Abstract
A novel helicobacter with the proposed name Helicobacter aurati (type strain MIT 97-5075c) has been isolated from the inflamed stomachs and ceca of adult Syrian hamsters. The new species is fusiform with multiple bipolar sheathed flagella and periplasmic fibers; it contains urease and gamma-glutamyl transpeptidase. By 16S rRNA sequencing and repetitive element PCR-based DNA fingerprinting, it was found that H. aurati represents a distinct taxon and clusters with Helicobacter muridarum, Helicobacter hepaticus, and Helicobacter sp. MIT 94-022. H. aurati was recovered from hamsters housed in various research and vendor facilities. Further studies are necessary to define its association with disease and other microbiota in hamsters, as well as its impact on research projects involving hamsters. H. aurati (GenBank accession number AF297868) can be used in animal experiments to define the factors that are important for gastric helicobacter pathogenesis.
A number of named Helicobacter species were first found in rodents, including H. muridarum (16), H. hepaticus (4), H. bilis (8), and H. rodentium (21) in mice; H. trogontum (17) in rats; and H. cholecystus (10) and H. mesocricetorum (22) in hamsters. The murine helicobacters in particular are being widely used in experiments to explore the basic biology of the genus Helicobacter, with a major objective of understanding the pathogenesis of H. pylori in the human gastric environment. For the most part, the murine helicobacters colonize extragastric sites, preferentially the large intestine and the liver. An exception is H. muridarum, usually a nonpathogenic inhabitant of the ileal and cecal mucosa, which will colonize the stomachs of mice and apparently induce inflammation (19). Attention is also being given to certain Helicobacter species in mice because these bacteria can confound experimental studies when their presence goes undetected, irrespective of the presence or absence of overt clinical disease (7, 13). Currently, most commercial facilities screen their mouse colonies on a routine basis to establish that they are free of H. hepaticus, since that bacterium was found to cause chronic hepatitis, hepatic cancer, and inflammatory bowel disease in susceptible mouse strains (1, 4, 6, 9, 15, 28).
Natural helicobacter infections in Syrian hamsters (Mesocricetus auratus) have been less well studied than those in mice, although the same potential exists for these microorganisms to interfere with research results or, alternatively, for using hamster helicobacters in animal model development. H. cholecystus has been cultured from the gallbladders of hamsters with histologically confirmed cholangiofibrosis and centrilobular pancreatitis; however, the bacteria were not visualized in affected tissues (10). Zoonotic risk has been attributed to hamsters after H. cinaedi, first recovered from homosexual men with colitis and proctitis (24), was described as a normal component of the intestinal microflora of hamsters (11). The microorganisms were cultured from hamster feces, but the ecological niche of H. cinaedi in hamsters is unknown. H. mesocricetorum has also been cultured from the feces of Syrian hamsters without gastrointestinal pathology (22). Other campylobacter-like bacteria have been identified from the ilea of healthy hamsters (23) and have been cultured from the intestines of hamsters with proliferative enteritis (12); the latter organism did not cause lesions typical of proliferative enteritis when inoculated into experimental hamsters.
Adult Syrian hamsters were submitted to our laboratory for diagnostic evaluation following a number of acute and subacute deaths in a research colony. A novel Helicobacter sp. was cultured from the inflamed stomachs and ceca of these hamsters, and the same microorganism was found in Syrian hamsters from several other facilities that were sampled. This paper describes the phenotypic characteristics and 16S rRNA analysis of the novel Helicobacter sp. with the proposed name Helicobacter aurati. In addition, repetitive sequence-based PCR was performed on the genomic DNA of the novel helicobacter isolates, as well as on related rodent helicobacters, in order to compare their DNA fingerprint patterns. Two other previously undescribed argyrophilic morphotypes were also isolated from the hamster gastrointestinal samples, but these bacteria are not addressed at length in the present report.
MATERIALS AND METHODS
Animals.
Thirty-five hamsters, between 7 and 12 months of age, were examined during the course of the study. Hamsters from two research colonies, the original group (Research Source 1 or RS1) and a second colony (RS2) experiencing similar mortalities, consisted of 12 animals (5 males, 7 females) and 6 animals (4 males, 2 females), respectively. The research hamsters were bred in-house but had historically come from a single commercial source (CS1), an exception being one RS2 male purchased directly from CS1. For comparative purposes, commercially reared hamsters were received from CS1 (1 male, 5 females), CS2 (3 males, 2 females), and CS3 (3 males, 3 females). After euthanasia with carbon dioxide, gastrointestinal samples were collected aseptically for microaerobic culture and PCR and were kept frozen at −70°C until processing. Complete necropsies were performed.
Culture techniques.
Stomach and cecal specimens were cultured from the first research group of hamsters (RS1), whereas in the other four groups, tissue samples of the antrum of the stomach, the body, the liver, and the cecum were cultured separately. Tissues rinsed with sterile phosphate-buffered saline were homogenized in sterile brucella broth (Difco Laboratories, Detroit, Mich.) with 5% fetal calf serum (Summit Biotechnology, Fort Collins, Colo.). Two selective media were used: 5% sheep blood agar with cefoperazone, vancomycin, and amphotericin B (CVA; Remel, Lenexa, Kans.) and 5% sheep blood agar with trimethoprim, vancomycin, and polymyxin (TVP; Remel). The remaining homogenate was put through a 0.45-μm-pore-size filter and was spread on nonselective tryptic soy (TSA) blood agar (Remel). Media were incubated at 37°C in vented jars under microaerobic conditions (10% H2; 10% CO2; 80% N2). The plates were maintained for up to 21 days.
PCR analysis of bacteria and tissues.
DNA was extracted from bacteria and tissues using the High Pure PCR template preparation kit (Roche Molecular Biochemicals, Indianapolis, Ind.). Helicobacter spp.-specific primer pairs C97 and C05 were used to generate 16S rRNA amplicons of 1,200 bases (3). Bacterial cultures were also analyzed using Campylobacter spp.-specific primer pairs, C98 (bp 681 to 698; reverse; 5′-GAT TTT ACC CCT ACA CCA-3′) and C99 (bp 402 to 419; forward; 5′-GCG TGG AGG ATG ACA CCT-3′), which amplified 297 bp fragments.
A PCR sample contained 10 μl of the DNA preparation added to a 90-μl reaction mixture of 1× Taq polymerase buffer, 0.5 μM concentrations of each of the two primers, 200 μM concentrations of each deoxynucleotide, and 200 μg of bovine serum albumin per ml. The samples were heated (94°C, 4 min), centrifuged briefly, and cooled to 58°C, and 0.5 μl of Taq polymerase (Roche Molecular Biochemicals) and 1 μl of Perfect Match PCR enhancer (Stratagene, La Jolla, Calif.) were added. Amplification conditions were as follows: denaturation (94°C, 1 min), annealing (58°C, 3 min), and elongation (72°C, 3 min). Thirty-five cycles were completed before a final elongation step (72°C, 8 min). A 15-μl aliquot of the PCR product was electrophoresed through a 1% agarose gel separation matrix prior to ethidium bromide staining.
Phenotypic characterization.
In addition to motility studies by phase microscopy and visualization by Gram staining, five isolates of the novel helicobacter underwent biochemical and morphological analyses, as described previously (17). The representative isolates were obtained from all four hamster sources where microaerobic culture was successful: MIT 97-5075cT and MIT 97-5310c from RS1, MIT 98-6169a from RS2, MIT 99-5036c from CS1, and MIT 98-6041a from CS2; the designation “a” indicates antral source, and “c” indicates cecal source. Nitrate reduction to nitrite was tested by inoculating the bacteria in a tube of nitrate broth (Remel) microaerobically for 5 days, followed by the addition of test reagents and a subsequent color change. The RapID NH system (Remel) was used according to the manufacturer's instructions to measure gamma-glutamyl transpeptidase and alkaline phosphatase activities; indoxyl acetate hydrolysis was determined using indoxyl acetate-impregnated disks (Remel).
Electron microscopy.
Isolate MIT 99-5036c was grown for 48 h on TSA blood agar plates. Centrifuged cells were suspended in 10 mM Tris-HCl buffer at an approximate concentration of 108 cells per ml. After negative staining with 1% (wt/vol) phosphotungstic acid for 20 to 30 s, the cells were examined with a JEOL model JEM-1200EX transmission electron microscope operating at 100 kV.
Histopathology.
Tissues from the gastrointestinal tract were placed in 10% neutral buffered formalin. After routine paraffin embedding, 5-μm sections were cut and stained with hematoxylin and eosin. Gastric and cecal sections were also stained with a Warthin-Starry silver stain for the identification of argyrophilic microorganisms.
Amplification of 16S rRNA cistrons by PCR and purification of PCR products.
Extracted genomic DNA from three isolates (MIT 97-5075cT, MIT 97-5310c, and MIT 98-6041a) was prepared for sequencing. The 16S rRNA cistrons were amplified with bacterial universal primers F24 (bp 9 to 27; forward; 5′-AGT TTG ATY MTG GCT CAG-3′) and F25 (bp 1525 to 1541; reverse; 5′-AAG GAG GTG WTC CAR CC-3′) as described earlier (2).
16S rRNA sequencing and data analysis.
Following PCR, purified DNA was sequenced using an ABI Prism cycle sequencing kit (BigDye Terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase, FS; PE Applied Biosystems, Foster City, Calif.). The sequencing primers and methods were as listed previously (2).
Sequence data were entered into the program RNA, a program set for data entry, editing, sequence alignment, secondary structure comparison, similarity matrix generation, and dendrogram construction for 16S rRNA in Microsoft QuickBasic for use with PC computers, and were aligned as before (18). Our Campylobacter and Helicobacter database contains over 250 sequences obtained in our laboratory or from GenBank. Dendrograms were constructed by the neighbor-joining method (20).
Fluorophore-enhanced rep-PCR-based DNA fingerprinting of H. aurati isolates.
The interspersed repetitive sequence REP was used as a target for repetitive element PCR (rep-PCR)-based chromosomal profiling (25, 26) on three isolates from different sources (MIT 97-5075cT, MIT 98-6041a, and MIT 98-6169a), as well as on the related species H. hepaticus and H. muridarum. The 18-mer degenerate primer pairs REP1R-Dt (5′-IIINCGNCGNCATCNGGC) and REP2-Dt (5′-NCGNCTTATCNGGGCCTAC) were each covalently linked with a fluorophore at the 5′ end (6-FAM) and were included in amplification reactions (Sigma Genosys, St. Louis, Mo.). Each reaction mixture (25-μl volume) contained 100 pmol of each primer, 200 ng of bacterial genomic DNA, 12.5 nM concentrations of each deoxynucleotide, and AmpliTaq DNA polymerase (2 U) (PE Applied Biosystems) in a reaction buffer with 10% (vol/vol) dimethyl sulfoxide (14). PCRs were performed in a DNA Thermocycler 480 (PE Applied Biosystems) with the following cycling conditions: initial denaturation (94°C, 7 min) followed by 30 cycles of denaturation (94°C, 30 s), annealing (40°C, 1 min) and extension (65°C, 8 min). A final extension (65°C, 16 min) completed the cycling protocol. PCR amplicons were visualized following electrophoresis in a 2.5% agarose gel and ethidium bromide staining.
Amplicons were purified with size exclusion columns (Quantum Prep PCR Kleen Spin columns; Bio-Rad, Hercules, Calif.). After purification, fluorescent amplicons (1 μl) were mixed with deionized formamide (12 μl) and TAMRA-labeled 2500-size standards (PE Applied Biosystems). Samples were heated (94°C, 5 min) and chilled on ice prior to loading. Each sample was fractionated by capillary electrophoresis (GS POP4 matrix; PE Applied Biosystems) in the ABI Prism 310 genetic analyzer (PE Applied Biosystems), and electrophoretograms (peak profiles) were analyzed with the GeneScan software system (PE Applied Biosystems).
Nucleotide sequence accession number.
The GenBank accession numbers for the strains used as references in this study were listed previously (3). The 16S rRNA sequence of Helicobacter aurati MIT 97-5075cT was deposited in GenBank under accession number AF297868.
RESULTS
Culture results.
In the RS1 group of hamsters, microaerobic bacteria compatible with helicobacter-like organisms were isolated from 2 of 12 stomach samples and 11 of 12 cecal samples (Table 1). Three of six antral and body samples were positive for microaerobic bacteria in the RS2 group, as were all six of the cecal samples. Positive microaerobic bacteria from antra and body samples, as well as from ceca, were also recovered from the commercial hamsters, except for CS3 hamsters, where no microaerobic organisms were isolated. Microaerobic bacteria were not cultured from any of the liver samples.
TABLE 1.
Summary of gastrointestinal microaerobic culture and PCR analysis from five hamster groupsa
| Hamster group | Antral tissue (by PCR) | Cultures
|
||
|---|---|---|---|---|
| Antrum | Body | Cecum | ||
| RS1 | 5/12b | 2/12b | NDb | 11/12 |
| RS2 | 5/6 | 3/6 | 3/6 | 6/6 |
| CS1 | 5/6 | 2/6 | 1/6 | 6/6 |
| CS2 | 4/5 | 3/5 | 1/5 | 5/5 |
| CS3 | 3/6 | 0/6 | 0/6 | 0/6c |
The denominator is the number of hamsters in the group; the numerator is the number of hamsters in the group with Helicobacter DNA by PCR or positive culture. A positive culture means that from one to three of the novel microaerobic phenotypes were cultured from a given tissue specimen.
In RS1, the stomach was sampled without division into antrum and body. ND, not determined.
By 1,200-bp Helicobacter spp.-specific PCR, 6 of 6 cecal tissue samples were positive.
More than one morphology were frequently observed in the bacteria cultured from a single tissue specimen. By light microscopy, three different morphological phenotypes, all gram negative and motile, were recovered in all. The most common phenotype was straight and fusiform, with a beaded appearance, while another phenotype was equally long but much more slender and curved. PCR of these two bacterial phenotypes demonstrated amplification with the 1,200-bp all-helicobacter primers. The third phenotype was short and curved; Campylobacter spp.-specific primers amplified isolates of this phenotype. Except for PCR analysis of tissues, Helicobacter sp. isolates with the fusiform phenotype represent the principal subjects in this study.
PCR tissue analysis.
PCR using the 1,200-bp Helicobacter spp.-specific primers was undertaken using antral tissue specimens from the hamsters. These data, given in Table 1, demonstrate that Helicobacter DNA was present in the antra of several hamsters with negative cultures, including the hamsters from CS3, a commercial source that had no positive cultures. Cecal tissue samples from this source were also processed for PCR, and all six samples were positive for Helicobacter spp.-specific DNA.
Culture and phenotypic characteristics.
Cultures on plated media were visible after incubation for 3 to 14 days in microaerobic conditions. Initial bacterial growth was most commonly seen on the TVP and/or CVA plates, as a thin, spreading film. Once pure cultures of the fusiform phenotype were isolated, subsequent passages yielded growth on blood agar plates by 48 h at 37°C; brucella broth with 5% fetal calf serum also supported growth at 37°C within 48 to 72 h.
Table 2 lists the phenotypic features of five isolates (from four separate hamster sources) of the fusiform hamster Helicobacter sp., along with those of other rodent helicobacter species. Urease and gamma-glutamyl transpeptidase activities were present in the fusiform helicobacter isolates. The fusiform isolates were positive for catalase and oxidase activities, indoxyl acetate hydrolysis, and growth at 42°C. The fusiform phenotype was susceptible to nalidixic acid and resistant to cephalothin.
TABLE 2.
Characteristics of hamster Helicobacter aurati compared to other, formally named Helicobacter spp. found in rodentsa
| Taxon | Catalase production | Nitrate reduction | Alkaline phosphatase hydrolysis | Urease | Indoxyl acetate hydrolysis | γ-Glutamyl transpeptidase | Growth at 42°C | Growth with 1% glycine | Susceptibility to:
|
Periplasmic fibers | No. of flagella | Distribution of flagella | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nalidixic acid (30-μg disk) | Cephalothin (30-μg disk) | ||||||||||||
| H. aurati | +(5/5) | −(0/5) | −(0/5) | +(5/5) | +(5/5) | +(5/5) | +(5/5) | −(0/5) | S(5/5) | R(5/5) | + | 7–10 | Bipolar |
| H. rodentium | + | + | − | − | − | − | + | + | R | R | − | 2 | Bipolar |
| H. trogontum | + | + | − | + | ND | + | + | ND | R | R | + | 5–7 | Bipolar |
| H. muridarum | + | − | + | + | + | + | − | − | R | R | + | 10–14 | Bipolar |
| H. hepaticus | + | + | ND | + | + | ND | − | + | R | R | − | 2 | Bipolar |
| H. bilis | + | + | ND | + | − | ND | + | + | R | R | + | 3–14 | Bipolar |
| ‘F. rappini’ | + | − | − | + | ND | + | + | − | R | R | + | 10–20 | Bipolar |
| H. cinaedi | + | + | − | − | − | − | − | + | S | I | − | 1–2 | Bipolar |
| H. cholecystus | + | + | + | − | − | − | + | + | I | R | − | 2 | Bipolar |
| H. mesocricetorum | + | + | + | − | ND | − | ND | − | S | R | − | 2 | Bipolar |
Modified from Versalovic and Fox (27) and Simmons et al. (22). Symbols and abbreviations: +, positive reaction; −, negative reaction; S, susceptible; R, resistant; I, intermediate; ND, not determined. Numbers in parentheses are the number of strains with the indicated result/number of strains tested.
Ultrastructure.
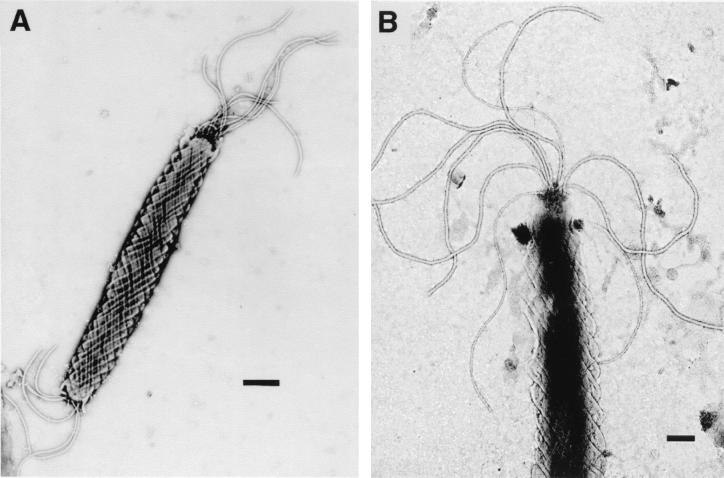
Transmission electron micrographs of the fusiform Helicobacter sp. are shown in Fig. 1. Individual cells contained periplasmic fibers and measured approximately 0.6 by 4 to 8 μm. Several sheathed flagella (7 to 10) were present at both ends of each cell.
FIG. 1.
Transmission electron micrographs of novel hamster microaerobic bacteria. (A) Fusiform Helicobacter sp., proposed name H. aurati, with periplasmic fibers. Bar, 500 nm. (B) Multiple sheathed flagella of H. aurati. Bar, 200 nm.
Sequencing and phylogenetic analysis of novel Helicobacter sp.
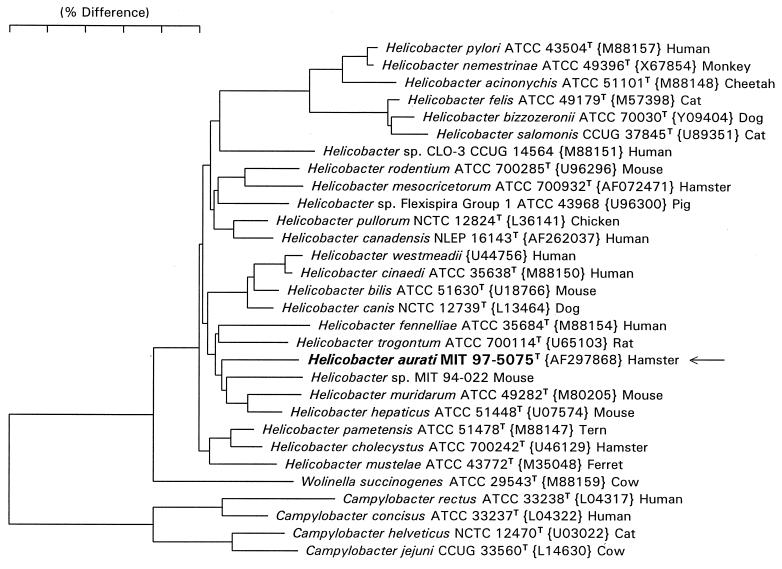
The essentially complete 16S rRNA sequences (bases 28 to 1524, Escherichia coli numbering) were determined for each of three fusiform isolates. A neighbor-joining phylogenetic tree for the sequence of MIT 97-5075cT and reference Helicobacter species is shown in Fig. 2. Fusiform isolates MIT 97-5075cT, MIT 97-5310c, and MIT 98-6041a have identical sequences and represent a novel Helicobacter species (proposed name Helicobacter aurati). The sequence of H. aurati differs by 3.5 to 5% from its close relatives H. muridarum, H. hepaticus, and Helicobacter sp. strain MIT 94-022, a species that infects mice.
FIG. 2.
Phylogenetic tree constructed from 16S rRNA sequence similarity values. Scale bar represents 5% difference in nucleotide sequences, determined by measuring the lengths of the horizontal lines connecting two species.
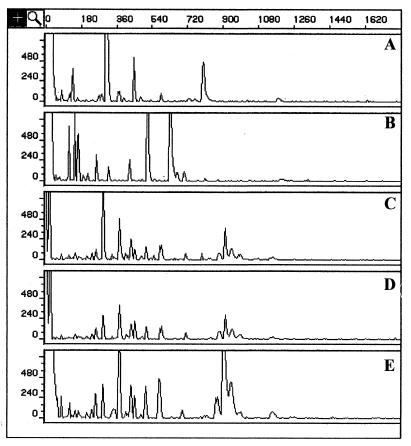
Consistent with these results, rep-PCR-based chromosomal DNA fingerprints for three H. aurati isolates were highly similar to each other and were distinct from DNA profiles obtained from the related rodent helicobacters H. hepaticus and H. muridarum (Fig. 3).
FIG. 3.
Electrophoretograms demonstrating rep-PCR-based chromosomal DNA profiles of rodent helicobacters H. hepaticus (A) and H. muridarum (B), compared to those of H. aurati isolates MIT 97-5075cT (C), MIT 98-6041a (D), and MIT 98-6169a (E). Horizontal axis units are base pairs; vertical axes display peak height in fluorescent units (PE Applied Biosystems).
Histopathology.
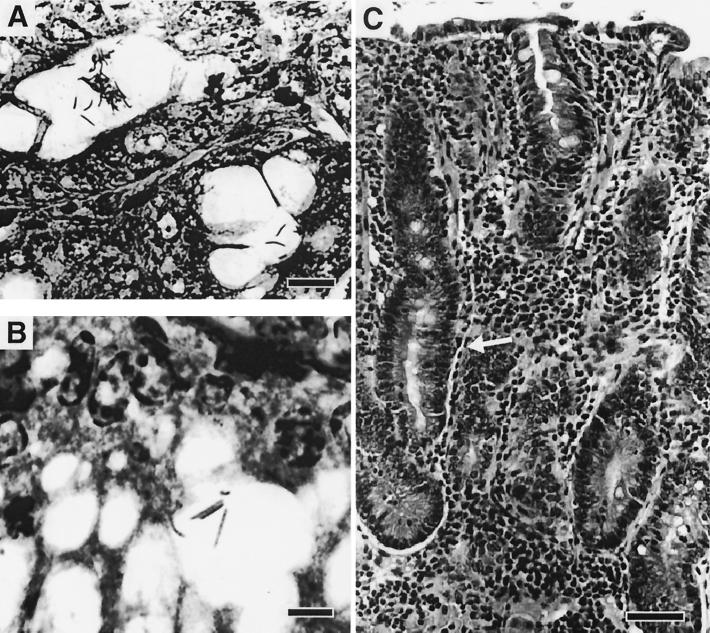
Hamster antra stained with silver stain reveal argyrophilic organisms compatible with the novel fusiform helicobacter isolated by microaerobic culture (Fig. 4). An example of the chronic, antral gastritis observed in several hamsters is also depicted.
FIG. 4.
Photomicrographs from gastric antra of hamsters. (A) Deep gastric mucosa colonized with numerous argyrophilic bacteria within lumina of gastric glands (Warthin-Starry stain). Bar, 10 μm. (B) Higher power showing the beaded appearance of the fusiform Helicobacter sp. H. aurati (Warthin-Starry stain). Bar, 6 μm. (C) Characteristic chronic gastritis in the antrum of hamsters colonized with the microaerobic bacteria. Note diffuse infiltration of lamina propria with mononuclear leukocytes and glandular hyperplasia (arrow) (hematoxylin and eosin stain). Bar, 80 μm.
DISCUSSION
Given the rapidity with which new Helicobacter species have been described in recent years and the large number of Helicobacter spp. found in mice, including formally unnamed ones (5), it is not surprising that we have documented a novel species in a different rodent, the Syrian hamster. We propose the name Helicobacter aurati (MIT 97-5075cT) for our fusiform, urease-positive helicobacter isolate. By 16S rRNA sequence analysis, it clusters with H. muridarum, H. hepaticus, and Helicobacter sp. MIT 94-022. rep-PCR-based DNA fingerprinting reinforces that the three H. aurati isolates are clearly distinguished from the related species H. hepaticus and H. muridarum. These isolates, cultured from hamsters obtained from three sources, share similar rep-PCR-based chromosomal profiles, and some subspecies genetic variation is demonstrated.
Unlike H. hepaticus, H. aurati has periplasmic fibers and multiple flagella, and it does not grow in 1% glycine or reduce nitrate. In contrast to H. muridarum, H. aurati is negative for alkaline phosphatase hydrolysis; also, it does not have the pronounced spiral-shape characteristic of H. muridarum (16). Of these three species, only H. aurati grows at 42°C and is sensitive to nalidixic acid. Various features, such as fusiform shape and periplasmic fibers, allow morphologic discrimination between H. aurati and the three other helicobacters that have thus far been identified in hamsters, H. cholecystus, H. cinaedi, and H. mesocricetorum. The presence of urease in H. aurati also distinguishes it from these three other Helicobacter species found in hamsters.
The isolation of a novel Helicobacter species from the hamster stomach is of interest, as no microaerobic bacteria have been reported previously from inflamed gastric tissue of hamsters. Because these bacteria were recovered from cecal samples more often than from antral samples, the lower gastrointestinal tract is more likely the primary site of infection, with subsequent spread to the stomach in selected animals. The coprophagic habits of hamsters probably play a role in gastric colonization by H. aurati. Stomach colonization by helicobacters is often linked to the urease enzyme, and another urease-positive helicobacter found in rodents, H. muridarum, has likewise been observed to inhabit both lower intestinal and gastric sites in mice (19). The presence of H. aurati in the stomach coincided with inflammatory cell infiltrates in the gastric mucosa in many of the infected hamsters we studied; comparable lesion development in the stomach has been reported in mice infected with H. muridarum (18a, 19).
Failure to culture the new microorganism from the gastrointestinal tracts of CS3 hamsters probably reflects a low microaerobic population density for unknown reasons. The presence of Helicobacter spp.-specific DNA amplified from antral and cecal tissues of these hamsters supports this hypothesis. Another unusual feature of these CS3 hamsters was the identification of yeastlike cells in most stomachs, both by culture and on histological examination. Whether the presence of these organisms selectively inhibits stomach colonization by microaerobes will require further investigation.
Additional studies also are required to define whether and how H. aurati contributes to disease in Syrian hamsters and its interaction with other microaerobic species in the hamster stomach. Regardless of when these questions are addressed, the occurrence of H. aurati in research and commercial hamster colonies may impact particular studies using hamsters. The suspected high prevalence of this helicobacter is exemplified by its recent isolation in our laboratory from the stomachs of hamsters that came from a third research colony (unpublished data). In the future, this new species can be used in comparative gastric helicobacter experiments designed to elucidate the factors important for gastric colonization and associated inflammation by H. pylori in humans.
Description of Helicobacter aurati sp. nov.
Helicobacter aurati (au.ra′ti. L. gen. masc. n. of the golden one, named after the Syrian golden hamster, Mesocricetus auratus). Cells are fusiform with periplasmic fibers and measure 0.6 by 4 to 8 μm. Bipolar, multiple sheathed flagella (7 to 10) account for the motility of the bacterium. Older cultures contain large coccoid forms. Growth on agar plates appears as a thin, spreading film; distinct colonies are absent. Microaerobic growth occurs at 37 and 42°C but not at 25°C. Brucella agar plates containing 1% glycine, 1.5% NaCl, 2.0% NaCl, and 3.0% NaCl do not support growth. No growth is seen under anaerobic conditions. Cells are positive for urease, catalase, oxidase, and gamma-glutamyl transpeptidase activities. Tests for indoxyl acetate hydrolysis were positive, but tests were negative for nitrate reduction and alkaline phosphatase hydrolysis. The bacteria are sensitive to nalidixic acid while resistant to cephalothin. Cells have been isolated from the stomachs and ceca of adult Syrian hamsters. The type strain, MIT 97-5075c, has been deposited with the American Type Culture Collection as ATCC BAA-1.
ACKNOWLEDGMENTS
This work was supported in part by NIH grants RRO7036 and RO1AI37750.
We thank Maggie Delano, Mark Whary, and Chuck Dangler for their clinical observations and initial diagnostic efforts.
REFERENCES
- 1.Cahill R J, Foltz C J, Fox J G, Dangler C A, Powrie F, Schauer D B. Inflammatory bowel disease: an immune-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewhirst F E, Chien C C, Paster B J, Ericson R L, Orcutt R, Schauer D B, Fox J G. Phylogeny of the defined murine microbiota (altered Schaedler flora) Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox J G, Dewhirst F E, Shen Z, Feng Y, Taylor N S, Paster B J, Ericson R L, Lau C N, Correa P, Araya J C, Roa I. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology. 1998;114:755–763. doi: 10.1016/s0016-5085(98)70589-x. [DOI] [PubMed] [Google Scholar]
- 4.Fox J G, Dewhirst F E, Tully J G, Paster B J, Yan L, Taylor N S, Collins M J, Gorelick P L, Ward J M. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox J G, Gorelick P L, Kullberg M C, Ge Z, Dewhirst F E, Ward J M. A novel urease-negative Helicobacter species associated with colitis and typhlitis in IL-10-deficient mice. Infect Immun. 1999;67:1757–1762. doi: 10.1128/iai.67.4.1757-1762.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox J G, Li X, Yan L, Cahill R J, Hurley R J, Lewis R, Murphy J C. Chronic proliferative hepatitis in A/JCr mice associated with persistent Helicobacter hepaticus infection: a model of helicobacter-induced carcinogenesis. Infect Immun. 1996;64:1548–1558. doi: 10.1128/iai.64.5.1548-1558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox J G, MacGregor J, Shen Z, Li X, Lewis R, Dangler C A. Comparison of methods to identify Helicobacter hepaticus in B6C3F1 mice used in a carcinogenesis bioassay. J Clin Microbiol. 1998;36:1382–1387. doi: 10.1128/jcm.36.5.1382-1387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox J G, Yan L, Dewhirst F E, Paster B J, Shames B, Murphy J C, Hayward A, Belcher J C, Mendes E N. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mouse strains. J Clin Microbiol. 1995;33:445–454. doi: 10.1128/jcm.33.2.445-454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox J G, Yan L, Shames B, Campbell J, Murphy J C, Li X. Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect Immun. 1996;64:3673–3681. doi: 10.1128/iai.64.9.3673-3681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin C L, Beckwith C S, Livingston R S, Riley L K, Gibson S V, Besch-Williford C L, Hook R L. Isolation of a novel species, Helicobacter cholecystus sp. nov., from the gallbladders of Syrian hamsters with cholangiofibrosis and centrilobular pancreatitis. J Clin Microbiol. 1996;34:2952–2958. doi: 10.1128/jcm.34.12.2952-2958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gebhart C J, Fennell C L, Murtaugh M P, Stamm W E. Campylobacter cinaedi is normal intestinal flora in hamsters. J Clin Microbiol. 1989;27:1692–1694. doi: 10.1128/jcm.27.7.1692-1694.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebhart C J, Kiehlbauch J A, Ward G E. A new species of Campylobacter isolated from the intestines of hamsters. Acta Gastro-Enterol Belg. 1993;56(Suppl.):26. [Google Scholar]
- 13.Hailey J R, Haseman J K, Bucher J R, Radovsky A E, Malarkey D E, Miller R T, Nyska A, Maronpot R R. Impact of Helicobacter hepaticus infection in B6C3F1 mice from twelve national toxicology program two-year carcinogenesis studies. Toxicol Pathol. 1998;26:602–611. doi: 10.1177/019262339802600503. [DOI] [PubMed] [Google Scholar]
- 14.Kogan S, Doherty M, Gitschier J. An improved method for prenatal diagnosis of genetic disease by analysis of amplified DNA sequences. New Engl J Med. 1987;317:985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- 15.Kullberg M C, Ward J M, Gorelick P L, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66:5157–5166. doi: 10.1128/iai.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee A, Phillips M W, O'Rourke J L, Paster B J, Dewhirst F E, Fraser G J, Fox J G, Sly L I, Romaniuk P J, Trust T J, Kouprach S. Helicobacter muridarum sp. nov., a microaerophilic helical bacterium with a novel ultrastructure isolated from the intestinal mucosa of rodents. Int J Syst Bacteriol. 1992;42:27–36. doi: 10.1099/00207713-42-1-27. [DOI] [PubMed] [Google Scholar]
- 17.Mendes E N, Quieroz D M M, Dewhirst F E, Paster B J, Moura S B, Fox J G. Helicobacter trogontum sp. nov., isolated from the rat intestine. Int J Syst Bacteriol. 1996;46:916–921. doi: 10.1099/00207713-46-4-916. [DOI] [PubMed] [Google Scholar]
- 18.Paster B J, Dewhirst F E. Phylogeny of Campylobacter, Wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int J Syst Bacteriol. 1988;38:56–62. [Google Scholar]
- 18a.Patterson, M. M., M. D. Schrenzel, Y. Feng, and J. G. Fox. Gastritis and intestinal metaplasia in Syrian hamsters infected with Helicobacter aurati and two other microaerobes. Vet Pathol., in press. [DOI] [PubMed]
- 19.Queiroz D M M, Contigli C, Coimbra R S, Nogueira A M M F, Mendes E N, Rocha G A, Moura S B. Spiral bacterium associated with gastric, ileal and caecal mucosa of mice. Lab Anim. 1992;26:288–294. doi: 10.1258/002367792780745760. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Shen Z, Fox J G, Dewhirst F E, Paster B J, Foltz C J, Yan L, Shames B, Perry L. Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int J Syst Bacteriol. 1997;47:627–634. doi: 10.1099/00207713-47-3-627. [DOI] [PubMed] [Google Scholar]
- 22.Simmons J H, Riley L K, Besch-Williford C L, Franklin C L. Helicobacter mesocricetorum sp. nov., a novel helicobacter isolated from the feces of Syrian hamsters. J Clin Microbiol. 2000;38:1811–1817. doi: 10.1128/jcm.38.5.1811-1817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stills H F, Hook R R, Kinden D A. Isolation of a Campylobacter-like organism from healthy Syrian hamsters (Mesocricetus auratus) J Clin Microbiol. 1989;27:2497–2501. doi: 10.1128/jcm.27.11.2497-2501.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Totten P A, Fennell C L, Tenover F C, Wezenberg J M, Perine P L, Stamm W E, Holmes K K. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis. 1985;151:131–139. doi: 10.1093/infdis/151.1.131. [DOI] [PubMed] [Google Scholar]
- 25.Versalovic J, Lupski J R. DNA fingerprinting of Neisseria strains by rep-PCR. Methods Mol Cell Biol. 1995;5:96–104. [Google Scholar]
- 26.Versalovic J, Kapur V, Koeuth T, Mazurek G H, Whittam T S, Musser J M, Lupski J R. DNA fingerprinting of pathogenic bacteria by fluorophore-enhanced repetitive sequence-based polymerase chain reaction. Arch Pathol Lab Med. 1995;119:23–29. [PubMed] [Google Scholar]
- 27.Versalovic J, Fox J G. Helicobacter. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 727–738. [Google Scholar]
- 28.Ward J M, Fox J G, Anver M R, Haines D C, George C V, Collins M J, Gorelick P L, Nagashima K, Gonda M A, Gilden R V, Tully J G, Russell R J, Benveniste R E, Paster B J, Dewhirst F E, Donovan J C, Anderson L M, Rice J M. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86:1222–1227. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]