Abstract
The major antigenic protein 2 (MAP2) homolog of Ehrlichia chaffeensis was cloned and expressed. The recombinant protein was characterized and tested in an enzyme-linked immunosorbent assay (ELISA) format for potential application in the serodiagnosis of human monocytic ehrlichiosis. The recombinant protein, which contained a C-terminal polyhistidine tag, had a molecular mass of approximately 26 kDa. The antigen was clearly identified by Western immunoblotting using antihistidine antibody. However, immune sera failed to react with the recombinant on immunoblots when the antigen was denatured by heat or reduced using β-mercaptoethanol. The recombinant MAP2 (rMAP2) was used in an ELISA format with 60 blinded serum samples. Twenty of the serum samples were previously demonstrated to contain antibodies reactive with E. chaffeensis by indirect immunofluorescence assays (IFAs). The remaining 40 samples were seronegative. All samples negative by IFA were also found to be negative for antibodies against the rMAP2 of E. chaffeensis by using the ELISA. Only 1 of 20 IFA-positive samples tested negative in the rMAP2 ELISA. There was 100% agreement using IFA-negative samples and 95% agreement using IFA-positive samples, resulting in a 97.5% overall agreement between the two assays. These data suggest that the rMAP2 homolog of E. chaffeensis may have potential as a test antigen for the serodiagnosis of human monocytic ehrlichiosis. To our knowledge, this recombinant is unique because it is thus far the only E. chaffeensis recombinant antigen that has been shown to work in an ELISA format.
Clinical findings associated with Ehrlichia chaffeensis infection (human monocytic ehrlichiosis [HME]) and infection with the human granulocytic ehrlichiosis agent are similar, consisting of fever, headache, and myalgia. Common laboratory findings for both diseases include leukopenia, thrombocytopenia, and elevated levels of hepatic enzymes (6). There is considerable antigenic cross-reactivity between the Ehrlichia spp. and other closely related species such as Cowdria ruminantium (4, 5, 8, 10, 13, 14, 19). Although a newly developed recombinant enzyme-linked immunosorbent assay (ELISA) for the diagnosis of human granulocytic ehrlichiosis appears to have good specificity (7), the presently available serologic tests for HME are unable to discriminate between infections caused by different Ehrlichia spp. Although several recombinant proteins have been evaluated for the serologic diagnosis of HME (16, 20, 21), the immunofluorescence assay (IFA), using in vitro-cultured, whole Ehrlichia organisms, is still the preferred laboratory method of serodiagnosis (18). Improved specificity for diagnosis of E. chaffeensis infection may be available by use of immunoblot analysis (3); however, DNA sequence analysis is presently the only reliable method for specific identification of Ehrlichia spp.
A 19-kDa protein, major surface protein 5 (MSP5), of Anaplasma marginale has been produced as a recombinant protein and is currently being used as a diagnostic test to identify A. marginale-infected cattle (9). This protein was found to be conserved in all recognized Anaplasma species, and when used with monoclonal antibodies in a competitive ELISA format, it was able to detect cattle persistently infected with A. marginale but not other closely related organisms (9, 17). A 21-kDa protein with 55.5% amino acid identity to MSP5 of A. marginale was identified in the closely related rickettsia C. ruminantium (11). The gene encoding this protein, major antigenic protein 2 (MAP2), was isolated, cloned, and expressed in Escherichia coli. The recombinant MAP2 (rMAP2) was shown to be recognized by sera from cattle, sheep, and goats infected with C. ruminantium (11). The map2 gene, like the msp5 gene of A. marginale, was also found to be well conserved between various isolates of C. ruminantium, and homologs of the map2 and msp5 genes from both Ehrlichia canis and E. chaffeensis were identified (2). Amino acid sequence analysis of the MAP2 homologs from E. chaffeensis and E. canis revealed 83.4 and 84.4% identities, respectively, with MAP2 from C. ruminantium (2).
In this study, we report the cloning and expression of the map2 homolog from E. chaffeensis and examine the potential value of the rMAP2 homolog for the serodiagnosis of HME.
MATERIALS AND METHODS
Source of E. chaffeensis organisms and DNA.
E. chaffeensis (Arkansas isolate) was kindly provided by Jacqueline E. Dawson and James G. Olsen, Centers for Disease Control, Atlanta, Ga. Organisms were grown in the canine macrophage cell line DH82 in Eagle's minimum essential medium containing 10% fetal bovine serum, 26 mM sodium bicarbonate, and 2 mM l-glutamine at 34°C. Cells were harvested when 90 to 100% of them were infected, and ehrlichiae were purified as previously described (5). Genomic DNA of E. chaffeensis was isolated by treatment of purified organisms with 5 mg of lysozyme per ml, 100 μg of proteinase K per ml, and 2% (wt/vol) sodium dodecyl sulfate (SDS), followed by phenol-chloroform extraction and ethanol precipitation (11).
Amplification of the map2 gene of E. chaffeensis.
Primers, which corresponded to the sequences encoding the predicted mature protein of the map2 gene, were synthesized by Genosys Biotechnologies Inc., The Woodlands, Tex. The forward primer ARA3 (5′ GGCAATTTTTCTAGGATATTCCTACGTAACA 3′) and reverse primer ARA4 (5′ GAGATATTGTTTTATTATAGATAGTAACTT 3′) were designed for in-frame insertion of amplicons into the pTrcHis2-TOPO vector (Invitrogen Corporation, Carlsbad, Calif.). The beginning of the predicted mature protein corresponds to nucleotide 46 of the open reading frame. The nucleotide sequence of the map2 gene of E. chaffeensis has been previously reported (2) and assigned GenBank accession number AF117731. Amplification was performed using Taq DNA polymerase in order to produce amplicons with the necessary 3′ A overhangs needed for ligation into the TOPO vector. Briefly, genomic DNA (10 ng) was amplified using 0.5 μM concentrations of primers ARA3 and ARA4 and 1.25 U of Taq DNA polymerase in 2 mM deoxynucleoside triphosphates–10 mM Tris-HCl (pH 8.8)–50 mM KCl–1.5 mM MgCl2. PCR assays were performed at 94°C for 3 min, followed by 10 cycles of denaturing at 94°C for 15 s, annealing at 43°C for 1 min, and extension at 72°C for 7 min. This was followed by 25 cycles of denaturing at 94°C for 15 s, annealing at 49°C for 1 min, and extension at 72°C for 7 min. A final extension step at 72°C was performed for 7 min. Amplicons were analyzed by gel electrophoresis on a 1% agarose gel in 1× TBE buffer (89 mM Tris, 89 mM boric acid, and 2 mM disodium EDTA).
Cloning and sequencing of E. chaffeensis map2.
Amplicons were inserted into the pTrcHis2-TOPO vector (Invitrogen Corporation) according to the manufacturer's recommendations. Recombinant plasmids were transformed into E. coli (One Shot cells; Invitrogen Corporation), and transformants were grown on Luria-Bertani (LB) agar plates in the presence of ampicillin (50 μg/ml). Colonies were selected and incubated in LB broth in the presence of ampicillin (50 μg/ml) overnight at 37°C with vigorous shaking. Plasmid DNA was extracted by a rapid miniprep method (22), reconstituted in Tris-EDTA buffer (pH 8.0) containing 1.0 μg of DNase-free RNase per ml, and analyzed on a 1% agarose gel. Recombinant clones containing the map2 homologs of E. chaffeensis were digested with restriction enzymes (NcoI and DraI) to ensure the correct orientation of the insert in the plasmid vector. Digested DNA was analyzed on a 1% agarose gel. The DNA sequences of both strands of the 570-bp insert of pTrcHis2-TOPO K1 were determined by the DNA Sequencing Core Laboratory (University of Florida, Gainesville) using forward and reverse primers based on vector sequences in flanking regions.
Production and purification of E. chaffeensis recombinant MAP2.
Transformed cells containing the map2 gene homologs of E. chaffeensis were incubated with vigorous shaking at 37°C in LB broth containing 50 μg of ampicillin per ml to an optical density at 600 nm of 0.6. Protein production was induced with 1 mM isopropyl-β-d-thiogalactopyranoside, and cells were incubated with vigorous shaking at 37°C for an additional 5 h. Recombinant proteins were purified by immobilized metal affinity chromatography (ProBond resin; Invitrogen Corporation) and isolated under native, nondenaturing conditions using 50, 200, 350, and 500 mM imidizole elution buffers according to the manufacturer's recommendations. Fractions containing the rMAP2 homolog were identified by SDS-polyacrylamide gel electrophoresis and staining with Coomassie blue. The rMAP2 protein contained a C-terminal polyhistidine tag for purification using affinity chromatography. The authenticity of the rMAP2 homolog of E. chaffeensis was evaluated by Western immunoblot and slot blot analysis using a horseradish peroxidase (HRP)-conjugated antihistidine antibody (Invitrogen Corporation) and known E. chaffeensis IFA-positive human serum. This serum sample was collected from an individual with a history of tick exposure, clinical and laboratory finding consistent with HME, and acute- and convalescent-phase E. chaffeensis, IFA titers of 1:160 and 1:320, respectively.
SDS-polyacrylamide gel electrophoresis.
The rMAP2 protein concentrations were determined by the Coomassie blue G dye-binding assay as previously described (15). The proteins were dissolved in a 3× sample buffer containing 0.1 M Tris (pH 6.8), 5% (wt/vol) SDS, 50% glycerol, and 0.00125% bromophenol blue, either with or without 7.5% β-mercaptoethanol. Samples were either heat denatured at 100°C for 3 min prior to electrophoresis or were electrophoresed without heat denaturation on SDS–10% (wt/vol) polyacrylamide gels. Proteins were electrophoretically transferred to nitrocellulose membranes (Hybond ECL; Amersham International plc, Little Chalfont, Buckinghamshire, England) and fixed in 25 mM Tris–191.8 mM glycine–20% methanol as described previously (1).
Antibodies and antisera.
HRP-labeled antihistidine antibody (Anti-His(C-term)-HRP; Invitrogen Corporation) was used as a positive control for the rMAP2 homolog on immunoblot assays. Sixty serum samples from the Wadsworth Center, New York State Department of Health, Albany, N.Y., and the University of Florida College of Medicine, Gainesville, Fla., were evaluated for antibodies to the rMAP2 homolog of E. chaffeensis. Investigators were blinded as to the immune reactivities of the samples. Twenty of the serum samples were previously demonstrated to contain antibodies reactive with E. chaffeensis (Arkansas strain) by IFA testing, and 40 samples were IFA negative. The IFA-positive samples were obtained from patients with presenting clinical signs consistent with human ehrlichioses. Ten of these samples had IFA titers of 1:160, and the other 10 had titers of 1:320 or greater. Of the 40 IFA-negative samples, 20 were from clinically ill patients, none of whom were diagnosed with HME, and 20 samples were from clinically normal individuals during well-patient visits.
Western immunoblot and slot blot analysis.
Nitrocellulose membranes containing electrophoretically transferred proteins or proteins directly blotted using a SlotBlot (Hoefer Scientific Instruments, San Francisco, Calif.) were blocked for 1 h with 5% (wt/vol) skim milk in 1× phosphate-buffered saline (PBS) with 0.25% Tween 20 and washed with 1% (wt/vol) milk in 1× PBS with 0.25% Tween 20 as described previously (1). The membranes were probed with either HRP-labeled antihistidine antibodies at a dilution of 1/15,000 or E. chaffeensis IFA-positive immune sera at a dilution of 1/300. As a negative control, noninfected human serum was used at a dilution of 1/300. Membranes were then washed with 1% (wt/vol) milk in 1× PBS as described previously (1) and reacted with a secondary antibody, HRP-conjugated goat anti-human immunoglobulin G (whole molecule; Sigma Chemical Co., St. Louis, Mo.), at a dilution of 1/40,000. Membranes were processed for enhanced chemiluminescence with detection reagents containing luminol (SuperSignal Substrate; Pierce, Rockford, Ill.) as a substrate and were exposed to X-ray film (Hyperfilm-MP; Amersham International plc) to visualize the bound antibody.
Indirect ELISA.
Polystyrene microtiter plates (Maxi Sorp; Nunc, Roskilde, Denmark) were coated with 100 μl (per well) of purified rMAP2 homolog of E. chaffeensis (2 μg/ml) in 0.05 M carbonate-bicarbonate buffer (pH 9.6) (Sigma Chemical Co.) and incubated overnight at 4°C. The wells were then washed four times with wash buffer containing 1× PBS and 0.5% (vol/vol) Tween 20 and blocked for 60 min at room temperature with 1% (wt/vol) bovine serum albumin (BSA) in 1× PBS. The plates were washed four times as described above and incubated for 60 min at room temperature with test sera at 1:100, 1:300, 1:1,000, 1:3,000, and 1:10,000 dilutions in 1.0% (wt/vol) BSA in 1× PBS (100 μl). The wells were again washed four times and incubated at room temperature for 60 min in the presence of alkaline phosphatase-conjugated goat anti-human immunoglobulin G (whole molecule; Sigma Chemical Co.) at a dilution of 1:5,000 in 1% (wt/vol) BSA in 1× PBS. The wells were again washed four times, and the substrate, p-nitrophenylphosphate (1 mg/ml) in 0.05 M carbonate-bicarbonate buffer (pH 9.6) (Sigma Chemical Co.), was added at 100 μl per well and incubated for 60 min at room temperature. Absorbance was measured at 405 nm using a Rainbow plate reader (Tecan U.S. Inc., Durham, N.C.). Serum samples from four clinically healthy individuals were used to establish the cutoff values for determining if a test sample was positive or negative. Negative controls were used each time an ELISA was performed. A test sample was considered positive if the absorbance reading was at least double that of the negative controls.
RESULTS
Analysis of the predicted amino acid sequence of the rMAP2 homolog indicated that it differed from the previously reported amino acid sequence of the E. chaffeensis MAP2 at two positions. Position 17 in the native E. chaffeensis MAP2 contains the polar amino acid serine, while the rMAP2 has a nonpolar amino acid, tryptophan, at this position. A conserved amino acid substitution also occurred at position 195 of the reported sequence, where an aspartic acid was replaced with asparagine in the recombinant protein. Both the native and recombinant map2 sequences were derived from the Arkansas strain of E. chaffeensis. We have not investigated whether these differences were the result of PCR amplification errors or whether they represent true polymorphism in regions of the gene.
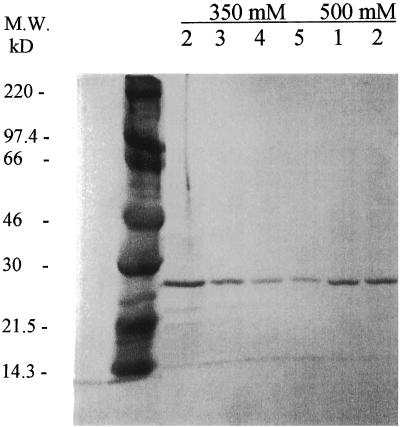
Coomassie blue-stained SDS-polyacrylamide gels were used to evaluate each of five fractions of 50, 200, 350, and 500 mM imidizole elution buffer for the presence of the rMAP2 homolog. A single protein with a molecular mass of approximately 26 kDa was identified in fractions 2 through 5 of the 350 mM imidizole buffer and fractions 1 and 2 of the 500 mM imidizole buffer (Fig. 1). This protein contains a fusion peptide, increasing the size of the recombinant seen on Coomassie blue-stained gels. Based on the amino acid sequence, the calculated mass of the rMAP2 is approximately 21 kDa.
FIG. 1.
Coomassie blue-stained gel of eluted fractions containing the rMAP2 homolog of E. chaffeensis. Lanes 2, 3, 4, and 5, respective fractions eluted with 350 mM imidizole elution buffer; lanes 1 and 2, respective fractions eluted with 500 mM imidizole buffer. Molecular size (M.W.) standards are in the unmarked lane on the left.
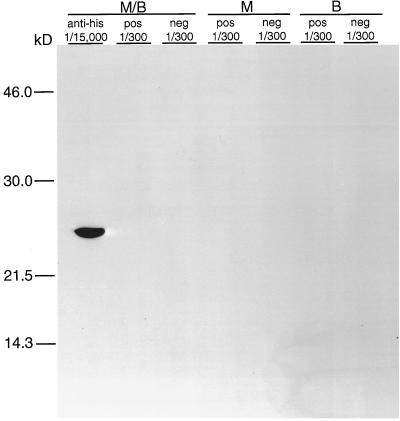
Western immunoblot analysis was done to evaluate reactivity of the rMAP2 homolog with immune serum from a patient who was seropositive for infection with E. chaffeensis. In these experiments, purified proteins were run on SDS-polyacrylamide gel electrophoresis under denaturing conditions. Normal human serum was used as a negative control on immunoblots, and HRP-labeled antihistidine antibody was used as a positive control for detection of the recombinant protein. The 26-kDa recombinant protein was clearly identified using the antihistidine antibody (Fig. 2). However, known positive immune serum failed to react with the recombinant antigen in Western immunoblots.
FIG. 2.
Immunoblot of the rMAP2 homolog of E. chaffeensis either reduced in sample buffer containing β-mercaptoethanol and heat denatured by boiling (M/B), reduced in sample buffer containing β-mercaptoethanol without boiling (M), or boiled in sample buffer without β-mercaptoethanol (B) and reacted with antihistidine antibodies (anti-his), E. chaffeensis IFA-positive immune serum (pos), or E. chaffeensis IFA-negative serum (neg). The fractions (1/300 and 1/15,000) indicate the dilutions of antibody or serum used. Molecular size standards are given on the left.
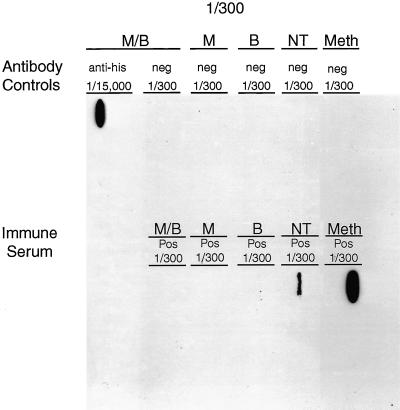
In order to determine if the denaturing conditions were affecting the protein reactivity, slot blotting was performed using the rMAP2 homolog in sample buffers with and without β-mercaptoethanol, with and without boiling, and with and without fixing the membrane in 20% methanol. rMAP2 was recognized by antihistidine antibody even when it was denatured by boiling in β-mercaptoethanol (Fig. 3). However, immune serum recognized rMAP2 only when it was prepared in sample buffer without β-mercaptoethanol and without boiling prior to blotting on the membrane. rMAP2 that was either heat denatured or reduced using β-mercaptoethanol failed to react with immune serum. Fixation of the protein to the membrane using 20% methanol did not abolish the reactivity of the protein with the immune serum but enhanced the signal visualized on the immunoblot (Fig. 3).
FIG. 3.
Slot blot of rMAP2 of E. chaffeensis either reduced in sample buffer containing β-mercaptoethanol and heat denatured by boiling (M/B), reduced in sample buffer containing β-mercaptoethanol without boiling (M), boiled in sample buffer without β-mercaptoethanol (B), in PBS without boiling or β-mercaptoethanol (NT), or in PBS without boiling or mercaptoethanol and with fixing in 20% methanol (Meth). Recombinant antigens were reacted with antibody controls (antihistidine antibodies [anti-his] or E. chaffeensis IFA-negative serum [neg]) or immune sera (E. chaffeensis IFA-positive immune serum [Pos]). The fractions (1/300 and 1/15,000) indicate the dilutions of antibody or serum used.
ELISAs were performed on 60 serum samples which were previously evaluated for antibodies against E. chaffeensis by IFA testing. All samples negative by IFA were also found to be negative for antibodies against the rMAP2 homolog using the ELISA (data not shown). A comparison of rMAP2 ELISA results with E. chaffeensis IFA-positive samples is presented in Table 1. Only 1 of 20 IFA-positive samples tested negative in the rMAP2 ELISA. Therefore, there was 100% agreement using IFA-negative samples and 95% agreement using IFA-positive samples, resulting in a 97.5% overall agreement between the two assays. In samples that were positive by both assays, the rMAP2 ELISA was usually able to detect antibodies at a higher dilution of serum (Table 1).
TABLE 1.
Comparison of rMAP2 ELISA results with 20 E. chaffeensis IFA-positive samples
| No. samples | rMAP2 ELISA titera | E. chaffeensis IFA titer |
|---|---|---|
| 1 | Negative | 1:320 |
| 1 | 1:100 | 1:160 |
| 2 | 1:100 | 1:320 |
| 1 | 1:100 | >1:640 |
| 4 | 1:300 | 1:160 |
| 1 | 1:1,000 | 1:160 |
| 1 | 1:1,000 | >1:640 |
| 4 | 1:3,000 | 1:160 |
| 2 | 1:3,000 | 1:320 |
| 3 | 1:3,000 | >1:640 |
A sample was considered positive in the rMAP2 ELISA when the absorbance reading of the test serum at a specific dilution was two or more times the absorbance reading of identical dilutions of standard negative control sera. A negative ELISA result indicates a titer of <1:100.
DISCUSSION
We have successfully cloned the map2 gene from E. chaffeensis and have purified rMAP2 translated from the open reading frame encoding the predicted mature protein. This recombinant is unique and to our knowledge is thus far the only E. chaffeensis recombinant antigen that has been shown to work in an ELISA format. The predicted amino acid sequence of this protein has significant homology with the sequences of the 21-kDa MAP2 protein of C. ruminantium and the 19-kDa MSP5 protein of A. marginale (2), and the protein is distinctly different in size and sequence from the 28-kDa outer membrane protein of E. chaffeensis, the p28 antigen. Genes encoding both the MAP2 of C. ruminantium and the MSP5 of A. marginale were found to be well conserved between various isolates of organisms within the respective species, and recombinant proteins developed from these genes have been used as diagnostic test antigens to identify infected animals (2, 9, 11, 17). Importantly, development of a competitive ELISA using a monoclonal antibody against MSP5 resulted in an assay that was capable of detecting both acutely and persistently infected cattle. This MSP5 ELISA did not cross-react with sera from cattle infected with closely related organisms (9, 17). Therefore, although these antigens are well conserved within a species, areas of sequence variability between species may provide a way to enhance test specificity.
Interestingly, we were not able to demonstrate reactivity of immune sera with rMAP2 of E. chaffeensis using Western immunoblot assays. Using slot blot assays, we further confirmed that heat denaturation or reduction of disulfide bonds by β-mercaptoethanol destroys important structural epitopes of the antigen needed for antibody recognition. Fixing the membrane-bound protein in 20% methanol did not inhibit reactivity with immune sera but resulted in enhancement of the signal, probably associated with increased amounts of protein being fixed to the nitrocellulose membrane. Similarly, conformationally dependent epitopes were also identified on MSP5 of A. marginale (12). In that study, reduction of disulfide bonds using dithiothreitol, followed by treatment with iodoacetamide to modify sulfhydryl groups, completely abolished monoclonal antibody binding to MSP5 in dot blot assays. In addition, Chen et al. identified 20 immunogenic proteins of E. chaffeensis using rabbit and human E. chaffeensis antisera in Western immunoblot assays (4) and showed that one protein of approximately 22 kDa was heat labile, while all of the other identified immunogens were heat stable. Based on its size and heat-labile nature, the 22-kDa protein could be identical to the MAP2 protein described here. In the same study (4), the 22-kDa protein of E. chaffeensis cross-reacted only with anti-E. canis immune serum, whereas the other major immunogenic antigens of 44, 55, and 66 kDa all cross-reacted with antisera against E. canis, Ehrlichia ewingii, Ehrlichia risticii, and Ehrlichia sennetsu. This gives further indication that the rMAP2 protein could be used as a sensitive and specific antigen in a diagnostic test.
ELISAs performed using the rMAP2 homolog of E. chaffeensis were in 100% agreement with the IFA test results when IFA-negative serum samples were evaluated. There was 95% agreement between these two assays using E. chaffeensis IFA-positive samples. Differences in reactivity between the ELISA and IFA could be explained by the use of a single recombinant protein versus the cultured, whole organisms used in the IFA. This could result in false-negative results using the rMAP2 ELISA. Alternately, IFA testing may result in false-positive results due to the difficulty in distinguishing true-positive reactions from nonspecific antibody binding and/or to an increased likelihood of cross-reactive antigens. In samples that were positive by both assays, the rMAP2 ELISA was usually able to detect antibodies at a higher dilution of serum. This may indicate enhanced sensitivity of the ELISA over the E. chaffeensis IFA and in the future may provide for a more sensitive assay.
These data, combined with information gathered from previous work involving the MSP5 of A. marginale and MAP2 of C. ruminantium, provide evidence that the MAP2 homolog of E. chaffeensis is a potential candidate for the serologic diagnosis of human ehrlichioses. Because of the antigenic similarities between organisms within the Ehrlichia species (4), and possibly closely related genera (8), it may be necessary to design a competitive ELISA based on the inhibition of monoclonal antibodies, similar to the assay developed using MSP5 of A. marginale. An assay such as this, although presently not available, would provide a convenient and cost-effective way to analyze large numbers of samples for clinical use and epidemiologic studies. Further investigation is needed to better characterize the MAP2 antigen of E. chaffeensis with regard to the sensitivity and specificity of this protein as a diagnostic antigen.
ACKNOWLEDGMENTS
This study was supported by a grant from the University of Florida, Division of Sponsored Research, project UPN 98062369. This work was supported in part by a grant from the Centers for Disease Control and Prevention (815-3478A) of the Public Health Service.
REFERENCES
- 1.Alleman A R, Barbet A F. Evaluation of Anaplasma marginale major surface protein 3 (MSP3) as a diagnostic test antigen. J Clin Microbiol. 1996;34:270–276. doi: 10.1128/jcm.34.2.270-276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowie M V, Reddy G R, Semu S M, Mahan S M, Barbet A F. Potential value of major antigenic protein 2 for serological diagnosis of heartwater and related ehrlichial infections. Clin Diagn Lab Immunol. 1999;6:209–215. doi: 10.1128/cdli.6.2.209-215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brouqui P, Lecam C, Olson J, Raoult D. Serologic diagnosis of human monocytic ehrlichiosis by immunoblot analysis. Clin Diagn Lab Immunol. 1994;1:645–649. doi: 10.1128/cdli.1.6.645-649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S M, Dumler J S, Feng H M, Walker D H. Identification of the antigenic constituents of Ehrlichia chaffeensis. Am J Trop Med Hyg. 1994;50:52–58. [PubMed] [Google Scholar]
- 5.Chen S M, Popov V L, Feng H M, Walker D H. Analysis and ultrastructural localization of Ehrlichia chaffeensis proteins with monoclonal antibodies. Am J Trop Med Hyg. 1996;54:405–412. doi: 10.4269/ajtmh.1996.54.405. [DOI] [PubMed] [Google Scholar]
- 6.Fishbein D B, Dawson J E, Robinson L E. Human ehrlichiosis in the United States, 1985 to 1990. Ann Intern Med. 1994;120:736–743. doi: 10.7326/0003-4819-120-9-199405010-00003. [DOI] [PubMed] [Google Scholar]
- 7.Ijdo J W, Caiyun W U, Magnarelli L A, Fikrig E. Serodiagnosis of human granulocytic ehrlichiosis by a recombinant HGE-44-based enzyme-linked immunosorbent assay. J Clin Microbiol. 1999;37:3540–3544. doi: 10.1128/jcm.37.11.3540-3544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly P J, Matthewman L A, Mahan S M, Peter T. Serologic evidence for antigenic relationships between Ehrlichia canis and Cowdria ruminantium. Res Vet Sci. 1994;56:176–174. doi: 10.1016/0034-5288(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 9.Knowles D P, Torioni de Echaide S, Palmer G H, McQuire T C, Stiller D, McElwain T. Antibody against an Anaplasma marginale MSP5 epitope common to tick and erythrocyte stages identifies persistently infected cattle. J Clin Microbiol. 1996;34:2225–2230. doi: 10.1128/jcm.34.9.2225-2230.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnarelli L A, Stafford K C, Mather T N, Yeh M T, Horn K D, Dumler J S. Hemocytic rickettsia-like organisms in ticks: serologic reactivity with antisera to Ehrlichiae and detection of DNA of agent of human granulocytic ehrlichiosis by PCR. J Clin Microbiol. 1995;33:2710–2714. doi: 10.1128/jcm.33.10.2710-2714.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahan S M, McGuire T C, Semu S M, Bowie M V, Jongejan F, Rurangirwa F R, Barbet A F. Molecular cloning of a gene encoding the immunogenic 21 kDa protein of Cowdria ruminantium. Microbiology. 1994;140:2135–2142. doi: 10.1099/13500872-140-8-2135. [DOI] [PubMed] [Google Scholar]
- 12.Munodzana D, McElwain T F, Knowles D P, Palmer G H. Conformational dependence of Anaplasma marginale major surface protein 5 surface-exposed B-cell epitopes. Infect Immun. 1998;66:2619–2624. doi: 10.1128/iai.66.6.2619-2624.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyindo M, Kakoma I, Hansen R. Antigenic analysis of four species of the genus Ehrlichia by use of protein immunoblots. J Am Vet Res. 1991;52:1225–1230. [PubMed] [Google Scholar]
- 14.Ohasi N, Unver A, Zhi N, Rikihisa Y. Cloning and characterization of multigenes encoding the immunodominant 30-kilodalton major outer membrane proteins of Ehrlichia canis and application of the recombinant protein for serodiagnosis. J Clin Microbiol. 1998;36:2671–2680. doi: 10.1128/jcm.36.9.2671-2680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Read S M, Northcote D H. Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal Biochem. 1981;116:53–64. doi: 10.1016/0003-2697(81)90321-3. [DOI] [PubMed] [Google Scholar]
- 16.Unver A, Rikihisa Y, Ohashi N, Cullman L C, Buller R, Storch G A. Western and dot blotting analyses of Ehrlichia chaffeensis indirect fluorescent-antibody assay-positive and -negative human sera by using native and recombinant E. chaffeensis and E. canis antigens. J Clin Microbiol. 1999;37:3888–3895. doi: 10.1128/jcm.37.12.3888-3895.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visser E S, McGuire T C, Palmer G H, Davis W C, Shkap V, Pipano E, Knowles D P. The Anaplasma marginale msp5 gene encodes a 19-kilodalton protein conserved in all recognized Anaplasma species. Infect Immun. 1992;60:5139–5144. doi: 10.1128/iai.60.12.5139-5144.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker D H. Diagnosing human ehrlichioses: current status and recommendations. ASM News. 2000;66:287–290. [Google Scholar]
- 19.Wong S J, Brady G S, Dumler J S. Serological responses to Ehrlichia equi, Ehrlichia chaffeensis, and Borrelia burgdorferi in patients from New York State. J Clin Microbiol. 1997;35:2198–2205. doi: 10.1128/jcm.35.9.2198-2205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X, Crocquet-Valdes P A, Cullman L C, Walker D H. The recombinant 120-kilodalton protein of Ehrlichia chaffeensis, a potential diagnostic tool. J Clin Microbiol. 1996;34:2853–2855. doi: 10.1128/jcm.34.11.2853-2855.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Crocquet-Valdes P A, Cullman L C, Popov V L, Walker D H. Comparison of Ehrlichia chaffeensis recombinant proteins for serologic diagnosis of human monocytic ehrlichiosis. J Clin Microbiol. 1999;37:2568–2575. doi: 10.1128/jcm.37.8.2568-2575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou C, Yang Y, Jong A Y. MiniPrep in ten minutes. BioTechniques. 1990;8:172–173. [PubMed] [Google Scholar]