Abstract
Mycobacterium tuberculosis converts pyrazinamide to its active form by using the enzyme pyrazinamidase. This enzyme is coded for on the pncA gene, and mutations in the pncA gene result in absence of active enzyme, conferring resistance to the drug pyrazinamide. We investigated 27 strains of Mycobacterium tuberculosis suspected of being multidrug resistant. Each isolate was tested for susceptibility to pyrazinamide by the BACTEC 460TB method, and 19 were pyrazinamide resistant. The presence of active pyrazinamidase enzyme was sought by using the Wayne assay, which was positive in all of the sensitive isolates and four of the resistant isolates. The pncA gene was amplified by PCR, and mutations were sought by single-strand conformation polymorphism (SSCP) analysis. We identified four isolates which were phenotypically resistant to pyrazinamide, but which had active pyrazinamide enzyme on the Wayne assay and normal pncA gene SSCP. MICs measured by BACTEC 460TB and susceptibility testing at a lower pH of 5.5 confirmed genuine resistance. The pncA gene was sequenced in these four isolates and found not to have any mutations. This implies that an alternative mechanism of resistance exists in these strains. We conclude that genotypic assessment of pyrazinamide resistance is unreliable, because it depends on the identification of a single resistance mechanism. Phenotypic methods such as the BACTEC 460TB technique remain the best methods for pyrazinamide susceptibility testing.
Pyrazinamide (PZA) is an important part of the therapeutic armory against tuberculosis (TB). It acts in the acidic extracellular microenvironment found in acute inflammation and kills at least 95% of bacilli during the first 2 weeks of treatment (6). Following its addition to anti-TB drug regimens, the length of a course of treatment has been reduced to 6 months, thus aiding compliance and decreasing the risk of development of multiple-drug-resistant (MDR) TB (1).
The target of action of PZA in Mycobacterium tuberculosis is thought to be fasI (17). It is known that M. tuberculosis converts PZA to its active form, pyrazinoic acid, by using the enzyme pyrazinamidase (PZAase) (2). As long ago as 1967, it was shown that PZA-resistant strains of M. tuberculosis lacked PZAase activity and were unable to convert the prodrug to its active form (2).
In 1996, the pncA gene, encoding PZAase, was cloned, and researchers showed that resistant strains of M. tuberculosis, and naturally resistant Mycobacterium bovis, had mutations in the pncA gene. Transformation of PZA-resistant strains with the PZA-sensitive gene led to their becoming susceptible to the drug again (10). In a study of 38 PZA-resistant clinical isolates, 33 had mutations in pncA, and the other five isolates were either falsely or borderline resistant (9).
Various laboratory indicators had been suggested in the past to assess susceptibility of M. tuberculosis to PZA. Traditional mycobacterial susceptibility testing methods, depending on growth of the organisms when exposed to drugs, are hampered by the requirement of a low pH for PZA activity (4), because this pH is inhibitory to the growth of M. tuberculosis (12). However, reliable pyrazinamide susceptibility testing on solid media has been achieved (14). Other approaches have attempted to detect the presence or absence of PZAase function as a marker of resistance, with resistant strains presumed to be lacking in the enzyme. These methods include the Wayne assay, which detects the presence of active PZAase enzyme by using the hydrolysis of PZA to pyrazinoic acid, as evidenced by a color change (15). A negative result implies no PZAase activity and hence resistance to PZA. Previous workers have commented on the difficulty of interpretation of this test. The description of the pncA gene opened the possibility of using a molecular marker for resistance determination. Single-stranded conformation polymorphism (SSCP) has been employed to detect point mutations in the pncA gene amplified by PCR, and sequencing of the pncA gene has also been suggested as a method of detecting PZA-resistant organisms (3, 10). In our study, we compared molecular methods with phenotypic resistance testing by the BACTEC 460TB culture method and with the Wayne assay.
MATERIALS AND METHODS
BACTEC 460TB.
We investigated 27 suspected MDR TB strains isolated in South Africa. Susceptibility to PZA was tested by the standard BACTEC 460TB technique according to the manufacturer's instructions (Becton Dickinson). Two BACTEC 460TB vials were inoculated at pH 6.0: one vial was drug free, and the other contained 100 mg of PZA per liter. PZA resistance was defined as a growth index of greater than 11% of that of the control vial when the control vial reached a growth index of greater than 200. The control vial had to remain below 200 for 4 days for the result to be acceptable (Becton Dickinson). Isolates with discrepant results (resistant by BACTEC 460TB, but Wayne assay positive) were selected for further susceptibility study. To confirm PZA resistance in these isolates, the susceptibility testing was repeated by BACTEC 460TB with concentrations of 300 and 900 mg of PZA per liter. To test whether susceptibility returned with lower pH, vials were acidified to pH 5.5 by the method described previously (8). The acidified vials were then used to test for PZA susceptibility by the BACTEC 460TB technique with 100 mg of PZA per liter.
PZAase assay.
The Wayne assay was performed by the technique described previously (15). Dubos agar butts containing 100 mg of PZA per liter were heavily inoculated and incubated for 4 days. At the end of the incubation period, 1 ml of ferrous ammonium phosphate was added to each tube, and the tube was examined for a pink band. The tubes were refrigerated for 4 h and reexamined for a pink band. The appearance of a pink band on diffusion of the ferrous salt was accepted as an indication of hydrolysis of PZA to free pyrazinoic acid. Tubes negative at 4 days were reincubated and reexamined at 7 days. The Wayne assay was read by two independent observers, whose results agreed for all of the test isolates.
PZAase gene PCR.
The PZAase gene (10) was amplified by PCR. DNA was extracted according to the method previously described (5). The primers employed were 5′ TGCGGGCGTTGATCATC and 5′ CAGGAGCTGCAAACCAACTC. The optimal reaction mixture contained 1.5 mM MgCl in KCl buffer, 150 nM deoxynucleoside triphosphates, 5 U of Taq polymerase enzyme, and 10 μl of primer at a 1 μM concentration for each PCR, in a total volume of 90 μl. An aliquot of 10 μl of M. tuberculosis DNA was added. The cycling conditions were as follows: 1 cycle at 95°C for 1 min and 35 cycles of 95°C for 1 min, 55°C for 2 min, and 72°C for 3 min. This was followed by strand elongation for 7 min at 72°C. This produced an amplicon of 559 bp.
SSCP.
The amplicons were analyzed by PCR-SSCP. The PCR product (6 μl) was denatured at 95°C for 10 min with 3 μl of SSCP loading buffer (0.1% sodium dodecyl sulfate, 10 mM EDTA) and 3 μl of stop dye (Promega, Southampton, United Kingdom). Samples were quenched on ice and then loaded directly onto a 0.5% mutation enhancement acrylamide analogue gel (Flowgen, Lichfield, United Kingdom). The gel was run for 18 h at 6 W with 0.6× Tris-borate-EDTA (Sigma, Poole, United Kingdom) at room temperature. A silver stain was used to visualize DNA bands (Sigma).
Sequencing was performed with six isolates. The PCR product was purified with the Wizard PCR kit according to the manufacturer's instructions (Promega). Sequencing was performed commercially by cycle sequencing with dye terminator technology (Cambridge Bioscience, Cambridge, United Kingdom).
RESULTS
BACTEC 460TB susceptibility testing.
BACTEC 460TB susceptibility testing was performed with all isolates. Nineteen isolates were found to be resistant by this method. The four isolates that were resistant with a positive Wayne assay (see below) had susceptibility testing repeated at a lower pH of 5.5. They remained resistant at the lower pH. MICs were also determined for these four isolates by the BACTEC 460TB method. This confirmed that these isolates were genuinely resistant, although the rate of growth decreased with increasing PZA concentration.
Wayne assay.
All eight sensitive strains were Wayne assay positive, but four of the resistant strains were also Wayne assay positive, as tested on two occasions, implying the presence of active PZAase enzyme. The other 15 resistant strains were Wayne assay negative.
SSCP analysis.
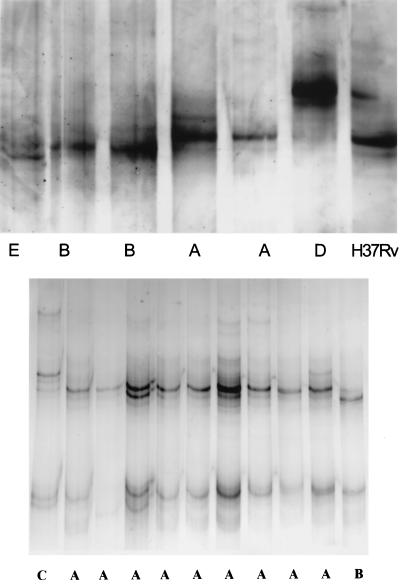
If the pncA gene contained mutations, then the SSCP pattern of the mutated gene should show variation from the susceptible gene (13). Thus, PZA-sensitive strains with a normal pncA gene would be expected to have the same SSCP pattern, while PZA-resistant strains should have diverse SSCP patterns, depending on the mutation they possess. Five different SSCP patterns were seen (Fig. 1). Not all PZA-resistant isolates had an abnormal SSCP pattern; 10 resistant isolates that were Wayne negative had the same SSCP pattern as H37Rv. Five of the resistant Wayne assay-negative isolates had SSCP patterns that differed from those of the H37Rv control. Of these, there were three unique patterns and a pair of strains with a similar pattern. The eight sensitive strains all had the same SSCP pattern as the reference strain, H37Rv. The four Wayne assay-positive resistant strains also had the same SSCP pattern as the susceptible reference strain.
FIG. 1.
Examples of the five SSCP patterns seen. Pattern A is similar to that of the susceptible reference strain H37Rv.
In summary, eight isolates were sensitive to PZA on BACTEC 460TB testing: all of these were positive on Wayne assay and had the same SSCP pattern as H37Rv. Nineteen isolates were resistant to PZA on BACTEC 460TB testing. Of these, 15 were Wayne assay negative with five SSCP patterns represented. However, four resistant isolates were positive on the Wayne assay and also had the same SSCP pattern as H37Rv.
Sequencing.
The pncA genes of two resistant isolates with a negative Wayne assay were sequenced and showed mutations. One was a point mutation at codon 58 resulting in leucine instead of phenylalanine, and the other was a point mutation at codon 139 resulting in alanine instead of valine. Similarly, the pncA genes of the four PZA-resistant isolates that were Wayne assay positive and demonstrated a “susceptible” SSCP pattern were sequenced. No mutations were found in the pncA genes of any of these four isolates, consistent with the result of the Wayne assays and normal SSCP patterns and implying the presence of an active PZAase enzyme.
DISCUSSION
Reproducible susceptibility testing is essential for effective management of TB. This is increasingly important with the rise of MDR worldwide. Conventional susceptibility tests are difficult to perform for PZA, because the drug is only active at low pH, which in itself inhibits the growth of mycobacteria. The Wayne assay directly measures the activity of PZAase thought to be essential for the activity of the drug, but this test is often difficult to perform and interpret. Molecular methods may be a solution, but depend on the hypothesis that mutations in the pncA gene are a reliable marker of PZA resistance. In this study, we were able to investigate the value of each of these methods when applied to a group of organisms likely to contain PZA resistance. When the Wayne assay was used, 4 of our 19 isolates that had been shown to be resistant by the BACTEC 460TB method were positive, implying the presence of an active PZAase. Thus, they would have been considered sensitive by this method. This suggests a sensitivity of about 79% when using the Wayne assay to identify resistant isolates. All four Wayne-positive resistant isolates had a susceptible SSCP pattern and wild-type pncA gene sequences. MICs determined by the BACTEC 460TB method confirmed that these isolates were genuinely resistant, although the rate of growth decreased with increasing concentrations of PZA, indicating some activity of PZA. In addition, susceptibility testing was repeated at a lower pH of 5.5 to see whether susceptibility returned: it did not, and the results of susceptibility testing were the same at both pH 6.0 and 5.5. Using SSCP, only 5 (26%) of the resistant strains would have been correctly identified. These results suggest that SSCP of the pncA gene is not a sensitive method of detecting PZA resistance in M. tuberculosis. Sequencing is not a satisfactory solution, and our data lend support to the idea that there are other mechanisms of PZA resistance besides loss of PZAase activity in this group of clinical strains. Sreevatsan et al. (11) found that all of 51 PZA-sensitive strains of M. tuberculosis had normal pncA genes, while 72% of 67 PZA-resistant strains had pncA mutations. This finding also suggested additional resistance mechanisms besides lack of PZAase activity.
A recent paper by Raynaud et al. (7) investigated uptake of PZA as a possible resistance mechanism in mycobacteria, including M. tuberculosis. The authors found that an ATP-dependent transport system is involved in the uptake of PZA. They investigated four naturally PZA-resistant species of mycobacteria as well as M. tuberculosis. M. tuberculosis was the only species displaying both an active PZA enzyme and an active PZA transport system. Of the other (resistant) species, M. bovis and Mycobacterium kansasii had neither PZAase activity nor PZA uptake, Mycobacterium smegmatis had functional PZAase, but no uptake system, and M. avium had PZA uptake similar to that of M. tuberculosis, but no PZAase activity. Their results indicate that both active PZAase and uptake systems are required for susceptibility to PZA, and our data support the hypothesis that not all PZA-resistant strains are deficient in PZAase activity or have pncA gene mutations.
The discrepancies between the various susceptibility testing methods are further confounded by the difficulty in defining susceptibility or resistance clinically in the case of treatment of tuberculosis. Clinical response can hardly be used as a measure of one drug's degree of resistance in a multidrug regimen. Failure of therapy is usually due to poor compliance or inadequate regimens (16). Our data suggest that phenotypic assays such as the BACTEC 460TB method probably remain the best available measure of resistance; this method has the advantage over the others tried here of not depending on detection of a single resistance mechanism.
REFERENCES
- 1.Fox W, Ellard G A, Mitchison D A. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council Tuberculosis Units, 1946–1986, with relevant subsequent publications. Int J Tuberc Lung Dis. 1999;3:S231–S279. [PubMed] [Google Scholar]
- 2.Konno K, Feldman F M, McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis. 1967;95:461–469. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- 3.Marttila H J, Marjamäki M, Vyshnevskaya E, Vyshnevskiy B I, Otten T F, Vasilyef A V, Viljanen M K. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from Northwestern Russia. Antimicrob Agents Chemother. 1999;43:1764–1766. doi: 10.1128/aac.43.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDermott W, Tomsett R. Activation of pyrazinamide and nicotinamide in acidic environment in vitro. Am Rev Tuberc. 1954;70:748–754. doi: 10.1164/art.1954.70.4.748. [DOI] [PubMed] [Google Scholar]
- 5.McHugh T D, Newport L E, Gillespie S H. IS6110 homologs are present in multiple copies in mycobacteria other than tuberculosis-causing mycobacteria. J Clin Microbiol. 1997;35:1769–1771. doi: 10.1128/jcm.35.7.1769-1771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchison D A. The action of antituberculosis drugs in short-course chemotherapy. Tubercle. 1985;66:219–225. doi: 10.1016/0041-3879(85)90040-6. [DOI] [PubMed] [Google Scholar]
- 7.Raynaud C, Laneelle M-A, Senaratne R H, Draper P, Laneelle G, Daffe M. Mechanisms of pyrazinamide resistance in mycobacteria: importance of lack of uptake in addition to lack of pyrazinamidase activity. Microbiology. 1999;145:1359–1367. doi: 10.1099/13500872-145-6-1359. [DOI] [PubMed] [Google Scholar]
- 8.Salfinger M, Heifets L B. Determination of pyrazinamide MICs for Mycobacterium tuberculosis at different pHs by the radiometric method. Antimicrob Agents Chemother. 1988;32:1002–1004. doi: 10.1128/aac.32.7.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–543. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 11.Sreevatsan S, Pan X, Zhang Y, Kreiswirth B N, Musser J M. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–640. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stottmeier K D, Beam R E, Kubica G P. Determination of drug susceptibility of mycobacteria to pyrazinamide in 7H10 agar. Am Rev Respir Dis. 1967;96:1072–1075. doi: 10.1164/arrd.1967.96.5.1072. [DOI] [PubMed] [Google Scholar]
- 13.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 14.Tripathy S P, Mitchison D A, Nair N G K, Radhakrishna S, Subbammal S. A comparison of various measures of susceptibility of Mycobacterium tuberculosis to pyrazinamide. Tubercle. 1970;51:375–388. doi: 10.1016/0041-3879(70)90003-6. [DOI] [PubMed] [Google Scholar]
- 15.Wayne L G. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974;109:147–151. doi: 10.1164/arrd.1974.109.1.147. [DOI] [PubMed] [Google Scholar]
- 16.WHO/IUATLD Global Project on Anti-Tuberculosis Drug Resistance Surveillance 1994–1997. Anti-tuberculosis drug resistance in the world. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 17.Zimhony O, Cox J S, Welch J T, Vilcheze C, Jacobs W R. Pyrazinamide inhibits the eukaryotic-like fatty acid synthase I (FASI) of Mycobacterium tuberculosis. Nat Med. 2000;6:1043–1047. doi: 10.1038/79558. [DOI] [PubMed] [Google Scholar]