Abstract
A combined PCR assay was developed for the detection and typing of human polyomavirus (huPoV) in clinical samples, consisting of (i) a qualitative seminested PCR assay (snPCR) to discriminate between huPoV BK and JC and (ii) a high-throughput, quantitative TaqMan PCR assay (TM-PCR) for the general detection of huPoV. The TM-PCR detects huPoV DNA in a linear range from 107 to 101 copies per assay. In reproducibility runs, the inter- and intra-assay variabilities were ≤60 and ≤50%, respectively. The snPCR assay uses a set of four primers for the same region of the BK and JC viral genomes. In the first round of amplification, two general primers were used; in the second round, one of these general primers and two additional, BK- or JC-specific primers were used simultaneously to produce amplicons of different sizes specific for BK virus (246 bp) and JC virus (199 bp), respectively. We tested different urine dilutions in order to determine the inhibitory effects of urine on PCR amplification. Furthermore, we compared the use of native urine with DNA purified by different preparation procedures. Our results show, that a 1:10 dilution of the urine led to complete reduction of the amplification inhibition found with 6% of undiluted urine samples. In a clinical study including 600 urine specimens, our assay turned out to be fast, cheap, and reliable in both qualitative and quantitative aspects.
Diagnosis of human polyomavirus (huPoV) infection is important in transplant medicine. Reactivation of huPoV under immunosuppression is suggested to be associated with hemorrhagic cystitis (HC), a common complication after bone marrow transplantation (BMT) (1, 7, 8, 12, 19). At present, the most commonly used treatment for HC is hydration and forced diuresis. Recently, however, successful attempts at antiviral therapy with, e.g., cidofovir have been reported (15). To control the efficacy of medical treatment of HC, a sensitive, fast diagnostic assay is required, which allows the quantification of the polyomaviral load. Finally, for epidemiological studies, differentiation between BK virus (BKV) and JC virus (JCV) is essential.
The methods presently used for the detection of huPoV in urine are cell culture (12), electron microscopy (2), and PCR (3, 10, 11). While cell culture is time-consuming and therefore not clinically relevant, electron-microscopic diagnosis can be fast but is hampered by low sensitivity (4). Presently established qualitative PCR assays for the detection and differentiation of huPoV DNA from urine or cerebrospinal fluid include various sample preparation steps followed by hybridization and/or digestion assays (3, 9, 10). Two recently published quantitative PCR assays for BKV and JCV are based on competitive amplification (5, 15); each test, however, can detect only one of the two virus species. Thus, the presently available huPoV detection methods either require extensive handling, have low sensitivity, or are expensive.
A new technique for quantitative PCR detection employs the commercially available ABI Prism 7700 SDS, an instrument which performs DNA amplification and simultaneously determines the amount of amplified DNA (real-time PCR) (13). Hence, we established a real-time PCR assay for the quantification of huPoV DNA, TaqMan PCR (TM-PCR), which profits from real-time quantification during the PCR log phase and the lack of post-PCR handling, thus providing high accuracy. Furthermore, the TM-PCR is fast, is specific, and displays a low detection limit.
The inhibitory effects of urine as a PCR template have been described for different diagnostic assays, as for the detection of cytomegalovirus or Chlamydia spp. (17, 20). However, amplification inhibition by urine was analyzed only on a qualitative basis, not with regard to quantitative aspects. We tried different methods as described in the literature to overcome the inhibitory effects of urine, e.g., dilution, heating, and DNA preparation. Subsequently we quantified the extent of inhibition with the TM-PCR. We could demonstrate, on a quantitative basis, that dilution of native urine is sufficient to reduce amplification-inhibitory effects.
In order to develop a complete diagnostic system, we established a qualitative seminested PCR (snPCR) to distinguish BKV from JCV. Finally, this combined assay, consisting of quantitative TM-PCR and quantitative differentiation snPCR, was applied to 600 clinical urine specimens in order to test the practical usefulness of this new diagnostic tool.
MATERIALS AND METHODS
Sample collection.
Midstream urine was obtained weekly from patients prior to and after allogeneic BMT and from control persons. Urine was transported at room temperature and stored at 4°C. In total, we assayed 600 samples from 103 BMT patients and 11 healthy individuals.
Urine manipulation.
Urine was used either undiluted (native) or diluted 1:10 or 1:100 with sterile double-distilled water. For DNA preparation we used a Qiagen DNA kit or digestion with 25 μl of a Tris-EDTA-buffered proteinase K solution (20 mg/ml) for 60 min at 56°C followed by enzyme denaturation at 95°C for 10 min and subsequent centrifugation at 20,000 × g.
The ability of prolonged heat denaturation to reduce the inhibitory effects of urine was determined: prior to the PCR, the urine was heated to 95°C for 1, 5, 10, 30, and 60 min.
For virus enrichment by sedimentation, ultracentrifugation was performed with 5 ml of urine for 60 min at 100,000 × g in a Beckman centrifuge. The supernatant was discarded, and the pellet was resuspended in 50 μl of sterile double-distilled water.
Purification of huPoV.
Virus was purified from urine by cesium chloride gradient centrifugation as described elsewhere (14). The gradient was fractionated, and the CsCl densities of the fractions were determined refractometrically; in addition, negative-staining electron microscopy was used to estimate the relative virus concentrations of the respective fractions. Finally, the virus suspension was dialyzed to remove the PCR-inhibiting salt.
Seminested polyomavirus PCR.
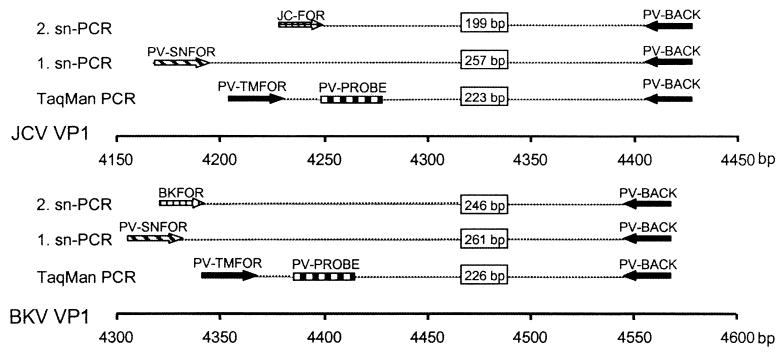
Primers for snPCR were located in the VP1 regions of the BKV and JCV genomes. Primer sequences are given in Table 1; a schematic representation of the locations and orientations of all oligonucleotides and corresponding amplicon sizes is given in Fig. 1. In a first amplification round, primers PV-SNFOR and PV-BACK were used, followed by a second amplification round using the general primer PV-BACK in combination with two variant specific primers for BKV and JCV. Each 30 μl of PCR mixture contained 5 μl of template, 333 nM concentrations of each primer, 50 μM each deoxynucleoside triphosphate (dNTP) (Gibco), 3 μl of 10× amplification buffer, and 1 U of Platinum Taq DNA polymerase (Gibco). After an initial denaturation for 3 min, the sample was subjected to 30 cycles of 94°C for 20 s and 53°C for 20 s, with a final extension for 5 min at 72°C, for the first amplification round. A 1-μl volume of the resulting PCR product was used as the template in a second amplification round under the same conditions, except that the annealing temperature was raised to 58°C.
TABLE 1.
Oligonucleotides used in this study
| Name | PCR for which useda | Sequence (5′ to 3′)b | Genome localization
|
||
|---|---|---|---|---|---|
| BKc | JCd | Human actine | |||
| PV-TMFOR | sn | TCTATTACTAAACACAGCTTACT | 4342–4365 | 4205–4228 | |
| PV-BACK | sn, TM | GGTGCCAACCTATGGAACAG | 4548–4567 | 4408–4427 | |
| PV-PROBE | TM | F-TGGAAAGTCTTTAGGGTCTTCTACCTT-p | 4387–4413 | 4250–4276 | |
| PV-SNFOR | sn | TCTTTAGTRGTATACACAGCAAA | 4307–4329 | 4170–4192 | |
| BK-FOR | sn | ACAGCAAAGCAGGCAAG | 4322–4338 | ||
| JC-FOR | sn | GAGGAATGCATGCAGATCTA | 4229–4248 | ||
| huAct se | Actin | TCACCCACAATGTGCCCATCT | 486–506 | ||
| huAct as | Actin | GTGAGGATCTTCATGAGGTAGTCAGTC | 565–591 | ||
| TMhuAct | Actin | F-ATGCCCTCCCCCATGCCATCCTGCGT-p | 516–541 | ||
Abbreviations: sn, snPCR; TM, TM-PCR.
T, 5-carboxytetramethylrhodamine (5-TAMRA) attached to 5-ethylamino-deoxythymidine; F, 6-carboxyfluorescein attached to the 5′ terminus (FAM); R, A/G (1:1); p, phosphate group attached to the 3′ terminus.
Nucleotide positions are based on the sequence of the huPoV BK genome (accession no. J02038).
Nucleotide positions are based on the sequence of the huPoV JC genome (accession no. J02226).
Nucleotide positions are based on sequence of the human β-actin gene (accession no. AB004047).
FIG. 1.
Schematic representation indicating spacing, positions, and orientations of primers and the exonuclease probe. Whereas the TM-PCR utilizes the same primer set and exonuclease probe for quantification of both huPoVs, the snPCR is based on different primers for BKV and JCV in the second amplification round (three-primer PCR).
Quantitative polyomavirus TM-PCR assay.
The TM-PCR primers and exonuclease probe are located in the VP1 regions of the huPoV BK and huPoV JC genomes and were selected to hybridize to BKV DNA as well as to JCV DNA. For oligonucleotide data, see Table 1 and Fig. 1. Each 50 μl of PCR mixture contained 5 μl of template, 500 nM concentrations of each primer (PV-TMFOR and PV-BACK), 100 nM exonuclease probe (PV-PROBE), 100 μM each dNTP (Gibco), 5 μl of 10× amplification buffer, 1 μM ROX, and 2 U of Platinum Taq DNA polymerase (Gibco). After an initial denaturation for 3 min, the sample was subjected to 45 cycles of 94°C for 30 s and 62°C for 30 s. Fluorescence intensity was read automatically during PCR cycling in an ABI Prism 7700 SDS. The total TM-PCR assay time is approximately 110 min.
Quantitative actin TM-PCR.
A primer set and exonuclease probe located in the human actin gene were used to quantify human genomic actin DNA (Table 1). Each 50 μl of PCR mixture contained 5 μl of template, 200 nM concentrations of each primer, 100 nM exonuclease probe, 200 μM each dNTP (Gibco), 5 μl of 10× amplification buffer, 1 μM 6-carboxy-x-rhodamine (ROX), and 2 U of Platinum Taq DNA polymerase (Gibco). After an initial denaturation for 3 min, the sample was subjected to 45 cycles of 94°C for 25 s and 65°C for 50 s. Fluorescence intensity was read automatically during PCR cycling in an ABI Prism 7700 SDS.
Plasmid preparation.
As a positive control for the snPCR, two plasmids were cloned, containing the amplicons of the first amplification round of the snPCR for BKV and JCV, respectively. The TOPO-TA Cloning Kit (Invitrogen) was used, and we obtained plasmids pPBK and pPJC. Since the TM-PCR assay detects BKV DNA as well as JCV DNA, we chose only plasmid pPJC for the construction of calibration curves.
RESULTS
snPCR.
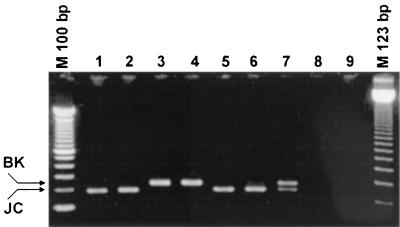
To provide an easy-to-perform PCR assay for the typing of BKV and JCV DNA, the snPCR assay was constructed. Using one general primer together with two variant specific primers in the second round of amplification, we obtained amplicon sizes of 246 bp for BKV and 199 bp for JCV DNA. The bands can easily be distinguished on a 2.5% agarose gel. Typical results of the snPCR are shown in Fig. 2.
FIG. 2.
snPCR. The ethidium bromide-stained 2.5% agarose gel shows the PCR products of six urine samples (lanes 1 to 6), a mix of plasmids pPBK and pPJC as a positive control (lane 7), and two no-template controls (lanes 8 and 9). The different amplicon sizes for BKV (246 bp) and JCV (199 bp) are clearly visible.
Quantitative huPoV TM-PCR assay.
For the quantitative assay, a set of general primers and an exonuclease probe were used to detect BKV DNA as well as JCV DNA. Since the amplicon sizes for BKV and JCV are 226 and 223 bp, respectively, and the two amplicons display similar base compositions, it is to be expected that BKV and JCV DNA are amplified with the same efficiency.
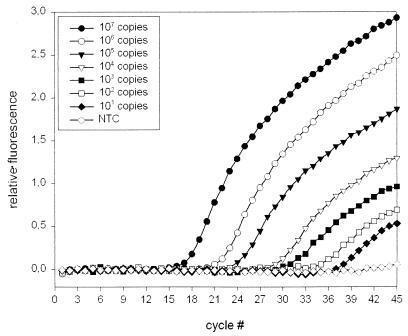
Initially, we examined the ability of the TM-PCR assay to detect huPoV DNA in urine. Serial 10-fold dilutions of plasmid pPJC were prepared in water, in six different huPoV DNA-negative native urine specimens, and in the same six urine specimens diluted 1:10 in water. Figure 3 shows amplification plots of plasmid pPJC in urine diluted 1:10. A range of 107 to 101 plasmids per assay could be detected; as expected, the no-template control (NTC) produced no detectable fluorescence signal. We observed comparable amplification plots for plasmid dilutions in water and for plasmid dilutions in five of the six native urine specimens. However, for one individual native urine specimen we obtained atypical amplification plots, and for low plasmid copy numbers the amplification even failed (data not shown).
FIG. 3.
TaqMan assay for the quantification of huPoV DNA. Amplification plots of serial dilutions from 107 to 101 copies of plasmid pPJC are shown. Relative fluorescence is plotted versus cycle number. Each amplification plot is the result of triplicate experiments.
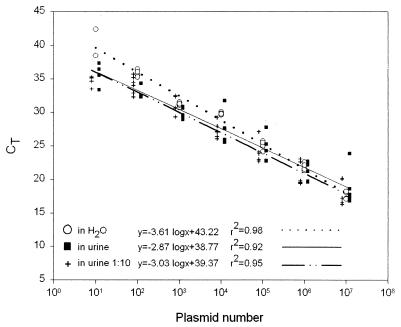
Looking at the corresponding calibration curves (Fig. 4), we observed the best linear correlation between log dilutions of plasmid and threshold cycle number (CT) for the plasmid dilution in water (r2 = 0.98). Although the calibration curves for the plasmid dilution in native urine and in urine diluted 1:10 possess similar characteristics, it is apparent that the use of native urine may lead to significantly increased scattering. Consequently, the use of native urine results in the lowest correlation between log dilutions of plasmid and CT (r2 = 0.92). The plasmid dilution in 1:10-diluted urine results in an acceptable linear correlation (r2 = 0.95).
FIG. 4.
Calibration curves were obtained by correlation of the CT value and the plasmid copy number. CT values were taken from amplifications of serial dilutions of plasmid pPJC in water (open circles), in six native urine specimens (solid squares), and in the same six urine specimens previously diluted 1:10 with water (crosses).
Urine dilution experiments.
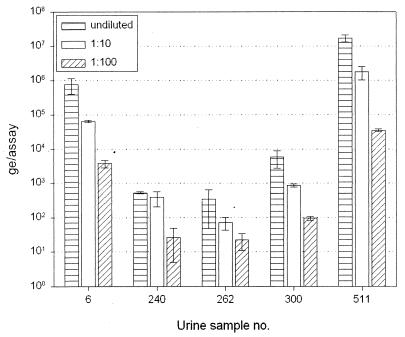
The amount of huPoV DNA in each urine dilution series was measured in triplicate. Results for five representative urine specimens are shown in Fig. 5. In most urine samples, dilution led to a corresponding 1-log-unit decrease in the level of viral DNA detected (Fig. 5), urine specimens 6, 262, 300, and 511). However, we observed a clear inhibitory effect in sample 240, for which the amounts of DNA detected were almost identical in undiluted urine and 1:10-diluted urine. As a further dilution of 1:100 led to the expected 1-log-unit decrease in the level of DNA detected in all specimens, diluting 1:100 seems not to be necessary. Overall, dilution was not necessary in 15 of the 16 (94%) urine specimens investigated. In 1 out of 16 (6%) urine specimens, 1:10 dilution of the native urine was required to dilute the amplification inhibitors. None of the 16 urine specimens investigated required a dilution higher than 1:10. These results indicate that in the majority of cases our quantitative TM-PCR would work with undiluted urine specimens. Nonetheless, 1:10 dilution guarantees sufficient dilution of amplification inhibitors; therefore, urine specimens were subsequently diluted by 1:10.
FIG. 5.
Urine dilution. HuPov DNA was quantified in native, 1:10-diluted, and 1:100-diluted urine. Each group of three bars represents one urine sample.
DNA preparation methods.
First, we compared DNA preparation by proteinase K digestion with the Qiagen DNA preparation kit for eight urine specimens. Although all specimens prepared with the commercial kit (8 of 8 [100%]) were positive for huPoV DNA, only in 3 of 8 (37.5%) specimens prepared by proteinase K digestion could huPoV DNA be detected. Moreover, with the proteinase K digestion, the amount of DNA detected was as much as 100,000-fold lower than the amount of kit-prepared DNA detected (data not shown).
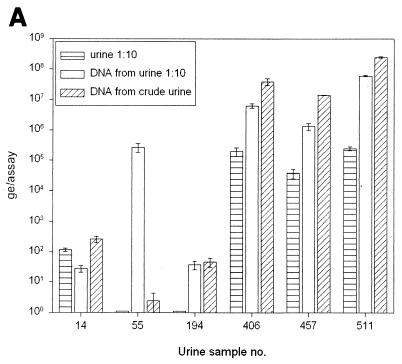
To find out whether Qiagen DNA preparation is superior to the use of diluted urine, we prepared DNA from 20 undiluted urine specimens and their 1:10 dilutions. Subsequently we compared the results with the amounts of DNA in the same urine specimens diluted 1:10, without any preceding DNA preparation step. Typical results are shown in Fig. 6A. For 12 of 20 (60%) urine specimens, DNA preparation from 1:10-diluted urine led to an increase in detectable huPoV DNA levels by a factor of 10, compared to those in the 1:10-diluted nonprepared urine. DNA preparation from undiluted urine could raise the detectable DNA amount by a factor of 100 (urine specimens 406, 457, and 511). However, these results could not be reproduced with every individual urine specimen. For specimen 14, DNA preparation could not improve DNA detectability. Further, for urine specimen 55, DNA preparation was necessary even to detect huPoV DNA, but there was significantly less DNA detectable when native urine was used for DNA preparation. For urine specimen 194, we found no difference in DNA detection between Qiagen preparations from 1:10-diluted and native urine.
FIG. 6.
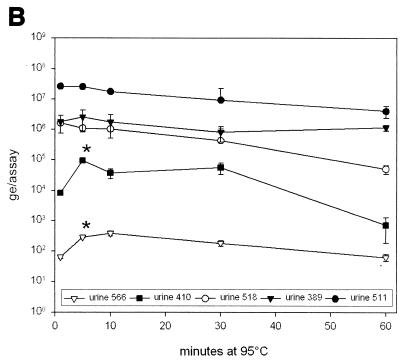
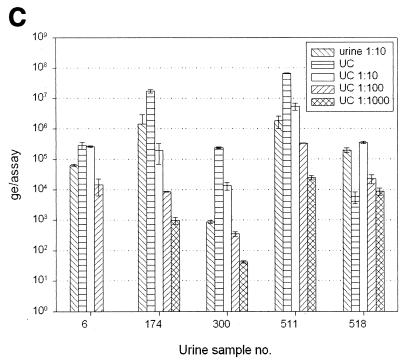
(A) Quantification of huPoV DNA in 1:10-diluted urine and in Qiagen DNA preparations from native urine and from 1:10-diluted urine. It is evident that DNA preparation from 1:10-diluted urine may be beneficial, whereas preparation from native urine can be affected by inhibitory factors, resulting in a decreased amount of detectable DNA (specimens 55 and 194). In most urine samples, DNA can be enriched at least by a factor of 10 by preparation (specimens 406, 457, and 511). (B) Urine was heated to 95°C for 1, 5, 10, 30, and 60 min immediately before use in PCR. Five different urine samples, covering a wide range of DNA contents, were investigated. There was no significant change in detectable DNA amounts as a result of heating longer than 5 min before PCR cycling. (C) Ultracentrifuge (UC) sedimentation of urine. The urine was sedimented and used for TM-PCR either pure or diluted with water. It is demonstrated that ultracentrifuge pretreatment of urine leads to enrichment of detectable huPoV DNA in some samples (samples 174, 300, and 511). In other samples PCR inhibitors seem to be enriched along with the virus (samples 5 and 518).
To determine the loss of viral DNA during DNA preparation, we diluted a purified huPoV suspension 1:10, 1:100, and 1:1,000. Thereafter, each dilution was divided into two aliquots; one aliquot was directly used for PCR, and the other was subjected to DNA preparation prior to PCR. In all three dilutions, we determined a loss of as much as 75% of DNA during preparation (data not shown).
Prolonged heat denaturation.
In order to inactivate amplification inhibitors in urine and improve virus disintegration, the initial heat denaturation step was prolonged. We chose five urine specimens (diluted 1:10) containing approximately 102 to 107 virus genome equivalents (ge)/assay. Figure 6B shows the correlation between additional heating time and quantified huPoV DNA. For low virus copy numbers, a significant increase in DNA detected could be observed by heating for as long as 5 min (urine specimens 410 [P = 0.021] and 566 [P = 0.004]). This step is usually performed as the initial denaturation step in PCR. However, heating longer than 10 min led rather to a decrease in detectable huPoV DNA levels in all samples.
Urine sedimentation.
To determine the enrichment factor that could be achieved by ultracentrifugation, 20 1:10-diluted urine specimens were sedimented. The resuspended pellet was either used undiluted or was diluted 1:10, 1:100, or 1:1,000 with double-distilled sterile water. The results of the DNA quantification are presented in Fig. 6C. For individual urine specimens, we obtained enrichment of huPoV DNA by a factor of 10 to 100 (urine specimens 174, 300, and 511), in agreement with the expected enrichment factor of approximately 100. For individual urine specimens, we observed decreases in amounts of detectable huPoV DNA after ultracentrifugation, which can be explained by concurrent enrichment of amplification inhibitors together with the virus (urine specimens 6 and 518). For example, the ultracentrifugation pellet of urine specimen 518 had to be diluted 1:10 in order to give the same result as the 1:10-diluted native urine.
Inhibition of actin amplification.
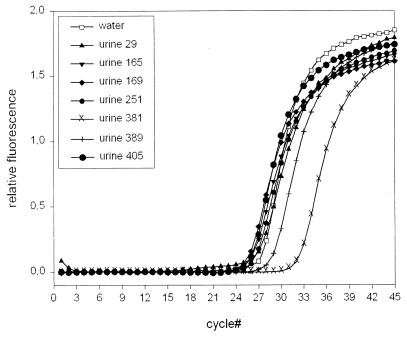
Finally, to determine the influence of the inhibitory factors present in native urine on a further quantitative PCR assay, we added 1 ng of human genomic DNA to undiluted urine specimens or to water as a control. In Fig. 7 the amplification plots of seven selected urine specimens spiked with human DNA are shown together with that of the control. Only in urine specimen 381 was the amplification of the human actin sequence significantly inhibited, as represented by a higher CT value; additionally, urine specimen 389 shows weak inhibition of amplification of the actin gene. All other urine specimens did not impair the actin PCR efficiency.
FIG. 7.
Inhibition of actin amplification caused by native urine. One nanogram of human genomic DNA was added to water and various native urine samples. Amplification plots are given for seven DNA-spiked urine samples and the DNA-spiked water control. The increased CT value for sample 381 reflects inhibition of actin amplification. The other samples display a nearly constant CT, indicating minor inhibitory influences.
Assay variability.
To determine the intra-assay precision of the huPoV TM-PCR assay, three 1:10-diluted urine specimens were simultaneously assayed four times. To determine the interassay precision, identical urine samples were assayed on 6 consecutive days in duplicate. The resulting standard deviations (SD) are given in Table 2. Using 103 copies of plasmid pPJC diluted in water, we determined the intra- and interassay variabilities to be <15 and <20%, respectively (data not shown).
TABLE 2.
TM-PCR assay performance characteristics
| Urine sample no. | No. of ge/assay | Variability (no. of ge [SDa])
|
|
|---|---|---|---|
| Intra-assay | Interassay | ||
| 511 | 4.58 × 106 | ±2.08 × 106 (45) | ±1.92 × 106 (42) |
| 651 | 1,940 | ±679 (35) | ±911 (47) |
| 468 | 902 | ±441 (49) | ±541 (60) |
Expressed as a percentage.
Clinical application.
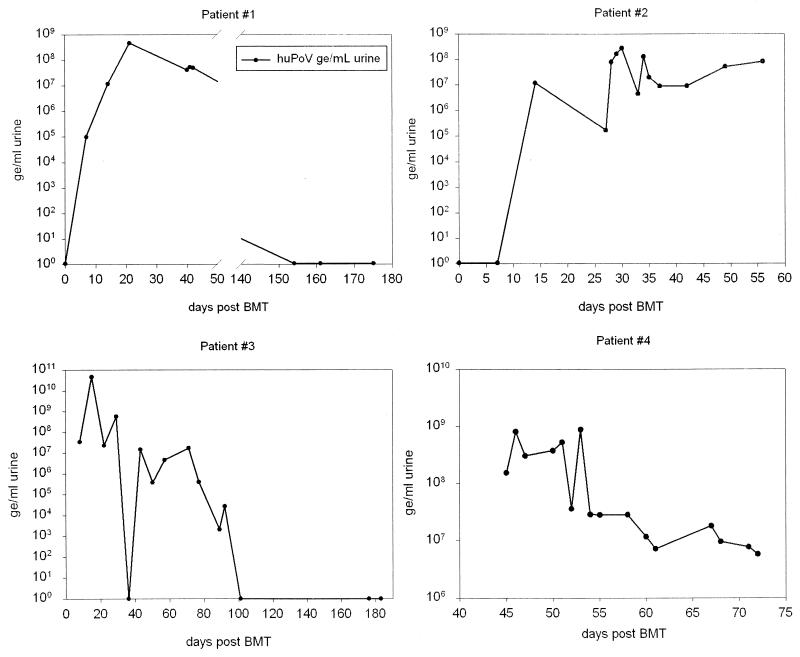
With the aim of demonstrating the clinical applicability of the huPoV TM-PCR assay, we studied consecutive urine samples of 103 patients after BMT. Four typical time courses are shown in Fig. 8. We were able to illustrate the progress of polyomaviral load in the urine of BMT patients during the posttransplantation period.
FIG. 8.
Monitoring of four representative BMT patients. Lines indicate huPoV DNA loads as determined by TM-PCR. As determined by snPCR, all four patients were positive for BKV only during the time course.
DISCUSSION
Most of the methods presently used for differentiation of huPoV utilize either species-specific hybridization, exonuclease digestion of the PCR product, or single PCRs for each species (3, 6, 9, 10). Using our qualitative snPCR assay, BKV and JCV DNA are amplified in one tube and can easily be distinguished by amplicon size only. This assay plainly simplifies the differential diagnosis of huPoV infection. The quantitative TM-PCR is the first real-time quantification assay published for huPoV DNA. It is fast, reproducible, and sensitive, allowing the quantification of huPoV DNA in a linear range between 107 and 101 ge per assay. Neither post-PCR handling nor competitive amplification of reference genes is required, which makes its handling clearly easier than the methods presently used. Moreover, since it is a real-time PCR detection method and quantifies during the log phase of the PCR, it allows an extremely accurate quantification compared to conventional PCR methods, which determine the amount of PCR product in the plateau phase of the PCR.
The TM-PCR assay was utilized to examine and compare different methods presently used for inactivation or elimination of the amplification inhibitors often found in urine. We could clearly demonstrate that the use of untreated 1:10-diluted native urine is sufficient for quantitative detection of huPoV DNA. No inhibitory effects are found in 94% of native urine specimens, which is in accordance with previously reported data (20). Moreover, omitting time- and money-consuming urine handling allows for cheap and fast diagnosis.
The detection of huPoV DNA in urine generally could not be improved either by heating or by proteinase K digestion. The still commonly used proteinase K digestion (3, 9, 18) resulted in a reduced amount of amplifiable DNA. However, the amount of amplifiable huPoV DNA could be increased by using a commercial kit for DNA preparation from native as well as from 1:10-diluted urine. But, interestingly, the yield of kit-prepared DNA was sometimes dramatically reduced, probably by inhibition of the preparation. Therefore we determined the general loss of DNA during DNA preparation from density gradient-purified virus particles to be 75%. This can be caused by the fact that the total amount of DNA in urine is too small to fit the optimal conditions for the commercial kit. Although for most urine samples the use of ultracentrifugation enrichment led to the expected virus enrichment by a factor of 10 to 100, in some specimens obviously a concurrent accumulation of amplification inhibitors occurred.
Summing up, since the inhibitory effects on amplification and DNA preparation described above are hardly predictable, we recommend the use of 1:10-diluted urine as the ideal compromise of effort on the one hand and reliability of results on the other hand. The assay presented here, as it stands, has proven useful for monitoring of BMT patients in general and especially for monitoring of antiviral treatment with cidofovir, as recently described (16). Moreover, it seems to be useful for shedding light on the connection between polyomavirus reactivation and HC following BMT, which has been the subject of controversy.
REFERENCES
- 1.Arthur R R, Shah K V, Baust S J, Santos G W, Saral R. Association of BK viruria with hemorrhagic cystitis in recipients of bone marrow transplants. N Engl J Med. 1986;315:230–234. doi: 10.1056/NEJM198607243150405. [DOI] [PubMed] [Google Scholar]
- 2.Arthur R R, Shah K V. Occurrence and significance of papovaviruses BK and JC in the urine. Prog Med Virol. 1989;36:42–61. [PubMed] [Google Scholar]
- 3.Arthur R R, Dagostin S, Shah K V. Detection of BK virus and JC virus in urine and brain tissue by the polymerase chain reaction. J Clin Microbiol. 1989;27:1174–1179. doi: 10.1128/jcm.27.6.1174-1179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azzi A, et al. Monitoring of polyomavirus BK viruria in bone marrow transplantation patients by DNA hybridization assay and by polymerase chain reaction: an approach to assess the relationship between BK viruria and hemorrhagic cystitis. Bone Marrow Transplant. 1994;14:235–240. [PubMed] [Google Scholar]
- 5.Azzi A, Zakrzewska K, Cesaro A, Fanci R, Bosi A. Monitoring of BKV viral load in urine of bone marrow transplantation patients and hemorrhagic cystitis. Acta Microbiol Immunol Hung. 1999;46:370–371. [Google Scholar]
- 6.Bedi A, Miller C B, Hanson J L, Goodman S, Ambinder R F, Charache P, Arthur R R, Jones R J. Association of BK virus with failure of prophylaxis against hemorrhagic cystitis following bone marrow transplantation. J Clin Oncol. 1995;13:1103–1109. doi: 10.1200/JCO.1995.13.5.1103. [DOI] [PubMed] [Google Scholar]
- 7.Biel S S, Gelderblom H R. Diagnostic electron microscopy is still a timely and rewarding method. J Clin Virol. 1999;13:105–119. doi: 10.1016/S1386-6532(99)00027-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogdanovic G, Brytting M, Cinque P, Grandien M, Fridell E, Ljungman P, Lönnqvist B, Hammarin A L. Nested PCR for detection of BK virus and JC virus DNA. Clin Diagn Virol. 1994;2:211–220. doi: 10.1016/0928-0197(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 9.Bogdanovic G, Ljungman P, Wang F, Dalianis T. Presence of human polyomavirus DNA in the peripheral circulation of bone marrow transplant patients with and without hemorrhagic cystitis. Bone Marrow Transplant. 1996;17:573–576. [PubMed] [Google Scholar]
- 10.Chan P K, Ip K W, Shiu S Y, Chiu E K, Wong M P, Yuen K Y. Association between polyomaviruria and microscopic haematuria in bone marrow transplant recipients. J Infect. 1994;29:139–146. doi: 10.1016/s0163-4453(94)90602-5. [DOI] [PubMed] [Google Scholar]
- 11.Chang D, Wang M, Ou W C, Tsai R T, Fung C Y, Hwang Y J. A simple method for detecting human polyomavirus DNA in urine by the polymerase chain reaction. J Virol Methods. 1996;58:131–136. doi: 10.1016/0166-0934(95)02001-2. [DOI] [PubMed] [Google Scholar]
- 12.Chernesky M A, Jang D, Sellors J, Luinstra K, Chong S, Castriciano S, Mahony J B. Urinary inhibitors of polymerase chain reaction and ligase chain reaction and testing of multiple specimens may contribute to lower assay sensitivities for diagnosing Chlamydia trachomatis-infected women. Mol Cell Probes. 1997;11:243–249. doi: 10.1006/mcpr.1997.0109. [DOI] [PubMed] [Google Scholar]
- 13.Childs R, Sanchez C, Engler H, Preuss J, Rosenfeld S, Dunbar C, van Rhee F, Plante M, Phang S, Barrett A J. High incidence of adeno- and polyomavirus-induced hemorrhagic cystitis in bone marrow allotransplantation for hematological malignancy following T cell depletion and cyclosporine. Bone Marrow Transplant. 1998;22:889–893. doi: 10.1038/sj.bmt.1701440. [DOI] [PubMed] [Google Scholar]
- 14.Garcia de Viedma D, Alonso R, Miralles P, Berenguer J, Rodriguez-Creixems M, Bouza E. Dual qualitative-quantitative nested PCR for detection of JC virus in cerebrospinal fluid: high potential for evaluation and monitoring of progressive multifocal leukoencephalopathy in AIDS patients receiving highly active antiretroviral therapy. J Clin Microbiol. 1999;37:724–728. doi: 10.1128/jcm.37.3.724-728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Held, T. K., S. S. Biel, A. Nitsche, A. Kurth, S. Chen, H. R. Gelderblom, and W. Siegert. Treatment of BK-virus-associated haemorrhagic cystitis and simultaneous CMV reactivation with cidofovir. Bone Marrow Transplant., in press. [DOI] [PubMed]
- 16.Minor P D. Concentration and purification of viruses. In: Cann A J, editor. Virus culture: a practical approach. Oxford, United Kingdom: Oxford University Press; 1999. pp. 61–80. [Google Scholar]
- 17.Nitsche A, Steuer N, Schmidt C A, Land O, Siegert W. Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin Chem. 1999;45:1932–1937. [PubMed] [Google Scholar]
- 18.Saitoh K, Sugae N, Koike N, Akiyama Y, Iwamura Y, Kimura H. Diagnosis of childhood BK virus cystitis by electron microscopy and PCR. J Clin Pathol. 1993;46:773–775. doi: 10.1136/jcp.46.8.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santis R, Azzi A. Duplex polymerase chain reaction for the simultaneous detection of the human polyomavirus BK and JC DNA. Mol Cell Probes. 1996;10:325–330. doi: 10.1006/mcpr.1996.0044. [DOI] [PubMed] [Google Scholar]
- 20.Toye B, Woods W, Bobrowska M, Ramotar K. Inhibition of PCR in genital and urine specimens submitted for Chlamydia trachomatis testing. J Clin Microbiol. 1998;36:2356–2358. doi: 10.1128/jcm.36.8.2356-2358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]