surgery | Pakistani |
| c.1137dupC | p.(Ser380Glnfs*88) | Heterozygous | Congenital Zonular Pulverulent 3 | Caucasian |
| c.1152dupG | p.(Ser385Glufs*83) | Heterozygous | Cataract 14 | Chinese |
| c.1189dupG | p.(Ala397Glyfs*71) | Heterozygous | Congenital coralliform cataract | Chinese |
| c.1200dupC | p.(Ala401Argfs*67) | Heterozygous | Nuclear | Chinese |
| c.1143_1165del23 | p.(Ser381Argfs*79) | Heterozygous | Congenital | Chinese |
* American College of Medical Genetics and Genomics classifications are given based on the https://Varsome.com (accessed on 15 November 2021) program predictions. Given in bold are the two known recessively inherited variants of GJA3.
As of August 2021, forty-nine different cataract-associated variants in GJA3 have been identified according to the Human Gene Mutation Database, and almost all of them cause cataract in a dominant fashion (Table 1; Supplementary Table S2). The only exception was the c.950dupG (p.(His318Profs*8) allele, which segregated in a recessive fashion in a Pakistani family [18]. Our study presents a second case of CC with a homozygous variant of GJA3 (Table 1). The clinical phenotype of the previously reported affected individuals with p.(His318Profs*8) homozygous variant include bilateral nuclear cataract, and secondary glaucoma after cataract surgery [18]. Similarly, the affected proband in our family also has nuclear cataract. She subsequently underwent uncomplicated cataract surgery with IOL implantation in both eyes, followed by yttrium aluminum garnet (YAG) laser capsulotomy in both eyes at cataract post-operative year 2. Following YAG capsulotomy, her visual acuity improved to 20/20 in each eye and has remained at this level at the most recent follow up post-operative year 4 (age 10).
Our study also represents the first reported case of GJA3 in an individual of African American ancestry (Table 1). Two studies previously reported heterozygous missense variants (p.(Asn188Thr), p.(Asn188Ile), present in second extracellular loop) in Chinese families [10,19], impacting the same GJA3 residue that is mutated in our case. However, in all three scenarios, the substituted amino acids are different (Table 1). The major clinical features of the reported dominant alleles (p.(Asn188Thr), p.(Asn188Ile)) include congenital nuclear pulverulent cataracts, dense coral-like opacities in nuclear region of the lens surrounded by blue dust-like opacities in the cortical zone [10,19]. However, in our case, the obligated carriers of the p.(Asn188Ser) variant were examined and had no cataracts or other eye problems. The second extracellular loop helps GJA3 anchor in hemichannel gap junctions. The p.Asn188Thr has been reported to have no impact on electrical assets of xenopus hemichannels. However, Asn188 is crucial for hemichannel docking by forming hydrogen bonds with amino acids arginine, threonine, and aspartic acid at positions 180, 189 and 191, respectively. Thus, it is reported that Asn188 replacement with threonine obstructs the anchorage of connexons to form gap junction channels [20]. However, it is worth mentioning that different alleles of GJA3 cause different phenotypes at different locations within the lens. It is possible that the genetic background modifies the resulting phenotype of GJA3 mutations, though differences between the alleles may directly account for the different outcomes.
PVS:1: Null variant (nonsense, frameshift, canonical ±1 or 2 splice sites, initiation codon, single or multiexon deletion) in a gene where LOF is a known mechanism of disease (Pathogenic, Very Strong).
PS1: Same amino acid change as a previously established pathogenic variant regardless of nucleotide change (Pathogenic, Strong).
PM1: Located in a mutational hot spot and/or critical and well-established functional domain (e.g., active site of an enzyme) without benign variation (Pathogenic, Moderate).
PM2: Absent from controls (or at extremely low frequency if recessive) in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium (Pathogenic, Moderate).
PM5: Novel missense change at an amino acid residue where a different missense change determined to be pathogenic has been seen before (Pathogenic, Moderate).
PP2: Missense variant in a gene that has a low rate of benign missense variation and in which missense variants are a common mechanism of disease (Pathogenic, Supporting).
PP3: Multiple lines of computational evidence support a deleterious effect on the gene or gene product (conservation, evolutionary, splicing impact, etc.) (Pathogenic, Supporting).
PP5: Reputable source recently reports variant as pathogenic, but the evidence is not available to the laboratory to perform an independent evaluation (Pathogenic, Supporting).
BS1: Allele frequency is greater than expected for disorder (Benign, Strong).
BS2: Observed in a healthy adult individual for a recessive (homozygous), dominant (heterozygous), or X-linked (hemizygous) disorder, with full penetrance expected at an early age (Benign, Strong).
BP4: Multiple lines of computational evidence suggest no impact on gene or gene product (conservation, evolutionary, splicing impact, etc.) (Benign, Supporting).
BP6: Reputable source recently reports variant as benign, but the evidence is not available to the laboratory to perform an independent evaluation (Benign, Supporting).
BP7: A synonymous (silent) variant for which splicing prediction algorithms predict no impact to the splice consensus sequence nor the creation of a new splice site AND the nucleotide is not highly conserved (Benign, Supporting).
4. Materials and Methods
4.1. Clinical Evaluation
Visual acuity was assessed by using the standard Snellen chart. Fundoscopy and slit lamp biomicroscopy were also performed. Axial length measurement and ultrasound biomicroscopy imaging were performed using the Aviso Ultrasound Platform A/B UBM with a 50 MHz linear transducer (Quantel Medical, Clermont-Ferrand, France). Blood samples were collected from all the participants for DNA extraction.
4.2. Exome Sequencing and Bioinformatic Analyses
Whole exome sequencing (WES) was performed on the proband DNA sample, and data were filtered using the criteria previously described [11]. Sanger sequencing was used for the segregation analysis of identified variants in the family. In silico analysis and three-dimensional (3D) molecular modeling were performed using various programs (see web resources). Finally, the Varsome.com online tool was used for the American College of Medical Genetics and Genomics classification of the GJA3 variants.
Acknowledgments
We would like to thank the participating patient, her family, and the health care professionals involved in their care. We also thank Muhammad A. Usmani, and Zeshan Tariq for their technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms23010240/s1.
Author Contributions
Conceptualization, O.J.S. and Z.M.A.; methodology, A.Y.H., S.Y., M.R.L., J.L.A. and Z.M.A.; software, A.Y.H. and S.Y.; validation, A.Y.H., S.Y., M.R.L., O.J.S., J.L.A. and S.R.; formal analysis, A.Y.H., S.Y., M.R.L. and J.L.A.; investigation, A.Y.H., S.Y., M.R.L. and Z.M.A.; resources, Z.M.A.; writing—original draft preparation, A.Y.H., S.Y., M.R.L., O.J.S., J.L.A. and Z.M.A.; writing—review and editing, S.R. and Z.M.A.; supervision, M.R.L. and Z.M.A.; project administration, O.J.S. and Z.M.A.; funding acquisition, Z.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Institute on Deafness and Other Communication Disorders/National Institutes of Health R01 DC016295 grant to Z.M.A. O.J.S. and J.L.A. are supported by a Career Development Awards from the National Eye Institute/National Institutes of Health (K23 EY025014; KL2 TR003099).
Institutional Review Board Statement
The current study was approved by Institutional Review Board Committees (HP00064793) at the University of Maryland School of Medicine, Baltimore, MD. All methods used in this study were in conjunction with the precepts of the Declaration of Helsinki. Informed written consent was obtained from all participants prior to inclusion in the study.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The variant has been submitted to the ClinVar database: Accession number: SCV001787094.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee C.M., Afshari N.A. The global state of cataract blindness. Curr. Opin. Ophthalmol. 2017;28:98–103. doi: 10.1097/ICU.0000000000000340. [DOI] [PubMed] [Google Scholar]
- 2.Berthoud V.M., Beyer E.C. Oxidative stress, lens gap junctions, and cataracts. Antioxid. Redox Signal. 2009;11:339–353. doi: 10.1089/ars.2008.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert C.E., Canovas R., Hagan M., Rao S., Foster A. Causes of childhood blindness: Results from west Africa, south India and Chile. Pt 1Eye. 1993;7:184–188. doi: 10.1038/eye.1993.39. [DOI] [PubMed] [Google Scholar]
- 4.Foster A., Gilbert C., Rahi J. Epidemiology of cataract in childhood: A global perspective. J. Cataract Refract Surg. 1997;23((Suppl. S1)):601–604. doi: 10.1016/S0886-3350(97)80040-5. [DOI] [PubMed] [Google Scholar]
- 5.Wirth M.G., Russell-Eggitt I.M., Craig J.E., Elder J.E., Mackey D.A. Aetiology of congenital and paediatric cataract in an Australian population. Br. J. Ophthalmol. 2002;86:782–786. doi: 10.1136/bjo.86.7.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nizami A.A., Gulani A.C. Cataract. StatPearls; Treasure Island, FL, USA: 2021. [Google Scholar]
- 7.White T.W. Unique and redundant connexin contributions to lens development. Science. 2002;295:319–320. doi: 10.1126/science.1067582. [DOI] [PubMed] [Google Scholar]
- 8.Gerido D.A., White T.W. Connexin disorders of the ear, skin, and lens. Biochim. Biophys. Acta. 2004;1662:159–170. doi: 10.1016/j.bbamem.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Addison P.K.F., Berry V., Holden K.R., Espinal D., Rivera B., Su H., Srivastava A.K., Bhattacharya S.S. A novel mutation in the connexin 46 gene (GJA3) causes autosomal dominant zonular pulverulent cataract in a Hispanic family. Mol. Vis. 2006;12:791–795. [PubMed] [Google Scholar]
- 10.Li Y., Wang J., Dong B., Man H. A novel connexin46 (GJA3) mutation in autosomal dominant congenital nuclear pulverulent cataract. Mol. Vis. 2004;10:668–671. [PubMed] [Google Scholar]
- 11.Riazuddin S., Hussain M., Razzaq A., Iqbal Z., Shahzad M., Polla D., Song Y., Van Beusekom E., A Khan A., Roca L.T., et al. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol. Psychiatry. 2017;22:1604–1614. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guleria K., Sperling K., Singh D., Varon R., Singh J.R., Vanita V. A novel mutation in the connexin 46 (GJA3) gene associated with autosomal dominant congenital cataract in an Indian family. Mol. Vis. 2007;13:1657–1665. [PubMed] [Google Scholar]
- 13.Scemes E., Suadicani S.O., Dahl G., Spray D.C. Connexin and pannexin mediated cell-cell communication. Neuron Glia Biol. 2007;3:199–208. doi: 10.1017/S1740925X08000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell S.J., Oluonye N., Harding P., Moosajee M. Congenital cataract: A guide to genetic and clinical management. Ther. Adv. Rare Dis. 2020;1:1–22. doi: 10.1177/2633004020938061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J.X., Goodenough D.A. Heteromeric connexons in lens gap junction channels. Proc. Natl. Acad. Sci. USA. 1996;93:1287–1291. doi: 10.1073/pnas.93.3.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon A.M., Goodenough D.A. Diverse functions of vertebrate gap junctions. Trends Cell Biol. 1998;8:477–483. doi: 10.1016/S0962-8924(98)01372-5. [DOI] [PubMed] [Google Scholar]
- 17.Verselis V.K., Ginter C.S., Bargiello T.A. Opposite voltage gating polarities of two closely related connexins. Nature. 1994;368:348–351. doi: 10.1038/368348a0. [DOI] [PubMed] [Google Scholar]
- 18.Micheal S., Niewold I.T.G., Siddiqui S.N., Zafar S.N., Khan M.I., Bergen A.A.B. Delineation of Novel Autosomal Recessive Mutation in GJA3 and Autosomal Dominant Mutations in GJA8 in Pakistani Congenital Cataract Families. Genes. 2018;9:112. doi: 10.3390/genes9020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Qu X., Su S., Guan L., Liu P. A novel mutation in GJA3 associated with congenital Coppock-like cataract in a large Chinese family. Mol. Vis. 2012;18:2114–2118. [PMC free article] [PubMed] [Google Scholar]
- 20.Schadzek P., Schlingmann B., Schaarschmidt F., Lindner J., Koval M., Heisterkamp A., Preller M., Ngezahayo A. The cataract related mutation N188T in human connexin46 (hCx46) revealed a critical role for residue N188 in the docking process of gap junction channels. Biochim. Biophys. Acta (BBA)-Biomembr. 2016;1858:57–66. doi: 10.1016/j.bbamem.2015.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variant has been submitted to the ClinVar database: Accession number: SCV001787094.