Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal form of cancer characterized by drug resistance, urging new therapeutic strategies. In recent years, protein kinases have emerged as promising pharmacological targets for the treatment of several solid and hematological tumors. Interestingly, cyclin-dependent kinase 1 (CDK1) is overexpressed in PDAC tissues and has been correlated to the aggressive nature of these tumors because of its key role in cell cycle progression and resistance to the induction of apoptosis. For these reasons, CDK1 is one of the main causes of chemoresistance, representing a promising pharmacological target. In this study, we report the synthesis of new 1,2,4-oxadiazole compounds and evaluate their ability to inhibit the cell growth of PATU-T, Hs766T, and HPAF-II cell lines and a primary PDAC cell culture (PDAC3). Compound 6b was the most active compound, with IC50 values ranging from 5.7 to 10.7 µM. Molecular docking of 6b into the active site of CDK1 showed the ability of the compound to interact effectively with the adenosine triphosphate binding pocket. Therefore, we assessed its ability to induce apoptosis (which increased 1.5- and 2-fold in PATU-T and PDAC3 cells, respectively) and to inhibit CDK1 expression, which was reduced to 45% in Hs766T. Lastly, compound 6b passed the ADME prediction, showing good pharmacokinetic parameters. These data demonstrate that 6b displays cytotoxic activity, induces apoptosis, and targets CDK1, supporting further studies for the development of similar compounds against PDAC.
Keywords: 1,2,4-oxadiazole; marine alkaloids; topsentin; CDK1 inhibitor; pancreatic cancer; PDAC; antiproliferative; apoptosis
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic cancer, is a highly lethal form of cancer, for which surgery is the only curative treatment [1]. However, only a small percentage of PDACs are eligible for resection. Polychemotherapy regimens, such as FOLFIRINOX (folinic acid, fluorouracil, irinotecan, and oxaliplatin) or gemcitabine plus nab-paclitaxel, are the standard therapies for most PDAC patients but are characterized by a high rate of toxicity and modest survival benefit, since almost all PDAC patients become or are already drug resistant [2,3,4]. The causes of such drug resistance are many, and recent omics studies, including phosphoproteomics, have shown aberrations in key functional signaling that could hopefully be used to identify new therapeutic strategies [5].
In recent years, particular attention has been paid to protein kinases, which emerged as promising pharmacological targets given their role in regulating fundamental cellular processes. Since the first kinase inhibitor, imatinib, was approved for the treatment of chronic myeloid leukemia in 2001, the Food and Drug Administration has approved more than 50 small-molecule kinase inhibitors, of which the majority are tyrosine kinase inhibitors, while some are serine/threonine kinase inhibitors [6,7,8,9].
Cyclin-dependent kinase 1 (CDK1) is a serine/threonine kinase which plays a crucial role in regulating the cell cycle and has recently emerged as a promising target for the treatment of PDAC. Indeed, CDK1 overexpression was correlated with the progression of this type of cancer [10,11]. Under physiological conditions, CDK1 tightly regulates the progression of the cell cycle. However, abnormal expression of CDK1 determines cell replication even in case of DNA damage, resulting in the proliferation of cancer cells. Moreover, the activity of CDK1 is strongly regulated by the gene TP53, which is mutated in 70% of PDAC [12]. Together, these observations suggest the potential of CDK1 inhibition as a novel promising strategy to treat PDAC. They prompted us to synthesize new bioactive compounds against this target.
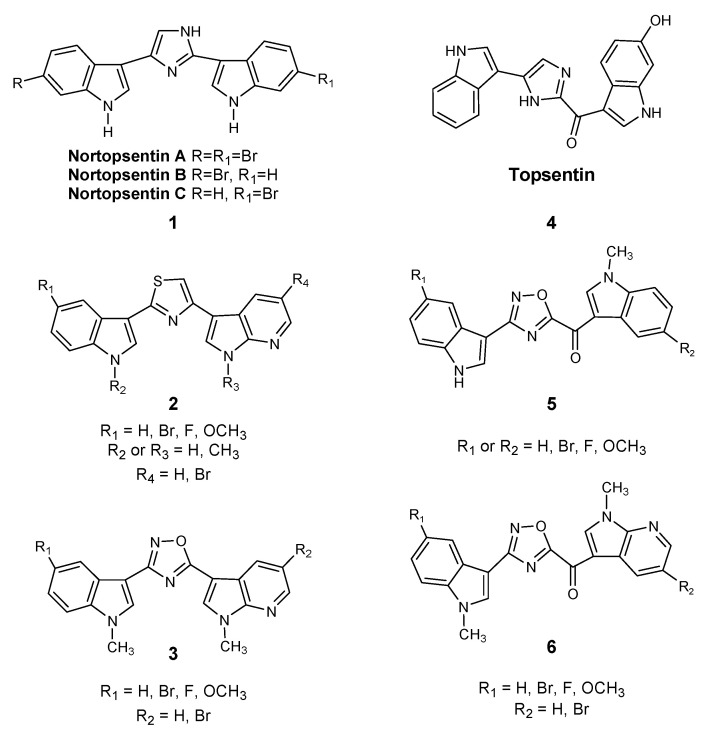
Considering the importance of marine microenvironments as an important resource of bioactive molecules containing different heterocyclic rings and different heteroatoms, our research group synthesized a number of small molecules obtained through the structural manipulation of nortopsentins 1, natural bis-indolyl alkaloids isolated from deep-sea sponges (Spongsorites ruetzleri), which are characterized by significant antiproliferative activity against the P388 murine leukemia cell line [13]. In particular, we produced many derivatives in which the modification of nortopsentin involved the central imidazole ring, which was replaced by several five-membered heterocycles, while an indole moiety was substituted with an azaindole portion. These compounds had a significantly improved antiproliferative activity against a wide range of tumor cell lines, with half of the maximal inhibitory concentration (IC50) in the micro-submicromolar range [14,15]. In particular, thiazole nortopsentin derivatives 2, determined CDK1 inhibition with IC50 values of 0.64–0.89 μM, which is comparable to the values reported for roscovitine and purvalanol A, two well-known CDK1 inhibitors. Moreover, a more recently synthesized indolyl-7-azaindolyl thiazole compound demonstrated its ability to reduce colorectal cancer stem cells (CR-CSCs), showing a synergistic effect with standard chemotherapy, such as oxaliplatin and 5-fluorouracil, as well as the ability of eradicating CR-CSCs when combined with the CHK1 inhibitor Rabusertinib [16]. Considering that, among nitrogen heterocycles, indole and oxadiazole rings are found in many molecules with significant biological activity, and especially antitumor activity [17,18,19], we also synthesized new 1,2,4-oxadiazole nortopsentin analogs 3, which were screened for their antiproliferative activity [14]. Then, we further investigated the bis-indolyl marine alkaloid topsentin 4, characterized by the presence of a carbonyl spacer group, which gives greater flexibility to the molecule to better adapt to the ATP-binding site of CDK1 and represents a hydrogen bonding acceptor, which could interact with the amino acid residues of the active site of CDK1. Topsentin was extracted from the sponge Topsentia genitrix and showed in vitro cytotoxic activity against P-388 murine tumor cells, with an IC50 of 8.8 μM, as well as at micromolar concentrations against several human cancer cell lines. Lastly, considering the promising results shown by [3-(1H-Indol-3-yl)-[1,2,4]oxadiazol-5-yl]-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-methanones 5 on PDAC cancer cell lines [20], we decided to synthesize a new series of topsentin analogs of the type 6 shown in Scheme 1, in which the imidazole central ring was replaced by 1,2,4-oxadiazole moiety, and one indole portion was converted into 7-azaindole ring (Scheme 1). In the present study, we also report the cytotoxic activity of these new derivatives against PDAC cells as well as their capability to inhibit CDK1 and to induce apoptosis.
Scheme 1.
Chemical structures of nortopsentin 1 and topsentin 4 alkaloids and their derivatives 2,3 and 5,6.
2. Results and Discussion
2.1. Chemistry
A new series of new 7-azaindolyl oxadiazole compounds 6a–f was efficiently synthesized as described in Scheme 2.
Scheme 2.
Synthesis of (1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-[3-(1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanones (6a–f). Reagents and conditions: (i) oxalyl chloride, diethyl ether, r.t., overnight, then −65 °C, sodium methoxide solution 25 wt. % in methanol, r.t., 2 h, 62–73%; (ii) dimethylsulfoxide, r.t., 30 min. 60–85%.
The key intermediates, (1-methyl-1H-pyrrolo[2,3-b]pyridine-3-yl)-oxo-acetic acid methyl esters 8a,b, were prepared in good yield (62% and 73%, respectively) by keeping the corresponding methylindole precursor of the type 7a,b, synthesized as previously described [14], with an excess of oxalyl chloride. The resulting non-isolated acetyl chloride intermediate was then converted in situ into methyl esters 8a,b using a sodium methoxide solution 25 wt. % in methanol.
The latter compounds were subjected to a coupling reaction with carboxamidines 9a–c, synthesized in turn from the corresponding 1-methyl-1H-indoles, converted to their carbonitriles, and had a successive reaction with hydroxylamine hydrochloride, as previously reported [21].
The azaindole substituent of the original oxo-acetic acid methyl ester is in position C(5), and the oxadiazole ring is obtained by means of the [4+1] synthetic route, as previously explained [21]. The coupling reaction, performed in anhydrous dimethylsulfoxide (DMSO) at r.t. for 30 min, proceeded without the isolation of an acylamidoxime intermediate, yielding, after purification by chromatography, the desired 7-azaindolyl oxadiazoles 6a–f (Scheme 2) in yields ranging from 60% to 85% (Table 1).
Table 1.
Yields of the synthesis of 6a–f; (1-Methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-[3-(1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanones.
| Compound | R1 | R2 | Yield (%) |
|---|---|---|---|
| 6a | H | Br | 60% |
| 6b | F | Br | 67% |
| 6c | OCH3 | Br | 85% |
| 6d | H | H | 77% |
| 6e | F | H | 68% |
| 6f | OCH3 | H | 80% |
2.2. Biological Studies
2.2.1. Antiproliferative Activity of the New 1,2,4-Oxadiazole Compounds 6a–f against PDAC3, PATU-T, Hs766T, and HPAF-II PDAC Cells
The in vitro antiproliferative activity of the new 7-azaindolyl oxadiazole compounds 6a–f was initially evaluated by the sulforhodamine-B assay (SRB) against PATU-T immortalized PDAC cell line. All compounds were tested at three different concentrations of 50.0, 5.0, and 0.5 µM.
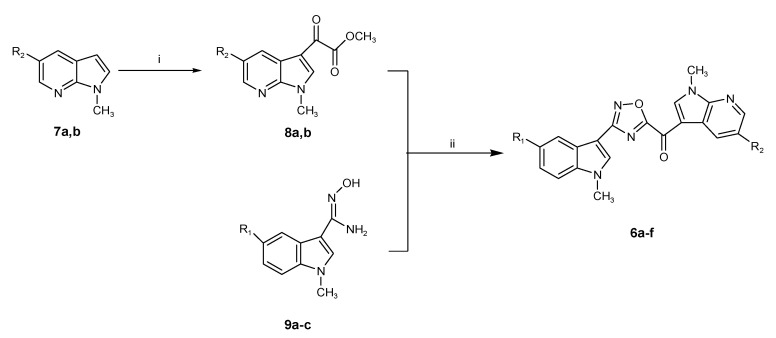
Among the tested compounds, derivative 6b showed the highest potency, exhibiting an IC50 value of 10.7 µM, while other compounds showed minimal cytotoxic effect with IC50s > 20 µM. In order to extend the antiproliferative evaluation of compound 6b towards other pancreatic cells, we assessed the inhibition of cell growth in primary pancreatic cell lines including both the immortalized HPAF-II and Hs766T cancer cell lines and the primary culture PDAC3. Interestingly, compound 6b was active against all these PDAC cells. The best result was observed against Hs766T, with an IC50 value of 5.7 µM. However, the compound 6b was also able to inhibit the viability of PDAC3 and HPAF-II with IC50 values of 6.9 and 9.8 μM, respectively (Table 2). A representative curve of cell growth inhibition of 6b in PDAC3 is reported in Figure 1. In parallel experiments we also evaluated the IC50 of the conventional anticancer drug gemcitabine, which was used as positive control and presented IC50 value below 1 µM. This result was in agreement with previous studies [22,23,24].
Table 2.
IC50s of 6b against Hs766T, HPAF-II, PDAC3, and PATU-T cells.
| IC50 a (µM) ± SEM | ||
|---|---|---|
| Compound | Cell Line | IC50 ± SEM |
| 6b | Hs766T | 5.7 ± 0.60 |
| PDAC3 | 6.9 ± 0.25 | |
| HPAF-II | 9.8 ± 0.70 | |
| PATU-T | 10.7 ± 0.16 | |
a Values (in µM) are reported as means ± SEM (Standard Error of the Mean) of three separate experiments.
Figure 1.
Representative growth inhibition curve of PDAC3 PDAC primary cells, treated for 72 h with the compound 6b. Points: mean values obtained from three independent experiments; bars: SEM.
From structure activity relationship (SAR) analysis on compounds of type 5 and 6 (Scheme 1), we observed that the introduction of a nitrogen atom at position 7 of the indole moiety does not significantly improve the antiproliferative activity, as well as the presence of a methyl group at the indole N-atom. Conversely, in accordance with previous results concerning [3-(1H-indol-3-yl)-1,2,4-oxadiazol-5-yl](1-methyl-1H-indol-3-yl) methanones 5 [20], in type 6 derivatives the presence of a halogen atom, e.g., fluorine or bromine, at 5 position of the indole or 7-azaindole ring seems to be essential for the cytotoxic activity.
2.2.2. Modulation of CDK1 Expression by ELISA
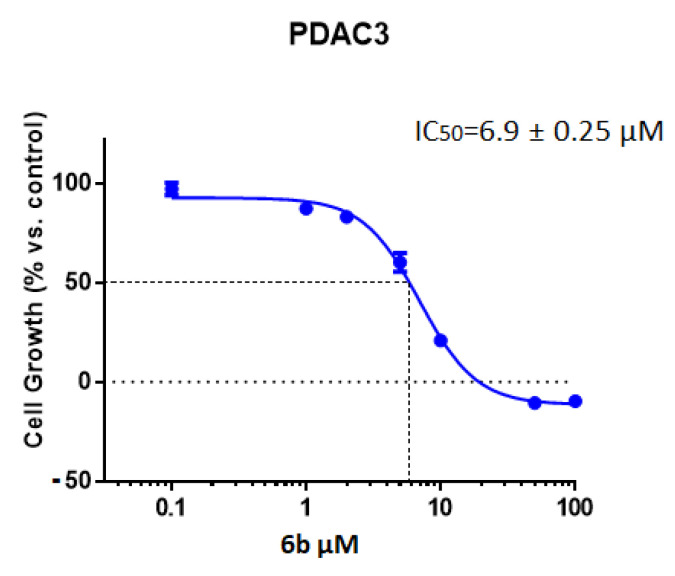
Considering the promising results obtained with compound 6b, further studies were carried out to investigate its mechanism of action. In order to evaluate whether compound 6b was able to reduce CDK1 expression, a specific ELISA assay was performed in the three immortalized PDAC cell lines, i.e., Hs766T, HPAF-II, and PATU-T, as well as in the PDAC3 cells. These cell models were treated with 10 μM 6b. As shown in Figure 2, we observed a reduction of CDK1 expression compared to control cells, supporting the potential role of the inhibition of CDK1 in the mechanism of action of this compound.
Figure 2.
ELISA-based quantitation of CDK1 expression in PDAC cells following a 24 h exposure to DMSO (control) or to 10 µM of compound 6b. CDK1 expression was quantified using Human cyclin-dependent kinase 1 (CDK1) ELISA Kit according to the manufacturer’s instructions. Columns: mean; bars: SEM (n = 3). *: p < 0.05, **: p < 0.01, ***: p < 0.001, significantly different compared with control.
2.2.3. Induction of Apoptosis
Considering that a number of previous studies reported that the inhibition of CDK1 activity contributes to the initiation of apoptotic process [25,26,27], we then evaluated the effect of 6b on the induction of apoptosis in pancreatic cancer cells. For this purpose, we measured the externalization of the plasma membrane phosphatidylserine, a reliable marker of cell apoptosis, which was quantified by measurement of fluorescence of annexin V by spectrophotometric and microscopy assays.
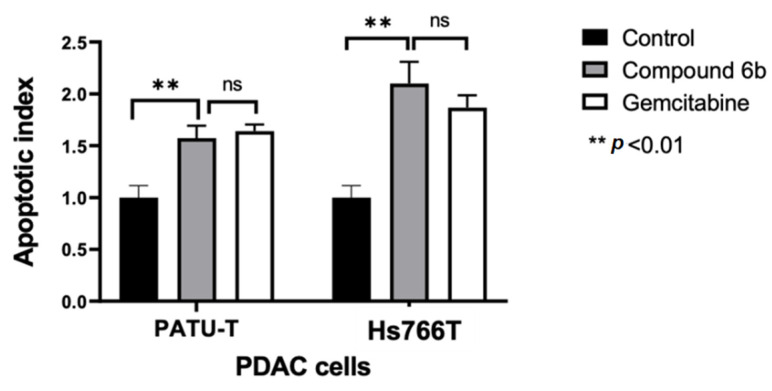
These experiments were performed on the most sensitive and most resistant models, i.e., Hs766T and PATU-T cells. After 24 h of treatment with 2*IC50 of 6b, a significant increase in the portion of apoptotic cells was observed. In particular, we noticed that the percentages of apoptotic Hs766T and PATU-T cells were comparable to the number of cells that underwent apoptosis after treatment with gemcitabine (at 2*IC50), which is a standard drug for the treatment of PDAC (Figure 3).
Figure 3.
Effects of compound 6b on apoptosis induction in PATU-T and Hs766T pancreatic cancer cells. The apoptotic index was calculated by evaluating the annexin V-FITC fold change compared with control cells after 24 h treatment. Gemcitabine was used as a positive control. Columns: mean; bars: SEM (n = 3). **: p < 0.01, significantly different compared with control; ns: not significant.
2.2.4. Molecular Modeling
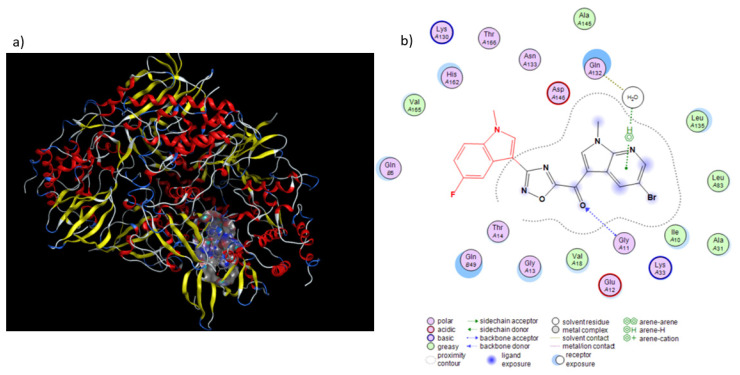
A potential binding mode for the most active compound 6b within the ATP binding site of CDK1 (PDB ID: 4YC6) is depicted in Figure 4. 6b is placed in a nucleotide pocket, establishing a hydrogen bond through the carbonyl group with the backbone of Gly11 residue.
Figure 4.
Proposed binding mode of compounds 6b with CDK1 (PDB ID: 4YC6). (a) Surface representation of 6b in CDK1 binding pocket. (b) H-bonds between the carbonyl group of our compound with the peptide nitrogen of Gly11 residue is shown with blue dashed arrow, while the water-mediated interaction between the nitrogen atom of the 7-azaindole portion adjacent to the oxadiazole ring and the Gln132 residue is depicted with green dashed lines.
Moreover, the nitrogen group of 7-azaindole moiety accepts a hydrogen bond from a water molecule interacting with Gln132, a residue well known to constitute the DFG motif and to establish water-mediated interaction with H-bond acceptors of CDK1 inhibitors (Figure 4) [28].
The molecular docking scores of compound 6b was found to be −6.999 Kcal/mol, indicating efficient binding to the active site of CDK1.
2.2.5. ADME Prediction
In order to evaluate and predict the fate of compound 6b within the human body, ADME and pharmacokinetic predictions were carried out using a freely available in silico ADME software [29]. Compound 6b, as shown in Table 3, displayed promising physicochemical and pharmacokinetic parameters in ADME prediction studies, showing 3 H-bond acceptors, 0 H-bond donors, 3 rotatable bonds, and log p < 5. Moreover, our compound respects the Lipinski rule of 5, a feature making it a promising hit compound in the drug discovery field. Compound 6b displayed a high gastrointestinal (GI) absorbance score but is not predicted to cross the blood brain barrier (BBB).
Table 3.
ADME predictions of compound 6b.
| Parameters | Score |
|---|---|
| n. H-bond acceptor | 6 |
| n. H-bond donor | 0 |
| n. rotable bonds | 3 |
| LogPo/w(iLOGP) | 3.67 |
| Lipinski’s rule violation | No |
| Bioavailability score | 0.55 |
| GI absorption | High |
| BBB permeation | no |
3. Materials and Methods
3.1. Chemistry
All melting points were taken on a Buchi-Tottoly capillary apparatus and were uncorrected. IR spectra were determined in bromoform using a Shimadzu FT/IR 8400S spectrophotometer. 1H and 13C NMR spectra were measured at 200 and 50 MHz, respectively, in DMSO-d6 solution, using a Bruker Avance II series 200 MHz spectrometer. Chromatography was performed with a MERK silica gel 230–400 mesh ASTM column or a FLASH40i Biotage or with a Buchi Sepacore chromatography module (prepacked cartridge reference). Elementary analyses (C, H, N) were within ± 0.4% of the theoretical values.
3.1.1. Synthesis of 1-methyl-1H-pyrrolo[2,3-b]pyridines (7a,b)
t-BuOK (0.38 g, 3.4 mmol) and TDA-1 (1 or 2 drops) were added to a cold solution of appropriate commercial 7-azaindoles (2.5 mmol) in anhydrous toluene (25 mL), at 0 °C. The reaction mixture was stirred at room temperature for 3 h, and then methyl iodide (2.5 mmol, 0.2 mL) was added at 0 °C. TLC analysis (cyclohexane/ethyl acetate 7:3) revealed that methylation was complete after 1 h. The solvent was evaporated under reduced pressure. The residue was treated with water, extracted with DCM (3 × 20 mL), dried (Na2SO4), evaporated, and purified by column chromatography using DCM/ethyl acetate (9/1) as the eluent, to give the desired product.
For 5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridine (7a) and 1-Methyl-1H-pyrrolo[2,3-b]pyridine (7b), analytical and spectroscopic data were in accordance to those reported in literature [14].
3.1.2. Synthesis of (1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-oxo-acetic Acid Methyl Esters (8a,b)
Oxalyl chloride (0.42 mL, 4.8 mmol) was added dropwise to a solution of appropriate 1-methyl-1H-pyrrolo[2,3-b]pyridine 7a,b (1 g, 4.8 mmol) in dry diethyl ether (8.5 mL) under a nitrogen atmosphere at 0 °C. The reaction mixture was stirred overnight at room temperature. The solution was cooled to −65 °C using an acetone bath with an immersion cooler, before adding a sodium methoxide solution, 25 wt. % in methanol (11 mL, 10.8 mmol, 2.3 eq.) dropwise. The reaction mixture was heated up to room temperature and stirred for 2 h. The reaction was quenched with brine (1 mL) and water (1 mL), and the obtained precipitate was filtered off. Upon filtering, the isolated solid was dried under high vacuum overnight.
-
(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-oxo-acetic acid methyl ester (8a)
Yield: 62%; light yellow solid; mp: 140.5–141.5 °C; IR (cm−1): 1731 (CO), 1653 (CO); 1H-NMR (200 MHz, DMSO-d6) δ: 3.91 (3H, s, CH3), 3.92 (3H, s, OCH3), 8.54 (1H, d, J = 2.2 Hz, H-4), 8.57 (1H, d, J = 2.2 Hz, H-6), 8.80 (1H, s, H-2); 13C NMR (50 MHz, DMSO-d6) δ: 32.0 (q), 52.8 (q), 109.2 (s), 114.7 (s), 131.4 (d), 143.0 (d), 145.1 (d), 146.6 (s), 162.6 (s), 176.8 (s), 177.6 (s); Anal. calculated for C11H9BrN2O3 (MW: 297.10): C, 44.47; H, 3.05; N, 9.43%. Found: C, 44.58; H, 3.24; N, 9.65%.
-
(1-Methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-oxo-acetic acid methyl ester (8b)
Yield: 73%; white solid; mp: 97.8–98.8 °C; IR (cm−1): 1729 (CO), 1650 (CO); 1H-NMR (200 MHz, DMSO-d6) δ: 3.91 (3H, s, CH3), 3.93 (3H, s, OCH3), 7.38 (1H, dd, J = 7.8, 4.8 Hz, H-4), 8.44–8.51 (2H, m, H-5 and H-6), 8.74 (1H, s, H-2); 13C NMR (50 MHz, DMSO-d6) δ: 31.8 (q), 52.7 (q), 109.7 (s), 118.3 (s), 119.3 (d), 129.8 (d), 141.8 (d), 144.9 (d) 148.1 (s), 163.1 (s), 176.8 (s), 178.0 (s); Anal. calculated for C11H10N2O3 (MW: 218.21): C, 60.55; H, 4.62; N, 12.84%. Found: C, 60.38; H, 4.78; N, 12.72%.
3.1.3. Synthesis of (1-methyl-1H-pyrrolo[2,3-b]pyridine-3-yl)-[3-(1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanones (6a–f)
A fine powder of sodium hydroxide (60 mg, 1.5 mmol) was quickly added to a solution of appropriate (1-methyl-1H-pyrrolo[2,3-b]pyridine-3-yl)-oxo-acetic acid methyl ester 8a,b (446 mg, 1.5 mmol) and a suitable carboxamidine 9a–c (190 mg, 1.0 mmol) in dry dimethylsulfoxide (DMSO) (2 mL). The resulting solution was allowed to stir at room temperature for 30 min. Then, water and ice were slowly added, and the obtained precipitate was filtered off. The residue was purified by column chromatography using cyclohexane/ethyl acetate (1/1) as the eluent.
-
(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-[3-(1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanone (6a)
Yield: 60%; yellow solid; mp: 258 °C (dec); IR (cm−1): 1630 (CO); 1H-NMR (200 MHz, DMSO-d6) δ: 3.90 (3H, s, CH3), 3.98 (3H, s, CH3), 7.21–7.33 (2H, m, H-5′ and H-6′), 7.56–7.60 (1H, m, H-7′), 7.99–8.06 (1H, m, H-4′), 8.37 (1H, s, H-2′), 8.56 (1H, d, J = 2.2 Hz, H-4), 8.68 (1H, d, J = 2.2 Hz, H-6), 9.24 (1H, s, H-2); Anal. calculated for C20H14BrN5O2 (MW: 436.26): C, 55.06; H, 3.23; N, 16.05%. Found: C, 55.18; H, 3.04; N, 15.89%.
-
(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-[3-(5-fluoro-1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanone (6b)
Yield: 67%; yellow solid; mp: 246 °C (dec); IR (cm−1): 1635 (CO); 1H-NMR (200 MHz, DMSO-d6) δ: 3.97 (3H, s, CH3), 4.04 (3H, s, CH3), 7.17–7.28 (1H, m, H-6′), 7.65–7.78 (2H, m, H-7′ and H-4′), 8.50 (1H, s, H-2′), 8.61 (1H, d, J = 1.8 Hz, H-4), 8.73 (1H, d, J = 1.8 Hz, H-6), 9.30 (1H, s, H-2); Anal. calculated for C20H13BrFN5O2 (MW: 454.25): C, 52.88; H, 2.88; N, 15.42%. Found: C, 55.80; H, 3.00; N, 15.55%.
-
(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-[3-(5-methoxy-1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanone (6c)
Yield: 85%; yellow solid; mp: 288 °C (dec); IR (cm−1): 1637 (CO); 1H-NMR (200 MHz, DMSO-d6) δ: 3.86 (3H, s, CH3), 3.92 (3H, s, CH3), 4.02 (3H, s, OCH3), 6.95–7.08 (1H, m, H-6′), 7.54–7.55 (2H, m, H-7′ and H-4′), 8.35 (1H, s, H-2′), 8.60 (1H, s, H-4), 8.72 (1H, s, H-6), 9.29 (1H, s, H-2); Anal. calculated for C21H16BrN5O3 (MW: 466.29): C, 54.09; H, 3.46; N, 15.02%. Found: C, 54.28; H, 3.30; N, 15.15%.
-
(1-Methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-[3-(1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanone (6d)
Yield: 77%; yellow solid; mp: 230 °C (dec); IR (cm−1): 1620 (CO); 1H-NMR (200 MHz, DMSO-d6) δ: 3.97 (3H, s, CH3), 4.05 (3H, s, CH3), 7.28–7.40 (2H, m, H-5′ and H-6′), 7.46 (1H, dd, J = 9.9, 4.8 Hz, H-4), 7.65 (1H, dd, J = 6.5, 2.1 Hz, H-7′), 8.11 (1H, dd, J = 6.2, 2.4 Hz, H-4′), 8.44 (1H, s, H-2′), 8.51 (1H, dd, J = 4.7, 1.3 Hz, H-6), 8.64 (1H, dd, J = 7.9, 1.3 Hz, H-5), 9.27 (1H, s, H-2); Anal. calculated for C20H15N5O2 (MW: 357.37): C, 67.22; H, 4.23; N, 19.60%. Found: C, 67.08; H, 4.04; N, 19.78%.
-
(1-Methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-[3-(5-fluoro-1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanone (6e)
Yield: 68%; yellow solid; mp: 257 °C (dec); IR (cm−1): 1638 (CO); 1H-NMR (200 MHz, DMSO-d6) δ: 3.98 (3H, s, CH3), 4.05 (3H, s, CH3), 7.18–7.28 (1H, m, H-6′), 7.42–7.50 (1H, m, H-4), 7.66–7.79 (2H, m, H-7′ and H-4′), 8.50 (1H, s, H-2′), 8.52–8.67 (2H, m, H-5 and H-6), 9.26 (1H, s, H-2); Anal. calculated for C20H14FN5O2 (MW: 375.36): C, 64.00; H, 3.76; N, 18.66%. Found: C, 64.20; H, 3.64; N, 18.50%.
-
(1-Methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-[3-(5-methoxy-1-methyl-1H-indol-3-yl)-[1,2,4]oxadiazol-5-yl]-methanone (6f)
Yield: 80%; yellow solid; mp: 231 °C (dec); IR (cm−1): 1623 (CO); 1H-NMR (200 MHz, DMSO-d6) δ: 3.87 (3H, s, CH3), 3.92 (3H, s, CH3), 4.04 (3H, s, OCH3), 6.95–7.08 (1H, m, H-6′), 7.40–7.63 (3H, m, H-7′, H-4′ and H-4), 8.31–8.67 (3H, m, H-2′, H-5 and H-6), 9.29 (1H, s, H-2); Anal. calculated for C21H17N5O3 (MW: 387.39): C, 65.11; H, 4.42; N, 18.08%. Found: C, 65.28; H, 4.60; N, 18.25%.
3.2. Biology
3.2.1. Drugs and Chemicals
The 1,2,4-oxadiazole compounds 6a–f were synthesized at the Department of Pharmacy, University of Palermo (Palermo, Italy). The drugs were dissolved in DMSO. The medium, fetal bovine serum (FBS), penicillin (50 IU mL−1), and streptomycin (50 µg mL−1) were from Gibco (Gaithersburg, MD, USA). All other chemicals were from Sigma (Zwijndrecht, the Netherlands).
3.2.2. Cell Cultures
HPAF-II, Hs766T, and PATU-T cell lines were purchased from the ATCC (American Type Culture Collection) (Manassas, VA, USA). The cell lines were tested for their authentication by STR–PCR (Short Tandem Repeat-Polymerase Chain Reaction). The primary cell line, PDAC3, was obtained from a patient undergoing pancreaticoduodenectomy, as described previously [30]. Additionally, all these cells were routinely tested for mycoplasma using PCR.
The cells were cultured in RPMI-1640 (Roswell Park Memorial Institute 1640) supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin or in DMEM (Dulbecco’s Modified Eagle’s Medium), supplemented with 10% heat-inactivated FBS and 1% HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid). The cells were kept in a humidified atmosphere of 5% CO2 and 95% air at 37 °C and harvested with trypsin-EDTA (Ethylenediaminetetraacetic acid).
3.2.3. Inhibition of Cell Growth
To evaluate the inhibitory effects of the 1,2,4-oxadiazole compounds 6a–f on cell growth, we performed a Sulforhodamine-B (SRB) assay, as previously described [31,32]. Cells were seeded into 96-well flat-bottom plates in triplicate at a density of 3 × 103 cells/well. Cells were incubated at 37 °C for 24 h to create a confluent monolayer, and then they were treated with 100 µL of increasing concentrations of the compounds dissolved in DMSO. After 72 h of treatment, the cells were fixed with 25 µL of 50% cold trichloroacetic acid and kept for at least 60 min at 4 °C. Then, the plates were washed gently with deionized water, dried at r.t. overnight, and stained with 50 µL of 0.4% SRB solution in 1% acetic acid for 15 min at r.t. The excess SRB stain was removed on dried tissues, and the plates were washed with 1% acetic acid and left to dry at r.t. overnight. The SRB was dissolved in 150 µL of tris(hydroxymethyl)aminomethane solution (pH = 8.8 (TRIS base)), and the optical density (OD) was detected at a wavelength of 490 nm and 540 nm. Cell growth inhibition was calculated as the percentage versus vehicle-treated cells (“negative control”), OD (corrected for OD before drug addiction). Finally, the half maximal inhibitory concentration (IC50) was calculated using a non-linear least squares curve fitting (GraphPad Prism 7, Intuitive Software for Science, San Diego, CA, USA).
3.2.4. Enzyme-Linked Immunosorbent Assay (ELISA) for CDK1 Expression
The expression of CDK1 was detected and quantified using an Enzyme-Linked Immunosorbent Assay (ELISA, Human cyclin-dependent kinase 1 (CDK1) ELISA Kit, Catalog Number: MBS707090) according to the manufacturer’s protocol and our previous studies [33]. Volumes of 100 μL of the standard and the sample were added for each well, and the plates were incubated for 2 h at 37 °C. Then, the liquid was removed, and 100 μL of Biotin-antibody was added to each well, the plates were then incubated for 1 h at 37 °C. The medium was removed, and the plates were washed with Wash Buffer (200 µL). Subsequently, 100 μL of HRP-avidin was added to each well, and the plates were incubated for 1 h at 37 °C. The washing process was repeated five times. At the end, 90 µL of TMB Substrate was added to each well, and the plates were incubated for 30 min at 37 °C in the dark. Finally, 50 μL of stop solution was added to each well. The optical density of each well was determined using a microplate reader set to 450 nm.
3.2.5. Apoptosis
Cells were seeded in 96-well plates (5 × 103 cells/well) and, after one day, treated with drugs at the indicated concentrations for 24 h. At the end of the treatments, cells were fixed with 4% paraformaldehyde in PBS at r.t. for 30 min, then washed twice with PBS and stained with annexin V-FITC in a binding buffer (10 mM HEPES/NaOH pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2) for 10 min at r.t. in the dark. Finally, cells were washed with binding buffer solution and fluorescence was measured using a multimode plate reader with excitation and emission filters at 485 nm and 535 nm, respectively. Parallel studies using the same method, counted the cells at the fluorescence microscopy and evaluated the apoptotic index. The values were normalized on cell proliferation using a crystal violet assay.
3.2.6. Statistical Analysis
All the SRB and ELISA assays were carried out in triplicate and repeated at least three times, whereas the percentages of apoptotic cells were calculated taking into account three biological replicates. The data were evaluated using a GraphPad Prism (GraphPad Software, San Diego, CA, USA). Data were expressed as mean values ± SEM and analyzed using a Student’s t-test.
3.3. In Silico Studies
3.3.1. Molecular Modelling and Docking
The molecular docking into CDK1 was performed for the compound 6b. The CDK1 (PDB: 4YC6) X-ray crystal structure with a resolution of 2.60 Å, R-value 0.227 (observed), was obtained from the protein data bank [34]. The ligand 6b was saved as a mol file, and the docking was performed using the Molecular Operating Environment (MOE) 2015.10.
3.3.2. ADME Studies
The ADME predictions were performed using SwissADME prediction software. The number of H-bond donors, H-bond acceptors, and rotatable bonds, as well as the bioavailability, GI absorption, and BBB permeation were evaluated.
4. Conclusions
In the present study we efficiently synthesized a new series of 1,2,4-oxadiazole derivatives 6a–f that showed promising antiproliferative activities in vitro against a series of PDAC cells.
PDAC is one of the most lethal forms of cancer, characterized by poor survival rates of 5 years, late diagnosis, and lack of effective treatment. The American Cancer Society estimated that 48,220 people will die of pancreatic cancer in the United States (US) in 2021 [35]. Due to the lack of specific symptoms, most patients are diagnosed at an advanced stage and cannot undergo surgical resection. The currently available therapeutic regimens include combinations of standard chemotherapy drugs, such as FOLFIRINOX [33,34]. However, PDACs are typically resistant to these treatments, and new therapeutic strategies are urgently needed.
Most PDACs harbor mutations linked to cell-cycle regulation [36], and TP53 is one of the most frequent and relevant driver genes of pancreatic tumorigenesis. When the TP53 gene is mutated, CDK1 is no longer inhibited and stimulates progression through the cell cycle [10]. In addition, recent studies have shown that CDK1 overexpression is associated with more advanced stages of significantly shorter survival of PDAC patients [37]. Thus, CDK1 inhibition seems a promising strategy for the treatment of PDAC, and there are multiple preclinical studies and clinical trials investigating the efficacy and tolerability of CDK1-inhibiting drugs.
Because of the key role of CDK1 in the regulation of apoptosis, we assessed the ability of compound 6b to induce apoptosis, which proved to be comparable to that observed for gemcitabine. As one of the major hallmarks of PDAC is its resistance to apoptosis induction by gemcitabine [38], these results support future investigation of this compound as an excellent candidate for combination with gemcitabine.
In addition, ADME predictions demonstrated that compound 6b possessed good pharmacokinetic properties, followed the Lipinski rule of five, and had a high level of gastrointestinal absorption, indicating that our compound could be formulated as an oral drug.
Overall, the results of cytotoxicity, modulation of CDK1 expression, and apoptosis obtained with compound 6b support its role as an interesting hit compound for further chemical modification and biological analysis in order to discover new compounds for the treatment of patients with PDAC.
Author Contributions
C.P., B.P. and S.C. performed chemical research and analyzed the data. C.P., A.P. and A.A. performed biological research and analyzed the data. P.D., G.J.P., E.G. and D.C. participated in the design of the research and the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the following grants: CCA Foundation 2015 and 2018 grants (to E.G.), KWF Dutch Cancer Society grants (KWF project#19571, #13598) (to E.G.), and AIRC IG grant 24444 (to E.G.); PRIN2017, Prot.No.2017E84AA4 (to P.D.) and European Union 2014–2020 PON Ricerca e Innovazione grant from the Italian Ministry of Education, University and Research, entitled “PROGEMA-Processi Green per l’Estrazione di Principi Attivi e la Depurazione di Matrici di Scarto e Non” (ARS01_00432) (to P.D.). EU Social Fund (FSE) and the Canary Islands ACIISI for a predoctoral grant TESIS2020010055 and a short internship grant EST2021010019 cofinanced 85% (to A.P.).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Conroy T., Desseigne F., Ychou M., Bouché O., Guimbaud R., Bécouarn Y., Adenis A., Raoul J.L., Gourgou-Bourgade S., de la Fouchardière C., et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 3.Caparello C., Meijer L.L., Garajova I., Falcone A., Le Large T.Y., Funel N., Kazemier G., Peters G.J., Vasile E., Giovannetti E. FOLFIRINOX and translational studies: Towards personalized therapy in pancreatic cancer. World J. Gastroenterol. 2016;22:6987–7005. doi: 10.3748/wjg.v22.i31.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Von Hoff D.D., Ervin T., Arena F.P., Chiorean E.G., Infante J., Moore M., Seay T., Tjulandin S.A., Ma W.W., Saleh M.N., et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Large T.Y.S., Bijlsma M.F., El Hassouni B., Mantini G., Lagerweij T., Henneman A.A., Funel N., Kok B., Pham T.V., de Haas R., et al. Focal adhesion kinase inhibition synergizes with nab-paclitaxel to target pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2021;40:91. doi: 10.1186/s13046-021-01892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roskoski R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2019;152:104609. doi: 10.1016/j.phrs.2019.104609. [DOI] [PubMed] [Google Scholar]
- 7.Roskoski R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2019;144:19–50. doi: 10.1016/j.phrs.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Wu P., Nielsen T.E., Clausen M.H. Small-molecule kinase inhibitors: An analysis of FDA-approved drugs. Drug Discov. Today. 2016;21:5–10. doi: 10.1016/j.drudis.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 9.Wu P., Nielsen T.E., Clausen M.H. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci. 2015;36:422–439. doi: 10.1016/j.tips.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Lim S., Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 11.Wijnen R., Pecoraro C., Carbone D., Fiuji H., Avan A., Peters G.J., Giovannetti E., Diana P. Cyclin Dependent Kinase-1 (CDK-1) Inhibition as a Novel Therapeutic Strategy against Pancreatic Ductal Adenocarcinoma (PDAC) Cancers. 2021;13:4389. doi: 10.3390/cancers13174389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelosi E., Castelli G., Testa U. Pancreatic Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells. Biomedicines. 2017;5:65. doi: 10.3390/biomedicines5040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakemi S., Sun H.H. Nortopsentins A, B, and C. Cytotoxic and antifungal imidazolediylbis[indoles] from the sponge Spongosorites ruetzleri. J. Org. Chem. 1991;56:4304–4307. doi: 10.1021/jo00013a044. [DOI] [Google Scholar]
- 14.Cascioferro S., Attanzio A., Di Sarno V., Musella S., Tesoriere L., Cirrincione G., Diana P., Parrino B. New 1,2,4-Oxadiazole Nortopsentin Derivatives with Cytotoxic Activity. Mar. Drugs. 2019;17:35. doi: 10.3390/md17010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pecoraro C., Carbone D., Aiello D., Carbone A. Synthesis and cytotoxic activity of 3-[2-(1H-Indol-3-yl)-1,3-thiazol-4-yl]-1H-pyrrolo[3,2-c]pyridine hydrobromides, analogues of marine alkaloid nortopsentin. Arkivoc. 2021 doi: 10.24820/ark.5550190.p011.640. [DOI] [Google Scholar]
- 16.Di Franco S., Parrino B., Gaggianesi M., Pantina V.D., Bianca P., Nicotra A., Mangiapane L.R., Lo Iacono M., Ganduscio G., Veschi V., et al. CHK1 inhibitor sensitizes resistant colorectal cancer stem cells to nortopsentin. iScience. 2021;24:102664. doi: 10.1016/j.isci.2021.102664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Petri G., Cascioferro S., El Hassouni B., Carbone D., Parrino B., Cirrincione G., Peters G.J., Diana P., Giovannetti E. Biological Evaluation of the Antiproliferative and Anti-migratory Activity of a Series of 3-(6-Phenylimidazo[2,1-b][1,3,4]thiadiazol-2-yl)-1H-indole Derivatives against Pancreatic Cancer Cells. Anticancer Res. 2019;39:3615–3620. doi: 10.21873/anticanres.13509. [DOI] [PubMed] [Google Scholar]
- 18.Cascioferro S., Li Petri G., Parrino B., El Hassouni B., Carbone D., Arizza V., Perricone U., Padova A., Funel N., Peters G.J., et al. 3-(6-Phenylimidazo [2,1-b][1,3,4]thiadiazol-2-yl)-1H-Indole Derivatives as New Anticancer Agents in the Treatment of Pancreatic Ductal Adenocarcinoma. Molecules. 2020;25:329. doi: 10.3390/molecules25020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitaku E., Smith D.T., Njardarson J.T. Analysis of the Structural Diversity, Substitution Patterns, and Frequency of Nitrogen Heterocycles among U.S. FDA Approved Pharmaceuticals. J. Med. Chem. 2014;57:10257–10274. doi: 10.1021/jm501100b. [DOI] [PubMed] [Google Scholar]
- 20.Carbone D., Parrino B., Cascioferro S., Pecoraro C., Giovannetti E., Di Sarno V., Musella S., Auriemma G., Cirrincione G., Diana P. 1,2,4-Oxadiazole Topsentin Analogs with Antiproliferative Activity against Pancreatic Cancer Cells, Targeting GSK3β Kinase. ChemMedChem. 2021;16:537–554. doi: 10.1002/cmdc.202000752. [DOI] [PubMed] [Google Scholar]
- 21.Parrino B., Carbone D., Cascioferro S., Pecoraro C., Giovannetti E., Deng D., Di Sarno V., Musella S., Auriemma G., Cusimano M.G., et al. 1,2,4-Oxadiazole topsentin analogs as staphylococcal biofilm inhibitors targeting the bacterial transpeptidase sortase A. Eur. J. Med. Chem. 2021;209:112892. doi: 10.1016/j.ejmech.2020.112892. [DOI] [PubMed] [Google Scholar]
- 22.Avan A., Caretti V., Funel N., Galvani E., Maftouh M., Honeywell R.J., Lagerweij T., Van Tellingen O., Campani D., Fuchs D., et al. Crizotinib inhibits metabolic inactivation of gemcitabine in c-Met-driven pancreatic carcinoma. Cancer Res. 2013;73:6745–6756. doi: 10.1158/0008-5472.CAN-13-0837. [DOI] [PubMed] [Google Scholar]
- 23.Maftouh M., Belo A.I., Avan A., Funel N., Peters G.J., Giovannetti E., Van Die I. Galectin-4 expression is associated with reduced lymph node metastasis and modulation of Wnt/β-catenin signalling in pancreatic adenocarcinoma. Oncotarget. 2014;5:5335–5349. doi: 10.18632/oncotarget.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Large T.Y., Mantini G., Meijer L.L., Pham T.V., Funel N., van Grieken N.C., Kok B., Knol J., van Laarhoven H.W., Piersma S.R., et al. Microdissected pancreatic cancer proteomes reveal tumor heterogeneity and therapeutic targets. JCI Insight. 2021;5:e138290. doi: 10.1172/jci.insight.138290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parry D., Guzi T., Shanahan F., Davis N., Prabhavalkar D., Wiswell D., Seghezzi W., Paruch K., Dwyer M.P., Doll R., et al. Dinaciclib (SCH 727965), a Novel and Potent Cyclin-Dependent Kinase Inhibitor. Mol. Cancer Ther. 2010;9:2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 26.Shendge A.K., Chaudhuri D., Mandal N. The natural flavones, acacetin and apigenin, induce Cdk-Cyclin mediated G2/M phase arrest and trigger ROS-mediated apoptosis in glioblastoma cells. Mol. Biol. Rep. 2021;48:539–549. doi: 10.1007/s11033-020-06087-x. [DOI] [PubMed] [Google Scholar]
- 27.Huang J., Chen P., Liu K., Liu J., Zhou B., Wu R., Peng Q., Liu Z.-X., Li C., Kroemer G., et al. CDK1/2/5 inhibition overcomes IFNG-mediated adaptive immune resistance in pancreatic cancer. Gut. 2020;70:890–899. doi: 10.1136/gutjnl-2019-320441. [DOI] [PubMed] [Google Scholar]
- 28.Navarro-Retamal C., Caballero J. Flavonoids as CDK1 Inhibitors: Insights in Their Binding Orientations and Structure-Activity Relationship. PLoS ONE. 2016;11:e0161111. doi: 10.1371/journal.pone.0161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rovithi M., Avan A., Funel N., Leon L.G., Gomez V.E., Wurdinger T., Griffioen A.W., Verheul H.M., Giovannetti E. Development of bioluminescent chick chorioallantoic membrane (CAM) models for primary pancreatic cancer cells: A platform for drug testing. Sci. Rep. 2017;7:44686. doi: 10.1038/srep44686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciarrillo R., Wojtuszkiewicz A., Kooi I.E., Gómez V.E., Boggi U., Jansen G., Kaspers G.J., Cloos J., Giovannetti E. Using RNA-sequencing to Detect Novel Splice Variants Related to Drug Resistance in In Vitro Cancer Models. J. Vis. Exp. 2016;118:54714. doi: 10.3791/54714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massihnia D., Avan A., Funel N., Maftouh M., van Krieken A., Granchi C., Raktoe R., Boggi U., Aicher B., Minutolo F., et al. Phospho-Akt overexpression is prognostic and can be used to tailor the synergistic interaction of Akt inhibitors with gemcitabine in pancreatic cancer. J. Hematol. Oncol. 2017;10:9. doi: 10.1186/s13045-016-0371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianco C., Giovannetti E., Ciardiello F., Mey V., Nannizzi S., Tortora G., Troiani T., Pasqualetti F., Eckhardt G., de Liguoro M., et al. Synergistic antitumor activity of ZD6474, an inhibitor of vascular endothelial growth factor receptor and epidermal growth factor receptor signaling, with gemcitabine and ionizing radiation against pancreatic cancer. Clin. Cancer Res. 2006;12:7099–7107. doi: 10.1158/1078-0432.CCR-06-0833. [DOI] [PubMed] [Google Scholar]
- 34.Brown N.R., Korolchuk S., Martin M.P., Stanley W.A., Moukhametzianov R., Noble M., Endicott J.A. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat. Commun. 2015;6:6769. doi: 10.1038/ncomms7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.American Cancer Society Key Statistics for Pancreatic Cancer. [(accessed on 2 November 2021)]. Available online: https://www.cancer.org/cancer/pancreatic-cancer/about/key-statistics.html.
- 36.Le Large T.Y.S., Bijlsma M.F., Kazemier G., van Laarhoven H.W.M., Giovannetti E., Jimenez C.R. Key biological processes driving metastatic spread of pancreatic cancer as identified by multi-omics studies. Semin. Cancer Biol. 2017;44:153–169. doi: 10.1016/j.semcancer.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Dong S., Huang F., Zhang H., Chen Q. Overexpression of BUB1B, CCNA2, CDC20, and CDK1 in tumor tissues predicts poor survival in pancreatic ductal adenocarcinoma. Biosci. Rep. 2019;39:BSR20182306. doi: 10.1042/BSR20182306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elnaggar M., Giovannetti E., Peters G.J. Molecular targets of gemcitabine action: Rationale for development of novel drugs and drug combinations. Curr. Pharm. Des. 2012;18:2811–2829. doi: 10.2174/138161212800626175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.