Abstract
Staphylococcus aureus coagulase type VII strains have been the strains most frequently isolated from staphylococcal food poisoning outbreaks in Tokyo, Japan. We applied pulsed-field gel electrophoresis (PFGE) of chromosomal DNA digested with SmaI to characterize 129 coagulase type VII strains. These were isolated from 129 cases occurring in outbreaks in 35 districts during a 16-year period (1980–1995). The 129 outbreak strains were classified into three types, designated A (n = 115), B (n = 10), and C (n = 4). Types A and C were further divided into 33 (A1 to A33) and 4 (C1 to C4) subtypes, respectively. Strains of the same subtypes were isolated from food poisoning cases in the same districts at time intervals of 1 or 2 to 5 years. PFGE typing appears to be a useful method for subdividing strains of S. aureus coagulase type VII. A combination of coagulase typing and PFGE typing would provide more detailed information than the former method alone in epidemiologic investigations of staphylococcal food poisoning.
Staphylococcal food poisoning, caused by enterotoxin-producing Staphylococcus aureus is an important food-borne infection in many countries, including Japan (9). Over a period of 20 years (1980–1999) in Japan, a total of 2,525 outbreaks of staphylococcal food poisoning were reported, which involved 59,964 persons and three deaths, according to the data of the Ministry of Health and Welfare of Japan. The foods most frequently involved in this type of food poisoning were typical Japanese-style processed foods composed mainly of rice, i.e., nigirimeshi (rice ball), inarizushi (fried bean curd stuffed with vinegared rice), chakinsushi (sushi wrapped in a layer of eggs), and benntou (delivered luncheon with rice) (12, 18, 27). The staphylococcal enterotoxins (SEs) most frequently detected were SEA and SEA plus SEB (12, 18).
In Japan, the coagulase typing method has been used successfully in epidemiological investigations of staphylococcal food poisoning. This method is based on the eight antigenic types (I to VIII) of coagulase (28). It has been reported that coagulase types VII (accounting for about 70% of the total outbreaks), III (12%), II (11%), and VI (3%) are, respectively, the first, second, third, and fourth most predominant types of coagulases responsible for food poisoning incidents throughout Japan (18). These four coagulase types have been almost exclusively implicated in outbreaks in the metropolitan city of Tokyo (12).
Recently, methods of direct analysis of the bacterial genome, such as pulsed-field gel electrophoresis (PFGE), ribotyping (1, 4, 20), and random amplified polymorphic DNA analysis (21, 29), have been used as alternative methods in the epidemiology of S. aureus infections. In particular, PFGE based on analysis of the whole genome by restriction endonuclease digestion has been shown to be a useful method for investigating the source, transmission, and spread of nosocomial infections and, more particularly, for epidemiologic typing and determination of the genetic relatedness of methicillin-resistant S. aureus strains (5, 6, 7, 10, 11, 16). Also, PFGE has proven valuable in epidemiological studies of methicillin-resistant S. aureus found in horses (22, 23) and dogs (19) in veterinary medicine. However, little information is available concerning the ability of the PFGE method to type S. aureus isolates from cases of food poisoning (25).
Therefore, in the current study we used PFGE of chromosomal DNA digested with SmaI to characterize the strains of S. aureus coagulase type VII most frequently involved in staphylococcal food poisoning outbreaks in Tokyo, Japan. We then conducted a molecular epidemiological analysis of coagulase type VII strains by using PFGE.
MATERIALS AND METHODS
Bacterial strains.
All 129 S. aureus coagulase type VII strains used in this study were from 129 incidents of staphylococcal food poisoning which occurred in 35 different districts of Tokyo, Japan, between 1980 and 1995. One hundred and twelve (86.8%) of the 129 strains were enterotoxigenic; of these, 66 produced SEA, 13 SEB, 2 SEC, 5 SED, and 26 SEA and SEB. The food-poisoning origin strains belonging to coagulase types II (3 strains; one each of SEA, SEB plus SED, and SED), III (3 strains, SEA), IV (5 strains, SEA), and VI (2 strains, SEA) were also used for references. Enterotoxins were detected by reversed passive latex agglutination (RPLA) using a commercial SET-RPLA kit (Denka Seiken, Tokyo, Japan). Enterotoxin types could not be distinguished by means of coagulase typing, since overlaps occurred. All strains were stored in 10% skim milk suspensions at −80°C until use.
Coagulase typing.
Coagulase typing was carried out according to the procedure of Ushioda et al. (28) using a coagulase typing kit (Denka Seiken) with neutralizing rabbit antisera specific to the eight coagulase types I to VIII. A 0.1-ml amount of each antiserum and normal rabbit serum (as a control) was added to 0.1 ml of the supernatant obtained from an overnight culture (Difco, Detroit, Mich.) of each test isolate, and this solution was incubated at 37°C for 1 h, after which 0.2 ml of rabbit plasma was added. Inhibition of coagulation after further incubation at 37°C for at least 1 h indicated the coagulase type. Strains whose coagulase activity was not neutralized by the set of antisera and strains that reacted to more than two specific sera were designated nontypeable.
PFGE typing.
The preparation of chromosomal DNA of S. aureus strains and the fragmentation of their genomic DNA with SmaI (New England Biolabs, Beverly, Mass.) were performed as described previously (2, 24). PFGE was performed with a 1% agarose gel using a CHEF-DR II system (Bio-Rad Laboratories, Inc., Hercules, Calif.) in a 0.5× Tris-borate-EDTA buffer at 14°C. The running parameters were as follows: initial pulse, 5 s; final pulse, 40 s; voltage, 6 V/cm; time, 22 h. After PFGE, the gel was stained with ethidium bromide, washed with distilled water, and photographed under UV light. Lambda DNA concatemers (New England Biolabs) for determining the size of SmaI-digested fragments were used as molecular size standards. PFGE patterns were interpreted based on the method of Tenover et al. (26). Isolates were considered the same strain if all bands matched, subtypes of the same strain if the patterns differed by one to three bands, and different strains if the patterns differed by four or more bands.
Phage typing.
Phage typing was performed as described previously (3) by using the international bacteriophage typing set of 23 phages (group I, 29, 52, 52A, 79, and 80; group II, 3A, 3C, 55, and 71; group III, 6, 42E, 47, 53, 54, 75, 77, 83A, 84, and 85; group V, 94 and 96; miscellaneous, 81 and 95). These phages were used at 100× routine test dilution.
RESULTS
PFGE analysis of coagulase type VII strains.
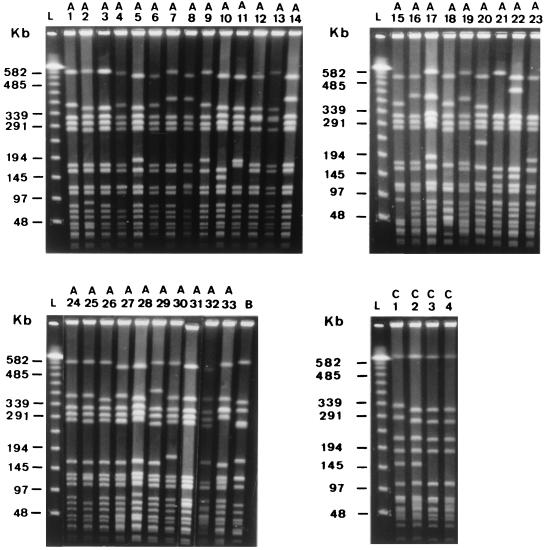
SmaI cut the chromosomal DNA into 10 to 13 fragments ranging in size from 48.5 to 650 kb. According to the criteria proposed by Tenover et al. (26), the 129 strains were classified into three types, arbitrarily designated A (n = 115), B (n = 10), and C (n = 4). Types A and C were further divided into 33 (A1 to A33) and 4 (C1 to C4) subtypes, respectively (Fig. 1).
FIG. 1.
PFGE of SmaI-digested genomic fragments of S. aureus coagulase VII strains isolated from staphylococcal food poisoning outbreaks in Tokyo, Japan. Subtypes A1 to A8, A24 to A29, and A33 and type B included 19, 4, 12, 2, 2, 2, 7, 2, 24, 3, 10, 4, 2, 2, 2, and 10 strains, respectively. Other subtypes included only one strain each. Lanes L, lambda ladder DNA concatemers used as molecular size markers.
PFGE patterns of strains belonging to other coagulase types.
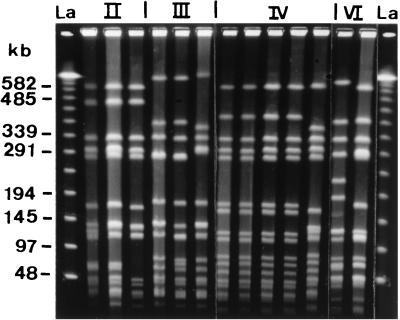
The PFGE patterns of coagulase type II (n = 3), III (n = 3), IV (n = 5), and VI (n = 2) strains differed from each other (Fig. 2). Coagulase type II, III, IV, and VI strains produced 3, 3, 2, and 2 subtypes, respectively. The patterns of strains belonging to these four coagulase types were different from those of coagulase type VII strains.
FIG. 2.
PFGE of SmaI-digested fragments of S. aureus coagulase type II, III, IV, and VI strains from staphylococcal food poisoning outbreaks in Tokyo, Japan. Lanes La, lambda ladder DNA concatemers used as molecular size markers.
Epidemiology of coagulase type VII strains.
Table 1 shows the geographical distribution of the PFGE patterns of the 129 coagulase type VII strains according to the districts in which they were isolated. Subtypes A1 (n = 19), A3 (n = 12), A7 (n = 7), A24 (n = 24), and A26 (n = 10) were most predominant, accounting for 55.8% of the total strains. These subtypes were isolated repeatedly from food poisoning incidents between 1980 and 1994, although they were isolated at different outbreak sites. For instance, subtype A24 was first found in 1981 and successively found in 1982 to 1985, 1987, 1989, 1990, and 1994 isolates. Also, subtype A1 was recovered from outbreaks in 1982 to 1987, 1989, 1991, and 1994. In contrast, type B (7.8% of the total strains) was isolated from eight districts in 1980 to 1983 and was not detected thereafter.
TABLE 1.
Distribution of PFGE types of S. aureus coagulase type VII strains isolated from staphylococcal food poisoning outbreaks which occurred in 35 districts of Tokyo, Japan, 1980 to 1995
| District | No. of outbreaks | PFGE type (yr of isolation) |
|---|---|---|
| Adachi-ku | 4 | B (1980), A1 (1986), A2 (1987), A24 (1990) |
| Arakawa-ku | 1 | A14 (1986) |
| Itabashi-ku | 6 | A2 (1982), A7 (1982), A24 (1982, 1983), A1 (1985), A27 (1990) |
| Edogawa-ku | 5 | A1 (1984), A24 (1984), A3 (1986), A18 (1988), A29 (1994) |
| Ohta-ku | 10 | A26 (1983), A1 (1984, 1987), A7 (1984), A24 (1984, 1985, 1994), A5 (1988), A6 (1989), A22 (1991) |
| Katsushika-ku | 4 | B (1982), A26 (1984), A1 (1986), A24 (1990) |
| Kita-ku | 2 | A7 (1982), A26 (1983) |
| Koutou-ku | 2 | A4 (1981), A24 (1984) |
| Shinagawa-ku | 4 | A7 (1983), A13 (1983), A21 (1985), A27 (1990) |
| Shibuya-ku | 3 | A12 (1981), A3 (1984), A26 (1989) |
| Shinjuku-ku | 7 | A3 (1980, 1982, 1987), A7 (1982), A8 (1982), A24 (1984, 1989) |
| Suginami-ku | 7 | A11 (1981), A24 (1981, 1983), C4 (1981), A19 (1984), A9 (1991), A3 (1994) |
| Sumida-ku | 3 | B (1980), A33 (1986), A1 (1987) |
| Setagaya-ku | 3 | A3 (1982), A24 (1984), A2 (1994) |
| Taitou-ku | 8 | A28 (1980), B (1981, 1981, 1982), A1 (1982), A26 (1987), A27 (1992), A3 (1993) |
| Chuo-ku | 5 | A3 (1980), A26 (1981), A24 (1985), A1 (1986), A32 (1986) |
| Chiyoda-ku | 6 | B (1980), A28 (1981), A4 (1982), A24 (1984), A20 (1986), A3 (1989) |
| Toshima-ku | 2 | A3 (1981), C3 (1994) |
| Nakano-ku | 5 | A26 (1981), A6 (1982), A15 (1982), A24 (1985), C1 (1995) |
| Nerima-ku | 4 | A26 (1981), A25 (1983), A1 (1987), A24 (1987) |
| Bunkyo-ku | 2 | A10 (1990), A29 (1992) |
| Minato-ku | 4 | B (1983), A16 (1989), A3 (1991), A7 (1991) |
| Meguro-ku | 2 | A1 (1991), A17 (1992) |
| Ome-shi | 3 | B (1981), A26 (1991, 1991) |
| Koganei-shi | 1 | A24 (1983) |
| Kodaira-shi | 1 | A1 (1984) |
| Tachikawa-shi | 3 | A7 (1986), C2 (1988), A1 (1989) |
| Hachioji-shi | 5 | A24 (1981, 1985), A30 (1981), A1 (1989, 1994) |
| Higashimurayama-shi | 3 | A24 (1983), A33 (1988), A31 (1993) |
| Hino-shi | 4 | A2 (1981), A24 (1982), A1 (1984), A5 (1988) |
| Fuchu-shi | 1 | A25 (1990) |
| Fusa-shi | 2 | A23 (1984), A1 (1989) |
| Machida-shi | 2 | A25 (1983), A27 (1990) |
| Mitaka-shi | 3 | B (1980), A24 (1982), A1 (1983) |
| Musashino-shi | 2 | A8 (1983), A1 (1985) |
Subtype A1, A3, A7, A24, and A26 strains were isolated from incidents which occurred in 17, 10, 6, 18, and 9 districts, respectively, indicating the wide geographic spread of these clones. Interestingly, strains of the same subtypes were isolated repeatedly from different outbreaks in the same districts, such as Itabashi-ku (subtype A 24, 1982 and 1983), Ohta-ku (A24, 1984, 1985, and 1994; A1, 1984 and 1987), Shinjuku-ku (A24, 1984 and 1989; A3, 1980, 1982, and 1987), Suginami-ku (A24, 1981 and 1983), and Hachioji-shi (A24, 1981 and 1985; A1, 1989 and 1994).
Phage typing.
One hundred and twenty-three (95.3%) of the 129 strains were typeable at 100× the routine test dilution. Two strains belonged to phage group I, 46 to group III, 1 to the miscellaneous (Misc) group, and 74 to mixed groups (I+III, 28 strains; I+Misc, 1; III+Misc, 25; and I+III+Misc, 20).
DISCUSSION
In evaluating a typing system for ecologic and epidemiological purposes, a series of attributes should be assessed, including ability to type strains, reproducibility and stability of patterns, and discriminatory power. In Japan, coagulase typing (28) has been used widely in epidemiologic investigations of staphylococcal infections and food poisoning. This method, which is based on antigenic differences between coagulases, has contributed greatly to our understanding of the source, transmission, and spread of food poisoning outbreaks. Unfortunately, this method has not been employed outside Japan. In the 1960s and 1970s, coagulase types causing food poisoning in Tokyo, Japan, were limited to four coagulase types, types II, III, VI, and VII. Since the 1980s, isolates of coagulase type VII have been a major cause of food poisoning. Phage typing (2, 3) has long been considered the “gold standard” for epidemiological tools throughout the world. The majority of strains isolated from foods implicated in food poisoning incidents are human phage typeable and susceptible to group I or III (17, 30). In the present study, we attempted phage typing of 129 coagulase type VII strains to clarify the relation between PFGE patterns and phage groups. Most (96.7%) of the typeable strains belonged to group III, I+III, III+Misc, or I+III+Misc. There was no significant relation between PFGE patterns and phage groups.
In this study, we applied the PFGE method to characterize the strains of S. aureus coagulase type VII most frequently involved in staphylococcal food poisoning outbreaks in Tokyo. By means of PFGE, we identified three type patterns and 37 subtype patterns among the 129 outbreak strains examined. These patterns were different from those presented by the strains of other coagulase types (II, III, IV, and VI). Each of the four coagulase types was further classified by PFGE. We therefore believe that coagulase typing is a useful epidemiological tool early in an epidemiological investigation and that PFGE analysis can be performed subsequently for further differentiation of the isolates. We thus recommend a combination of phenotypic and genotypic typing methods for more comprehensive epidemiology of staphylococcal food poisoning.
On the basis of genomic typing by PFGE, we performed an epidemiological investigation of food poisoning caused by coagulase type VII strains. In Tokyo, subtype A24 was most frequently observed, followed by subtypes A1, A3, A26, and A7, suggesting that these subtypes were widely involved in food poisoning. These subtypes were isolated repeatedly from food poisoning incidents throughout the investigation. Interestingly, strains of the same genotypes (subtypes A1, A3, and A24) were isolated from food poisoning incidents in the same districts at time intervals of 1 or 2 to 5 years. To further characterize coagulase type VII strains, the PFGE patterns of these types of isolates from food poisoning incidents which occurred in other districts outside Tokyo must be compared.
In the past 20 years, there has been an increased incidence of food poisoning caused by coagulase type VII, which has now become the most predominant type in Japan. The exact reason why food poisoning with coagulase VII strains has occurred in Japan is not known. Where organisms of this type live remains unknown. We are now starting a survey of the distribution of this type of strain in humans, animals, food, and the natural environment.
Staphylococcal food poisoning is caused by S. aureus producing enterotoxins. However, enterotoxigenic strains of the coagulase-positive species Staphylococcus intermedius have also been isolated frequently from dogs (8, 13, 14, 15). Khambaty et al. (15) reported the isolation of S. intermedius (enterotoxin A production) from a food poisoning outbreak involving butter-blend products in the western United States. They also demonstrated the usefulness of the PFGE method in distinguishing the outbreak strains of S. intermedius. Cases of food poisoning due to this organism have not yet been reported in Japan.
In conclusion, the results of the present study suggest that PFGE analysis might be a useful tool for subdividing coagulase types within S. aureus. PFGE analysis together with coagulase typing should be useful in detailed epidemiological studies.
REFERENCES
- 1.Aarestrup F M, Wegener H C, Rosdahl V T. Evaluation of phenotypic and genotypic methods for epidemiological typing of Staphylococcus aureus isolates from bovine mastitis in Denmark. Vet Microbiol. 1995;45:139–150. doi: 10.1016/0378-1135(95)00043-a. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair J E, Williams R E O. Phage typing of staphylococci. Bull WHO. 1961;24:771–784. [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg H M, Rimland D, Kiehlbauch J A, Terry P M, Wachsmuth I K. Epidemiologic typing of Staphylococcus aureus by DNA restriction fragment length polymorphisms of rRNA genes: elucidation of the clonal nature of a group of bacteriophage-nontypeable, ciprofloxacin-resistant, methicillin-susceptible S. aureus isolates. J Clin Microbiol. 1992;30:362–369. doi: 10.1128/jcm.30.2.362-369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couto I, Melo-Cristino J, Fernandes M L, Garcia T, Serrano N, Salgado M J, Torres-Pereira A, Santos Sanches I, de Lencastre H. Unusually large number of methicillin-resistant Staphylococcus aureus clones in a Portuguese hospital. J Clin Microbiol. 1995;33:2032–2035. doi: 10.1128/jcm.33.8.2032-2035.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Lencastre H, de Lencastre A, Tomasz A. Methicillin-resistant Staphylococcus aureus isolates recovered from a New York City hospital: analysis by molecular fingerprinting techniques. J Clin Microbiol. 1996;34:2121–2124. doi: 10.1128/jcm.34.9.2121-2124.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Adhami W, Roberts L, Vickery A, Inglis B, Gibbs A, Stewart P R. Epidemiological analysis of a methicillin-resistant Staphylococcus aureus outbreak using restriction fragment length polymorphisms of genomic DNA. J Gen Microbiol. 1991;137:2713–2720. doi: 10.1099/00221287-137-12-2713. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda S, Tokuno H, Ogawa H, Sasaki M, Kishimoto T, Kawano J, Shimizu A, Kimura S. Enterotoxigenicity of Staphylococcus intermedius strains isolated from dogs. Zentbl Bakteriol Mikrobiol Hyg A. 1984;258:360–367. doi: 10.1016/s0176-6724(84)80054-1. [DOI] [PubMed] [Google Scholar]
- 9.Genigeorgis C A. Present state of knowledge on staphylococcal intoxication. Int J Food Microbiol. 1989;9:327–360. doi: 10.1016/0168-1605(89)90100-1. [DOI] [PubMed] [Google Scholar]
- 10.Givney R, Vickery A, Holliday A, Pegler M, Benn R. Evolution of an endemic methicillin-resistant Staphylococcus aureus population in an Australian hospital from 1967 to 1996. J Clin Microbiol. 1998;36:552–556. doi: 10.1128/jcm.36.2.552-556.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichiyama S, Ohta M, Shimokata K, Kato N, Takeuchi J. Genomic DNA fingerprinting by pulsed-field gel electrophoresis as an epidemiological marker for study of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1991;29:2690–2695. doi: 10.1128/jcm.29.12.2690-2695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igarashi H. Current trends on staphylococcal food poisoning outbreaks in Tokyo. Food Sanit Res. 1993;43:31–45. . (In Japanese.) [Google Scholar]
- 13.Kaji Y, Kato E. Occurrence of enterotoxigenic staphylococci in household and laboratory dogs. Jpn J Vet Res. 1980;28:86–94. [PubMed] [Google Scholar]
- 14.Kato E, Kaji Y, Kaneko K. Enterotoxigenic staphylococci of canine origin. Am J Vet Res. 1978;39:1771–1773. [PubMed] [Google Scholar]
- 15.Khambaty F M, Bennett R W, Shah D B. Application of pulsed-field gel electrophoresis to the epidemiological characterization of Staphylococcus intermedius implicated in a food-related outbreak. Epidemiol Infect. 1994;113:75–81. doi: 10.1017/s0950268800051487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemaitre N, Sougakoff W, Masmoudi A, Fievet M-H, Bismuth R, Jarlier V. Characterization of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus involved in nosocomial spread. J Clin Microbiol. 1998;36:81–85. doi: 10.1128/jcm.36.1.81-85.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niskanen A, Koiranen L. Correlation of enterotoxin and thermonuclease production with some physiological and biochemical properties of staphylococcal strains isolated from different sources. J Food Prot. 1977;40:543–548. doi: 10.4315/0362-028X-40.8.543. [DOI] [PubMed] [Google Scholar]
- 18.Oda T. A review of staphylococcal food poisoning in Japan. J Food Hyg Soc Jpn. 1998;39:J179–185. . (In Japanese.) [Google Scholar]
- 19.Pak S-I, Han H-R, Shimizu A. Characterization of methicillin-resistant Staphylococcus aureus isolated from dogs in Korea. J Vet Med Sci. 1999;61:1013–1018. doi: 10.1292/jvms.61.1013. [DOI] [PubMed] [Google Scholar]
- 20.Prevost G, Jaulhac B, Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992;30:967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saulnier P, Bourneix C, Prevost G, Andremont A. Random amplified polymorphic DNA assay is less discriminant than pulsed-field gel electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:982–985. doi: 10.1128/jcm.31.4.982-985.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seguin J C, Walker R D, Caron J P, Kloos W E, George C G, Hollis R J, Jones R N, Pfaller M A. Methicillin-resistant Staphylococcus aureus outbreak in a veterinary teaching hospital: potential human-to-animal transmission. J Clin Microbiol. 1999;37:1459–1463. doi: 10.1128/jcm.37.5.1459-1463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimizu A, Kawano J, Yamamoto C, Kakutani O, Anzai T, Kamada M. Genetic analysis of equine methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis. J Vet Med Sci. 1997;59:935–937. doi: 10.1292/jvms.59.935. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu A, Kawano J, Yamamoto C, Kakutani O, Fujita M. Comparison of pulsed-field gel electrophoresis and phage typing for discriminating poultry strains of Staphylococcus aureus. Am J Vet Res. 1997;58:1412–1416. [PubMed] [Google Scholar]
- 25.Suzuki Y, Saito M, Ishikawa N. Restriction fragment length polymorphism analysis by pulsed-field gel electrophoresis for discrimination of Staphylococcus aureus isolates from foodborne outbreaks. Int J Food Microbiol. 1999;46:271–274. doi: 10.1016/s0168-1605(98)00193-7. [DOI] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Guest commentary. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terayama T, Ushioda H, Shingaki M, Inaba M, Kai A, Sakai S. Coagulase types of Staphylococcus aureus from food poisoning outbreaks and a kind of incriminated foods. Annu Rep Tokyo Metropolitan Res Lab Public Health. 1977;28:1–4. . (In Japanese with English summary.) [Google Scholar]
- 28.Ushioda H, Terayama T, Sakai S, Zen-Yoji H, Nishiwaki M, Hidano A. Coagulase typing of Staphylococcus aureus and its application in routine work. In: Jeljaszewicz J, editor. Staphylococci and staphylococcal infections, Zentbl. Bakteriol. Suppl. 10. Stuttgart, Germany: Gustav Fischer Verlag; 1981. pp. 77–83. [Google Scholar]
- 29.Van Belkum A, Kluytmans J, Van Leeuwen W, Bax R, Quint W, Peters E, Fluit A D, Vandenbroucke-Grauls C, Van Den Brule A, Koeleman H, Melchers W, Meis J, Elaichouni A, Vaneechoutte M, Moonens F, Maes N, Struelens M, Tenover F, Verbrugh H. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J Clin Microbiol. 1995;33:1537–1547. doi: 10.1128/jcm.33.6.1537-1547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wieneke A. Enterotoxin production by strains of Staphylococcus aureus isolated from foods and human beings. J Hyg (Cambridge) 1974;73:255–262. doi: 10.1017/s0022172400024104. [DOI] [PMC free article] [PubMed] [Google Scholar]