Abstract
Alcohol is a well-known risk factor for hepatocellular carcinoma. Autophagy plays a dual role in liver cancer, as it suppresses tumor initiation and promotes tumor progression. Transcription factor EB (TFEB) is a master regulator of lysosomal biogenesis and autophagy, which is impaired in alcohol-related liver disease. However, the role of TFEB in alcohol-associated liver carcinogenesis is unknown. Liver-specific Tfeb knockout (KO) mice and their matched wild-type (WT) littermates were injected with the carcinogen diethylnitrosamine (DEN), followed by chronic ethanol feeding. The numbers of both total and larger tumors increased significantly in DEN-treated mice fed ethanol diet than in mice fed control diet. Although the number of tumors was not different between WT and L-Tfeb KO mice fed either control or ethanol diet, the number of larger tumors was less in L-Tfeb KO mice than in WT mice. No differences were observed in liver injury, steatosis, inflammation, ductular reaction, fibrosis, and tumor cell proliferation in DEN-treated mice fed ethanol. However, the levels of glypican 3, a marker of malignant hepatocellular carcinoma, markedly decreased in DEN-treated L-Tfeb KO mice fed ethanol in comparison to the WT mice. These findings indicate that chronic ethanol feeding promotes DEN-initiated liver tumor development, which is attenuated by genetic deletion of hepatic TFEB.
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved lysosomal degradation pathway that involves the formation of double membrane autophagosomes, which carry the autophagic cargos and fuse with lysosomes wherein the cargos are degraded. Autophagy is critical for the clearance of long-lived proteins and insoluble protein aggregates, invading microbes as well as damaged organelles to maintain cellular homeostasis at both basal and stress conditions.1, 2, 3 The liver is one of the most dynamic organs in mammals and humans, which is critical in regulating the metabolism of carbohydrates, lipids, proteins, amino acids, and xenobiotoics. It is well accepted that autophagy plays crucial roles in liver physiology and pathology,4, 5, 6 and that dysregulation of autophagy is associated with various liver diseases, including alcohol-related liver disease (ALD),7 nonalcoholic fatty liver disease (NAFLD),8,9 nonalcoholic steatohepatitis,10 viral hepatitis,11,12 and hepatocellular carcinoma (HCC).13
HCC was the third leading cause of cancer deaths worldwide in 2020, which claims >830,000 deaths annually.14 In the United States, the mortality rate of HCC increased approximately 40% between 2000 and 2016.15 It has long been known that chronic alcohol consumption causes ALD, which is characterized with simple steatosis that can progress to fibrosis and cirrhosis, resulting in the development of HCC.7,16,17 Alcohol consumption results in an estimated 18,200 to 21,300 cancer deaths, or 3.2% to 3.7% of all US cancer deaths.18 Several mechanisms have been implicated in alcohol-induced liver carcinogenesis, including direct DNA damage by acetaldehyde, increased hepatic iron deposition, and oxidative stress, as well as epigenetic changes.19
Recent evidence suggests that autophagy plays a dual role in tumorigenesis and cancer progression. During the tumor initiation stage, autophagy acts as a tumor suppressor to inhibit tumorigenesis, which is likely achieved by maintaining cellular homeostasis via removing damaged organelles, and mitigating DNA damage and genome instability induced by oxidative stress. However, transformed cancerous cells use autophagy as a survival mechanism by providing nutrients against the harsh microenvironment. Therefore, boosting autophagy activity is generally beneficial for tumor prevention, whereas a combination of chemotherapeutic drugs with autophagy inhibitors has been considered a promising therapeutic strategy for tumor treatment. Transcription factor EB (TFEB) belongs to the microphthalmia-associated transcription factor (MITF) basic helix-loop-helix leucine-zipper transcription factor superfamily,20 which is a master regulator of lysosomal biogenesis and autophagy-related gene expression.21,22 In vertebrates, the MITF family includes four evolutionarily conserved and closely related members: MITF, TFEB, transcription factor E3 (TFE3), and transcription factor EC (TFEC).23 TFEB and TFE3 were originally reported to be translocated in chromosomes, which results in gene fusions. The substitution of TFEB promoter leads to increased TFEB protein level and nuclear translocation in a subset of renal tumors in children.24 Subsequent studies revealed that genetic alterations of TFEB are associated with different types of human cancers, including renal carcinomas,25 alveolar sarcomas,26 non–small-cell lung cancer,27,28 melanomas,29 pancreatic ductal adenocarcinoma,30, 31, 32 prostate cancer,33 colorectal cancer,34 gastric carcinoma,35 and breast cancer.36,37 In addition to its oncogenic activity, TFEB also controls cancer cell-autonomous responses and participates in regulating cellular functions, such as cell cycle and metabolism, as well as the tumor microenvironment, such as epithelial-mesenchymal transition.38 Alcohol feeding plus binge decreases nuclear TFEB in hepatocytes, resulting in insufficient autophagy, and promotes alcohol-induced liver injury in mice.39,40 A recent study reported that TFEB also plays a role in liver regeneration on injury through the regulation of SRY-Box transcription factor 9 (SOX9) expression and liver cell fate.41 TFEB-mediated up-regulation of SOX9 expression promotes cholangiocarcinoma development by inducing hepatic progenitor cell proliferation after liver injury.41 However, the role of TFEB in human HCC, especially in alcohol-associated HCC, is unknown. Although there is a strong association of alcohol consumption with HCC, few animal models have been established to mimic the pathogenesis of alcohol-associated HCC, which has significantly limited our understanding on how alcohol promotes HCC. Herein, hepatic carcinogen diethylnitrosamine (DEN) injection plus chronic alcohol feeding was used to establish an alcohol-associated liver tumor model in L-Tfeb knockout (KO) and matched wild-type (WT) mice, and further investigate the role of TFEB in alcohol-associated HCC development and progression.
Materials and Methods
Animal Experiments
Tfeb flox/flox (Tfebflox/flox; C57BL/6N) mice were generated, as described previously,42 and were crossed with albumin Cre (Alb-Cre; C57BL/6J; The Jackson Laboratory, Bar Harbor, ME) to generate liver-specific Tfeb KO (Tfebflox/flox, Alb-Cre+, L-Tfeb KO) mice. Tfebflox/flox, Alb-Cre mice were further crossed with Tfebflox/flox mice (6N background) more than six times. The Tfebflox/flox, Alb-Cre and their matched Alb-Cre-negative littermates were used for the experiments. All animals received humane care. Mice were specific pathogen free and maintained in a barrier rodent facility under standard experimental conditions. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Kansas Medical Center (Kansas City, KS). For alcohol-associated liver cancer model, 2-week-old male mice were injected with one dose of carcinogen DEN (10 mg/kg, intraperitoneally). Eleven weeks later, mice were acclimated to the Lieber-DeCarli liquid control diet (F1259SP; Bio-Serv, Flemington, NJ) for 1 week, followed by further feeding with the liquid control diet or 5% ethanol diet (F1258SP; Bio-Serv) for 24 weeks. The volume of control diet given to mice was matched to the volume of ethanol diet consumed. For Western diet-associated liver cancer model, 2-week–old male mice were given one dose of carcinogen DEN (10 mg/kg, intraperitoneally). Two weeks later, mice were fed either a control low-fat diet (LFD) or Western diet (WD), containing 42% fat calories and 0.2% cholesterol for 22 or 34 weeks, respectively. Mice were then euthanized, and blood and liver tissues were collected. Liver tumor numbers and sizes were measured. Liver injury was determined by measuring serum alanine aminotransferase (ALT). Liver cryosections were obtained, and hematoxylin and eosin staining as well as confocal microscopy were performed as described previously.43
Antibodies
The following antibodies were used for Western blot analysis and immunohistochemistry staining: TFEB (A303-673A; Bethyl, Montgomery, TX), glyceraldehyde-3-phosphate dehydrogenase (2118; Cell Signaling, Danvers, MA), alpha smooth muscle actin (α-SMA; ab5694; Abcam, Cambridge, UK), glypican 3 (ab66596; Abcam), cytokeratin 19 (CK19; AB_2133570; Developmental Studies Hybridoma Bank, Iowa City, IA), Ki-67 (GTX16667; Gene Tex, Irvine, CA), F4/80 (14-4801; Invitrogen, Carlsbad, CA), proliferating cell nuclear antigen (PCNA; sc-56; Santa Cruz, Dallas, TX), lysosome-associated membrane protein 1 (Lamp1; 1D4B; Developmental Studies Hybridoma Bank), Lamp2 (ABL-93; Developmental Studies Hybridoma Bank), autophagy related gene 5 (Atg5; 12994; Cell Signaling), β-catenin (610,154; BD Biosciences, Franklin Lakes, NJ), p62 (H00008878-M01; Abnova, Taipei, Taiwan), TFE3 (HPA023881; Sigma-Aldrich, St. Louis, MO), and SOX9 (ab5535; Millipore, Burlington, MA). The rabbit polyclonal anti-microtubule–associated protein 1A/1B-light chain 3 (LC3B) antibody was made using a peptide representing the NH2-terminal 14 amino acids of human LC3B and an additional cysteine (PSEKTFKQRRTFEQC). The rabbit anti-Vatp6v1a was a gift from Dr. Dennis Brown (Harvard Medical School, Boston, MA). Horseradish peroxidase–conjugated secondary antibodies used for Western blot analysis and immunohistochemistry staining were from Jackson ImmunoResearch (West Grove, PA) and Vector Laboratories (Burlingame, CA), respectively.
Western Blot Analysis
Total liver lysates were prepared using radioimmunoprecipitation assay buffer [1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl (lauryl) sulfate]. Protein (30 μg) was separated by a 12% SDS-PAGE gel before transfer to a polyvinylidene difluoride membrane. Membranes were probed using appropriate primary and secondary antibodies, and were developed with SuperSignal West Pico chemiluminescent substrate (34080; Life Technologies, Carlsbad, CA).
Histology and Immunohistochemistry
Paraffin-embedded liver sections were stained with hematoxylin and eosin. In addition, immunostaining for indicated antibodies was performed as previously described.44
Lipid Droplet Staining
For Oil Red O staining, liver tissue cryosections were washed twice with phosphate-buffered saline and then incubated with 60% isopropanol for 1 minute. Tissue cryosections were dried in a 37°C incubator for approximately 10 minutes before incubating with Oil Red O solution. Oil Red O solution was prepared by adding 0.35 g Oil Red O (0625; Sigma-Aldrich) to 100 mL of 100% isopropanol, which was further diluted 1.7 times in water and filtered immediately before use. Slides were incubated with Oil Red O solution for 15 minutes. The Oil Red O solution was aspirated from the slides, and 60% isopropanol was added to the slides for several minutes to remove any residual Oil Red O. Slides were then washed in phosphate-buffered saline and stained for 30 seconds with hematoxylin (GHS132; Sigma-Aldrich), followed by more washes in distilled water. Slides were mounted with a mounting medium [glycerol in phosphate-buffered saline (6:1)], and then microscopy was performed.
Sirius Red and Reticulin Staining
Paraffin-embedded liver tissue sections were dewaxed and rehydrated followed by staining sections with Sirius Red Solution. Reticulin staining was conducted according to the manufacturer's instructions, as previously described (Dako, Santa Clara, CA).45
Electron Microscopy
Liver tissues were fixed with 2% glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.4) followed by 1% OsO4. After dehydration, thin sections were stained with uranyl acetate and lead citrate. Images were acquired digitally by using a JEM 1016CX electron microscope (JEOL, Peabody, MA). The sizes of lipid droplets were quantified using ImageJ software version 1.44p (NIH, Bethesda, MD; http://imagej.nih.gov/ij).
Statistical Analysis
All experimental data are expressed as means ± SEM and were subjected to one-way analysis of variance with Bonferroni post hoc test or t-test where appropriate. P < 0.05 was considered significant.
Results
Deletion of Hepatic TFEB Does Not Have a Significant Impact on the Tumor Burden Induced by DEN and Chronic Alcohol Feeding in Mice
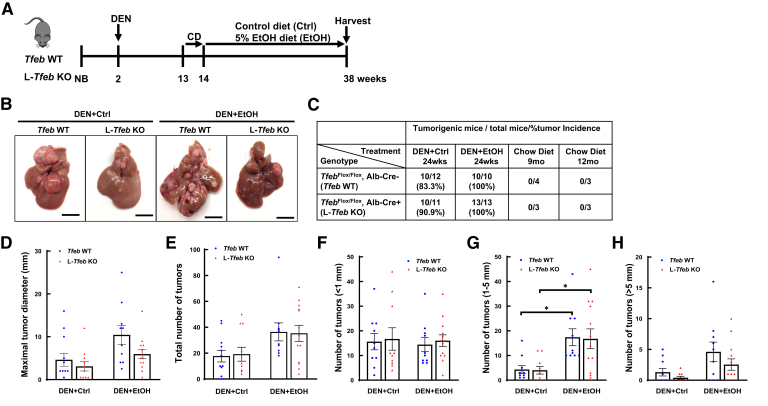
To determine the role of TFEB in alcohol-associated liver tumorigenesis, L-Tfeb KO mice and WT mice were injected with DEN, then fed either a liquid control diet or ethanol diet for 24 weeks (Figure 1A). Most mice fed a control diet (83% of WT and 91% of L-Tfeb KO mice) developed liver tumors, whereas all ethanol-fed mice developed liver tumors regardless of the genotype (Figure 1, B and C). Moreover, the liver tumor size and number in ethanol-fed mice were higher than in mice fed the control diet in both WT and L-Tfeb KO mice. There was no difference in the total number of tumors between WT and L-Tfeb KO mice, although the number of large size tumors decreased approximately 30% in L-Tfeb KO mice fed either control or ethanol diet (Figure 1, D and E). The tumor size was further divided into different categories, and the numbers of small tumors (<1 mm) were found to be similar in both WT and L-Tfeb KO mice fed with either control or ethanol diet (Figure 1F). The number of bigger size tumors (between 1 and 5 mm diameter) was also similar between WT and L-Tfeb KO mice, but was significantly higher in ethanol-fed mice than mice fed with the control diet regardless of the genotype (Figure 1G). The number of tumors >5 mm in diameter increased more than fourfold in ethanol-fed mice compared with control diet–fed mice in both WT and L-Tfeb KO mice, but the number of these larger tumors decreased around 50% in L-Tfeb KO mice, compared with WT mice fed with either control or ethanol diet (Figure 1H). The data suggest that chronic alcohol consumption promotes liver tumor progression. Hepatic loss of TFEB did not significantly affect DEN or DEN and alcohol feeding-induced liver tumorigenesis in mice, although there was a trend towards loss of TFEB decreasing the tumor numbers and size.
Figure 1.
Loss of hepatic transcription factor EB (TFEB) dampens diethylnitrosamine (DEN)–alcohol–induced hepatocellular carcinoma (HCC) in mice. A: The scheme of DEN-alcohol–Associated HCC model. Male mice at the age of 2 weeks were given one dose of carcinogen DEN (10 mg/kg, intraperitoneally). Eleven weeks later, mice were acclimated to the Lieber-DeCarli liquid control (Ctrl) diet for 1 week, followed by further feeding with the liquid Ctrl or 5% ethanol diet (EtOH) for 24 weeks. B: Representative gross images of livers from Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice from different treatments are shown. C: Summary of tumor formation in Tfeb WT and L-Tfeb KO mice from different treatments. D and E: Maximal tumor diameter (D) and total number of tumors (E) of each mouse from different treatments. F–H: Number of tumors within different size categories of each mouse from different treatments. Data are presented as means ± SEM (D–H). n = 10 to 13; n = 12 Tfeb WT DEN+Ctrl; n = 10 Tfeb WT DEN+EtOH; n = 11 L-Tfeb KO DEN+Ctrl; n = 13 L-feb KO DEN+EtOH. ∗P < 0.05 (one-way analysis of variance with Bonferroni post hoc test). Scale bars = 10 mm (B). Alb-Cre, albumin Cre; CD, control diet; NB, newborn.
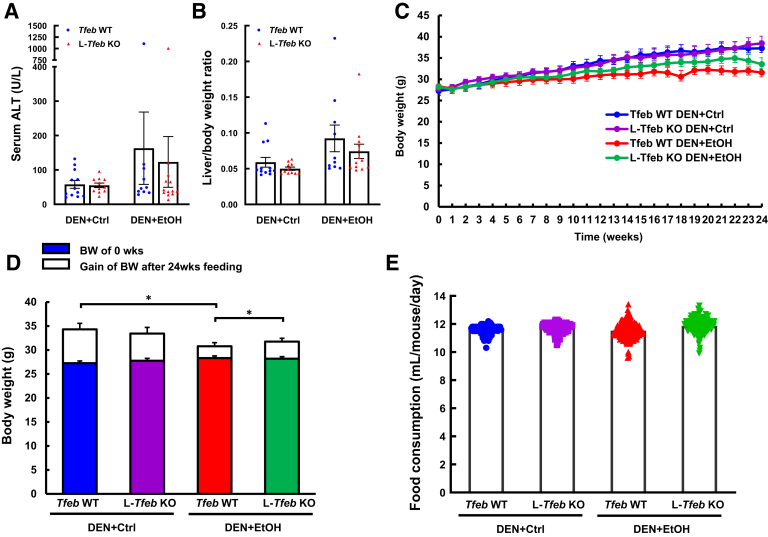
Liver injury and hepatomegaly in mice was examined by analyzing serum ALT activity and liver/body weight ratio. Except for one outlier mouse in WT and L-Tfeb KO group that had high ALT values for around 1000 U/L after ethanol feeding, there was no significant difference of serum ALT activities in ethanol-fed mice compared with that in control diet–fed mice (Figure 2A). Ethanol feeding caused slightly increased liver/body weight ratio (Figure 2B), which is likely due to the increased tumor burden in ethanol-fed mouse livers. Although there was a trend towards decrease in both serum ALT levels and liver/body weight ratio in L-Tfeb KO mice (Figure 2, A and B), they were not statistically different. Body weight and food consumption was monitored during the entire period of experiments. WT mice fed ethanol gained less body weight than the mice fed the control diet. Ethanol-fed L-Tfeb KO mice also had more body weight gain than ethanol-fed WT mice (Figure 2, C and D). However, there was no difference in the food intake among all the experimental groups (Figure 2E), suggesting that the decreased body weight gain in ethanol-fed WT mice is not due to the amount of food intake, but is likely because of the poorer health condition as a result of the tumor development. Taken together, these data indicate that chronic ethanol feeding induced higher tumor incidence and increased tumor size in DEN-injected mouse livers. Overall, genetic deletion of Tfeb in mouse livers did not have a significant impact on DEN and chronic alcohol feeding-induced tumor burden in mice.
Figure 2.
Chronic ethanol (EtOH) feeding increases liver injury and hepatomegaly, and reduces body weight gain in diethylnitrosamine (DEN)–treated mouse livers. Male Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice were treated with a DEN-alcohol–associated hepatocellular carcinoma model. Serum alanine aminotransferase (ALT) activity (A) and liver/body weight (BW) ratio (B) were measured. Mouse body weight was monitored weekly (C and D), and volume of liquid food consumption was recorded daily (E). Data are presented as means ± SEM (A–E). n = 10 to 13 (A–E). ∗P < 0.05 (one-way analysis of variance with Bonferroni post hoc test). Ctrl, control.
Western Diet Promotes DEN-Initiated Liver Tumorigenesis, which Is Attenuated by Genetic Deletion of TFEB
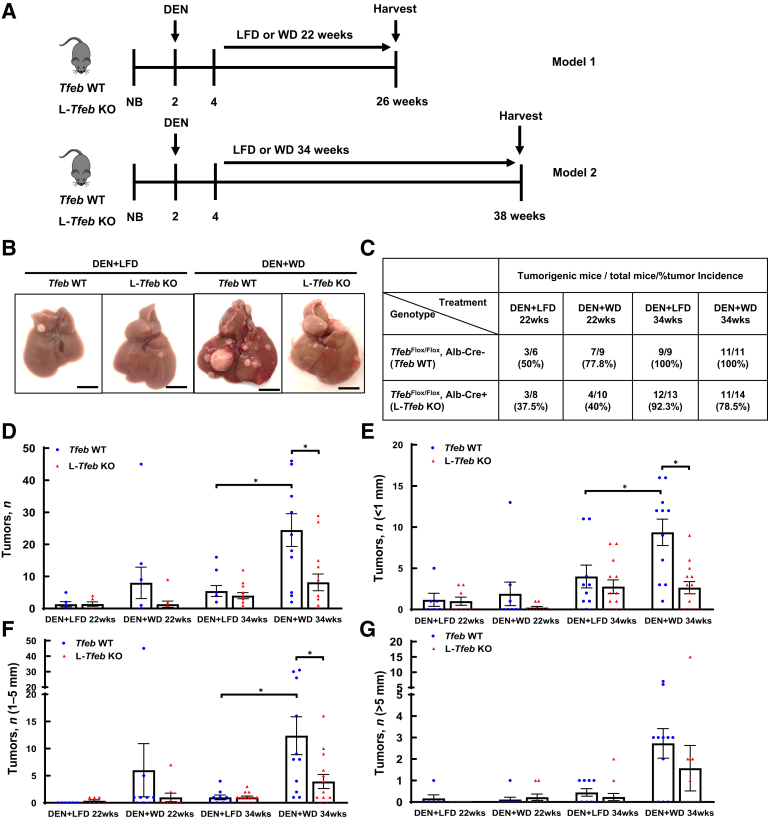
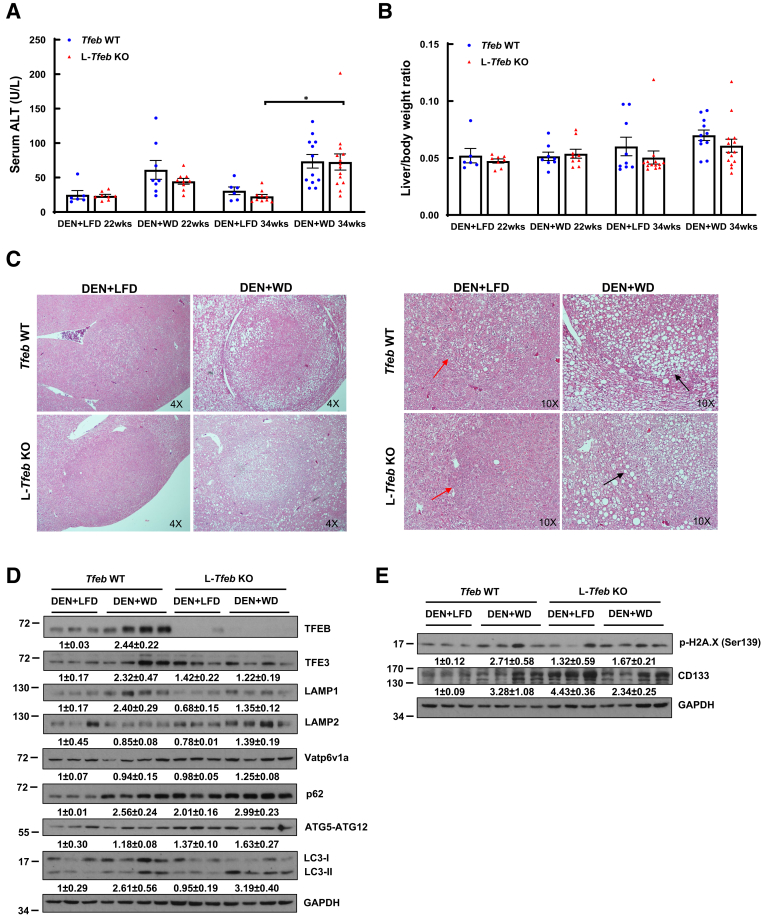
As NAFLD is becoming the fastest growing cause of HCC worldwide,46 the role of TFEB in Western diet–associated liver tumorigenesis was examined. L-Tfeb KO mice and WT mice were injected with DEN and then fed either a WD, containing 42% fat calories and 0.2% cholesterol, or a control LFD for either 22 or 34 weeks (Figure 3A). Interestingly, 77.8% DEN-treated WT mice fed WD for 22 weeks developed liver tumors, whereas only 50% LFD-fed WT mice developed liver tumors. In contrast, the tumor incidence decreased in DEN-treated L-Tfeb KO mice fed either WD (40%) or LFD (37.5%). After feeding for 34 weeks, all the DEN-treated WT mice fed either LFD or WD developed liver tumors, but approximately 10% and 20% L-Tfeb KO mice fed LFD or WD had still not developed tumors (Figure 3, B and C). The number of total liver tumors increased slightly in DEN-treated mice fed WD for 22 weeks but increased approximately fivefold in WT mice fed WD for 34 weeks, compared with WT mice fed LFD. However, the number of liver tumors only marginally increased in DEN-treated L-Tfeb KO mice fed WD for 22 or 34 weeks compared with those in LFD-fed L-Tfeb KO mice. The number of small size (<1 mm), medium size (1 to 5 mm), and large size (>5 mm) tumors also increased in DEN-treated mice fed WD in both WT and L-Tfeb KO mice, but this increase was more dramatic in WT mice compared with that in L-Tfeb KO mice. Moreover, compared with WT mice, the overall tumor numbers (including small and larger size) in DEN-treated L-Tfeb KO mice fed either LFD or WD were lower than those in WT mice (Figure 3, D–G). The serum ALT values were slightly higher in DEN-treated WT and L-Tfeb KO mice fed WD for either 22 or 34 weeks than in mice fed LFD (Figure 4A). No significant difference of liver/body weight ratio was found among all the groups of mice (Figure 4B). Hematoxylin and eosin staining results showed well-differentiated HCC cells with increased nuclei/cytoplasm ratio in DEN-treated WT and L-Tfeb KO mice fed LFD (Figure 4C). WD-fed mice not only had increased steatosis in normal hepatocytes but also in well-differentiated HCC cells in DEN-treated WT and L-Tfeb KO mice (Figure 4C). The changes of several autophagy- and lysosome-related proteins in WT and L-Tfeb KO mice were next determined. Results from the Western blot analysis indicated that DEN-treated and WD-fed WT mice had increased hepatic levels of TFEB, TFE3, LAMP1, p62, and LC3-II, but LAMP2, Vatp6v1a, and ATG5-ATG12 levels were not affected. Although the levels of hepatic TFEB were almost undetectable in L-Tfeb KO mice, the levels of other autophagy (p62, LC3-II, and ATG5-ATG12) and lysosomal (LAMP1, LAMP2, and Vatp6v1a) proteins were almost comparable between the WT and L-Tfeb KO mice (Figure 4D). This is likely due to the compensatory effects of other MITF family proteins, such as TFE3, in the L-Tfeb KO mice, as previously reported.39 WD feeding also increased the hepatic levels of p-H2A.X (DNA damage marker) and CD133 (cancer stem cell marker) in DEN-treated mice, which were not affected significantly by the deletion of hepatic Tfeb (Figure 4E). Taken together, these data indicate that WD promotes DEN-initiated liver tumorigenesis, which is likely potentiated by TFEB.
Figure 3.
Western diet (WD) promotes diethylnitrosamine (DEN)–initiated liver tumorigenesis, which is attenuated by genetic deletion of transcription factor EB (TFEB) in mice. A: The scheme of DEN–Western diet–associated hepatocellular carcinoma model. Male mice at the age of 2 weeks were given one dose of carcinogen DEN (10 mg/kg, intraperitoneally). Two weeks later, mice were fed a control low-fat diet (LFD) or WD containing 42% fat calories and 0.2% cholesterol for 22 or 34 weeks, respectively. B: Representative images of livers from Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice from different treatments are shown. C: Summary of tumor formation in Tfeb WT and L-Tfeb KO mice from different treatments. D: Total number of tumors of each mouse from different treatments. E–G: Number of tumors with different sizes of each mouse from different treatments. Data are presented as means ± SEM (D–G). n = 6 to 14; n = 6 Tfeb WT DEN+LFD 22wks; n = 9 Tfeb WT DEN+WD 22wks; n = 9 Tfeb WT DEN+LFD 34wks; n = 11 Tfeb WT DEN+WD 34wks; n = 8 L-Tfeb KO DEN+LFD 22wks; n = 10 L-Tfeb KO DEN+WD 22wks; n = 13 L-Tfeb KO DEN+LFD 34wks; n = 14 L-Tfeb KO DEN+WD 34wks. ∗P < 0.05 (one-way analysis of variance with Bonferroni post hoc test). Scale bar = 10 mm (B). Alb-Cre, albumin Cre; NB, newborn.
Figure 4.
Western diet (WD) increases liver injury in diethylnitrosamine (DEN)–treated mice. Male Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice were treated with a DEN-WD–associated hepatocellular carcinoma (HCC) model. A and B: Serum alanine aminotransferase (ALT) activity (A) and liver/body weight ratio (B) were measured. C: Representative images of hematoxylin and eosin staining are shown. Red arrows denote differentiated HCC cells with increased nuclei/cytoplasm ratio. Black arrows denote steatotic HCC cells. D and E: Total liver lysates were subjected to Western blot analysis. Data are presented as means ± SEM (A and B). n = 6 to 14 (A and B); n = 3 to 4 (D and E). ∗P < 0.05 (one-way analysis of variance with Bonferroni post hoc test). Original magnifications, ×4 (C, left panels); ×10 (C, right panels). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LFD, low-fat diet; TFEB, transcription factor EB.
Chronic Ethanol Feeding and Loss of Hepatic TFEB Do Not Further Exacerbate Steatosis and Ductular Reaction in DEN-treated Mice
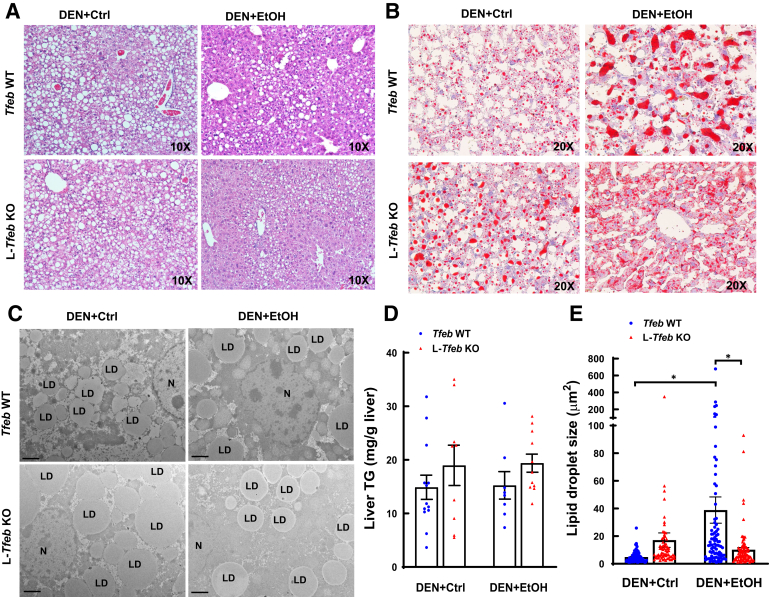
Results from hematoxylin and eosin staining and electron microscopy analysis of liver tissues indicated that DEN-treated WT and L-Tfeb KO mice fed ethanol diet for 24 weeks markedly increased accumulation of lipid droplets in hepatocytes, suggesting that chronic ethanol feeding induces hepatic steatosis. Surprisingly, DEN-treated WT and L-Tfeb KO mice fed the liquid control diet for 24 weeks also developed massive hepatic steatosis, which was not different from that in the ethanol-fed mice (Figure 5, A and B). Increased steatosis in DEN-treated WT and L-Tfeb KO mice fed either a control diet or an ethanol diet was further confirmed by Oil Red O staining of liver tissues, electron microscopy analysis, and quantification of hepatic triglyceride levels (Figure 5, B–D). Moreover, the size of lipid droplets slightly increased in L-Tfeb KO hepatocytes compared with that in the matched WT mice fed a control diet. More interestingly, compared with the mice fed control diet, the size of lipid droplets in hepatocytes significantly increased in ethanol diet-fed WT mice but not in L-Tfeb KO mice, although the overall hepatic triglyceride levels were similar (Figure 5E). These data indicate that long-term liquid control diet feeding induces steatosis in DEN-treated mice. Ethanol feeding and loss of hepatic TFEB do not further exacerbate steatosis in DEN-treated mice.
Figure 5.
Chronic ethanol (EtOH) feeding and loss of hepatic transcription factor EB (TFEB) do not further increase steatosis in diethylnitrosamine (DEN)-treated mice compared with that in liquid control (Ctrl) diet–fed DEN–treated mice. Male Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice were subjected to the DEN-alcohol–associated hepatocellular carcinoma model. A–C: Representative images of hematoxylin and eosin staining (A), Oil Red O staining (B), and electron microscopic images (C) are shown. D: Liver triglyceride (TG) was measured. E: Size of lipid droplets (LDs). The size (area) of >60 LDs was quantified from at least 10 different images. Data are presented as means ± SEM (D) or means ± SD (E). n = 8 to 13 (D); n = 67 to 87 (E). ∗P < 0.05 (one-way analysis of variance with Bonferroni post hoc test). Scale bars = 2 μm (C). Original magnifications, ×10 (A); ×20 (B). N, nucleus.
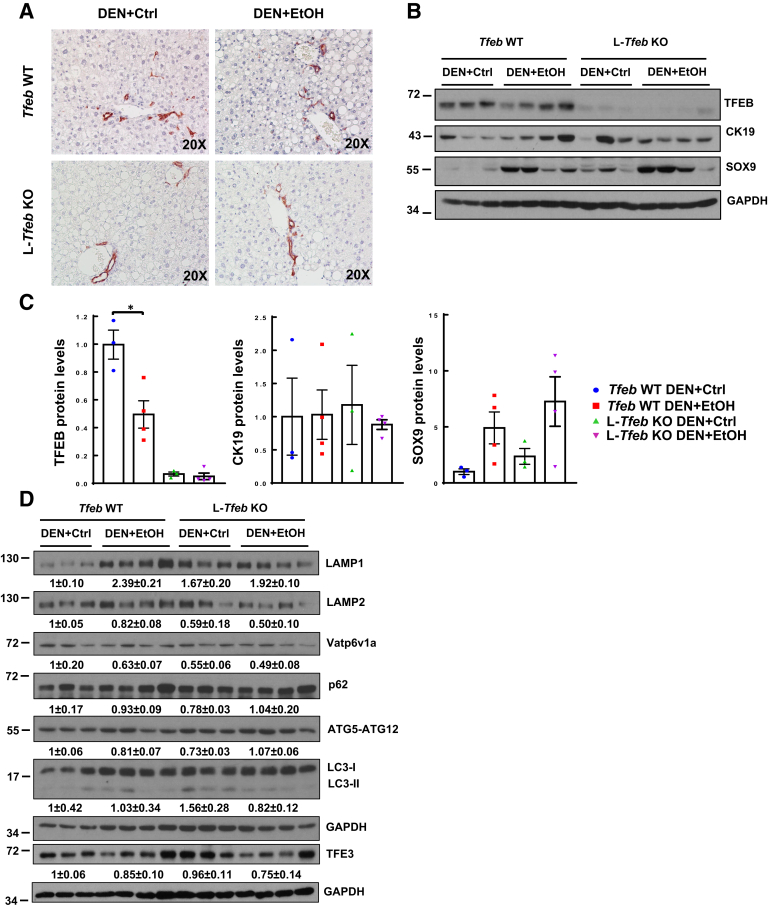
Increased ductular reaction is a typical feature in human severe ALD. Whether chronic alcohol feeding would increase ductular reaction in DEN-treated mouse livers was next investigated. Mice treated with DEN and fed ethanol diet for 24 weeks did not increase CK19-positive ductular cells and structures, as assessed by immunohistochemistry staining of CK19 (Figure 6A) and Western blot analysis of hepatic CK19 (Figure 6, B and C) when compared with both WT and L-Tfeb KO DEN-treated mice that were fed a control diet. Hepatic levels of TFEB markedly decreased in L-Tfeb KO mice compared with those in WT mice, confirming the successful deletion of TFEB in L-Tfeb KO mice livers. The low residual levels of TFEB detected by Western blot analysis in L-Tfeb KO mice are likely owing to the TFEB expression in nonparenchymal cells. DEN-treated and ethanol-fed mice also had decreased hepatic TFEB levels compared with DEN-treated and control diet–fed mice (Figure 6, B and C), which is consistent with the previous findings in chronic plus binge alcohol-fed mice.39 Interestingly, the levels of SOX9, a hepatic progenitor cell marker, increased in DEN-treated and ethanol-fed WT and L-Tfeb KO mice compared with WT and L-Tfeb KO DEN-treated and control diet–fed mice. Notably, the levels of SOX9 were higher in L-Tfeb KO mice than in WT mice treated with DEN and fed the control diet (Figure 6, B and C). For the lysosomal markers, hepatic levels of LAMP1 but not LAMP2 and Vatp6v1a increased in DEN-treated and ethanol-fed mice in comparison with DEN-treated and control diet–fed mice. In contrast, in L-Tfeb KO mice, the hepatic levels of LAMP1 slightly increased, whereas the levels of LAMP2 and Vatp6v1a slightly decreased in DEN-treated mice fed either the ethanol or the control diet in comparison with those in WT mice. In csae of the autophagy-related genes, there were no obvious changes on hepatic p62, ATG5-ATG12, and LC3-II among all the experimental groups (Figure 6D). Together, the data suggest that long-term liquid diet feeding induces hepatic steatosis in DEN-treated mice, which is not further increased by long-term ethanol feeding. Long-term ethanol feeding does not increase ductular reaction compared with liquid control diet in DEN-treated mice. L-Tfeb KO mice do not significantly alter the hepatic autophagy and lysosomal proteins regardless of DEN treatment or alcohol feeding.
Figure 6.
Chronic ethanol (EtOH) feeding and loss of hepatic transcription factor EB (TFEB) do not further increase ductular reaction compared with liquid control (Ctrl) diet–fed diethylnitrosamine (DEN)–treated mouse livers. Male Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice were treated with a DEN-alcohol–associated hepatocellular carcinoma model. A: Representative images of CK19 immunohistochemistry staining are shown. B: Total lysates from mouse livers were subjected to Western blot. C: Densitometry analysis of B. D: Total liver lysates were subjected to Western blot. Data are presented as means ± SEM (C). n = 3 to 4 (C and D). ∗P < 0.05 (one-way analysis of variance with Bonferroni post hoc test). Original magnification, ×20 (A). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Chronic Ethanol Feeding and Loss of Hepatic TFEB Do Not Increase Fibrosis and Inflammation in DEN-Treated Mouse Livers
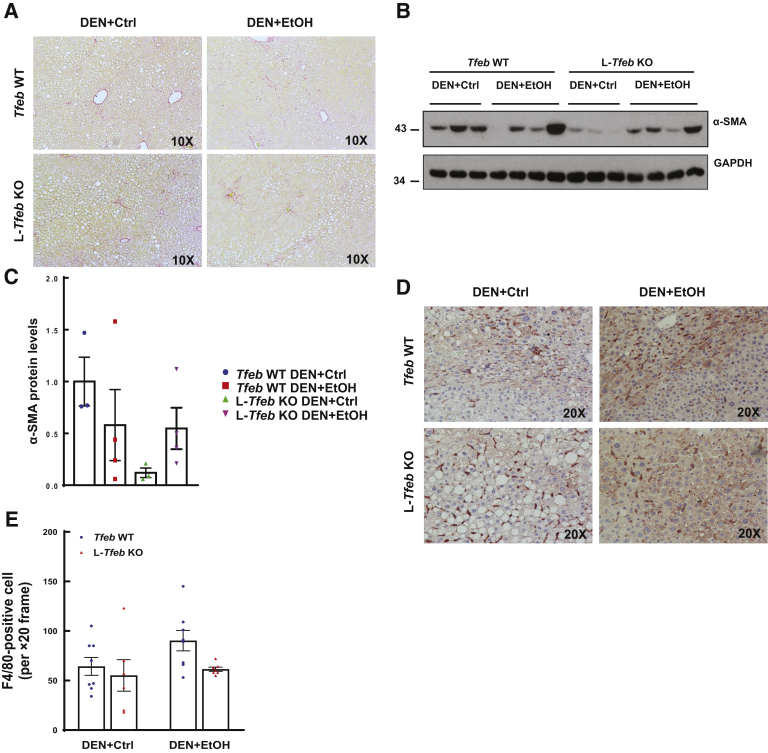
Fibrosis is another typical feature of human ALD, so the next set of experiments evaluated whether chronic ethanol feeding in DEN-treated mice would develop liver fibrosis. Results from Sirius red staining for hepatic collagen revealed no obvious increase in positive staining in DEN-treated mice fed either liquid control diet or ethanol diet (Figure 7A). The hepatic levels of α-smooth muscle actin were varied among the DEN-treated mice fed ethanol, but the overall levels of α-smooth muscle actin were lower in DEN-treated mice fed ethanol than in mice fed the control diet (Figure 7, B and C). In DEN-treated mice fed the control diet, the protein levels of α-smooth muscle actin in L-Tfeb KO mice were lower than in WT mice. However, the levels of α-smooth muscle actin were similar between DEN-treated WT and L-Tfeb KO mice fed ethanol (Figure 7, B and C).
Figure 7.
Long-term ethanol (EtOH) feeding and loss of hepatic transcription factor EB (TFEB) do not increase fibrosis or inflammation in diethylnitrosamine (DEN)–treated mouse livers. Male Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice were subjected to the DEN-alcohol–associated hepatocellular carcinoma model. A: Representative images of Sirius Red staining are shown. B: Total lysates from mouse livers were subjected to Western blot. C: Densitometry analysis of B. D: Representative images of F4/80 immunohistochemistry staining are shown. E: Numbers of F4/80-positive cells were counted from eight different fields (×20) from each mouse in a blinded manner (M.H.). Data are presented as means ± SEM (C and E). n = 3 to 4 (C); n = 4 to 8 (E). Original magnifications, ×10 (A); ×20 (D). Ctrl, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; α-SMA, α-smooth muscle actin.
Hepatic inflammation in DEN-treated mice fed either the liquid control diet or the ethanol diet was next determined. Compared with DEN-treated mice fed the control diet, the number of F4/80 (marker of hepatic resident macrophage/Kupffer cells)-positive cells only slightly increased in mice fed ethanol (Figure 7, D and E). Compared with DEN-treated WT mice fed ethanol, there were fewer F4/80-positive Kupffer cells in L-Tfeb KO mouse livers after ethanol feeding, but these changes did not reach a statistical significant difference (Figure 7, D and E). Together, the data indicate that ethanol feeding did not promote fibrosis and inflammation in this DEN-alcohol–associated HCC mouse model.
Loss of Hepatic TFEB Attenuates Proliferation of Liver Tumor Cells but Not Normal Cells in DEN-Treated Mice Fed Control Diet Only
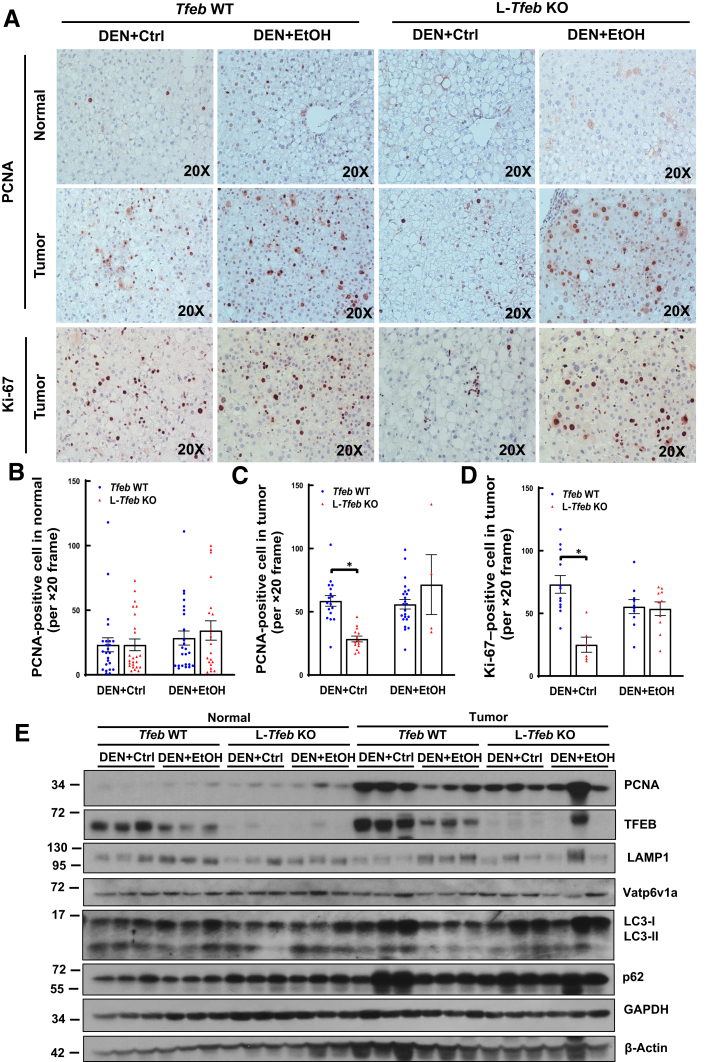
Cell proliferation in normal and tumor cells was next examined by using immunostaining of PCNA and Ki-67 as well as immunoblotting of protein levels of PCNA. Tumor cells had higher amounts of PCNA-positive cells than adjacent normal cells, but ethanol feeding did not further increase the number of PCNA-positive cells in either normal or tumor cells in WT mice (Figure 8, A–C). Interestingly, the number of PCNA-positive tumor cells, but not normal cells, was lower in DEN-treated L-Tfeb KO mice fed the control diet compared with those in WT mice. However, the number of PCNA-positive cells was not different between DEN-treated WT and L-Tfeb KO mice fed ethanol (Figure 8, A–C). Similar to the PCNA staining, the number of Ki-67–positive tumor cells was also lower in DEN-treated L-Tfeb KO mice compared with those in WT mice fed control diet, but this difference was abolished in ethanol-fed mice (Figure 8D). Results from Western blot analysis also revealed increased PCNA levels in tumors compared with adjacent normal tissues, but the PCNA levels were not different between WT and L-Tfeb KO mice, and ethanol feeding did not further increase PNCA levels (Figure 8E). Ethanol feeding decreased hepatic TFEB but not LAMP1 and Vatp6v1a in both normal and tumor tissues of DEN-treated mice. The hepatic levels of p62 were much higher in tumor tissues than those of normal tissues of both WT and L-Tfeb KO mice, which were not affected by ethanol feeding or deletion of Tfeb. The levels of hepatic LC3-II were not significantly different among all the experimental groups (Figure 8E). The data suggest that TFEB may promote DEN-initiated tumor cell proliferation in control diet– but not ethanol-fed mice. Lack of hepatic TFEB does not have profound effects on autophagy-related and lysosomal proteins in both normal and tumor tissues of DEN-treated mice.
Figure 8.
Loss of hepatic transcription factor EB (TFEB) attenuates proliferation of liver tumor but not normal cells in diethylnitrosamine (DEN)–treated mice fed control (Ctrl) diet but not ethanol (EtOH) diet. Male Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice were subjected to the DEN-alcohol–associated hepatocellular carcinoma model. A: Representative images of proliferating cell nuclear antigen (PCNA) and Ki-67 immunohistochemistry staining are shown. B–D: Numbers of PCNA- and Ki-67–positive cells were counted from eight different fields (×20) from each mouse in a blinded manner (M.H.). E: Total lysates from mouse liver tumors and adjacent normal liver tissues were subjected to Western blot analysis. Data are presented as means ± SEM (B–D). n = 4 (B–D); n = 3 (E). ∗P < 0.05 (one-way analysis of variance with Bonferroni post hoc test). Original magnification, ×20 (A). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
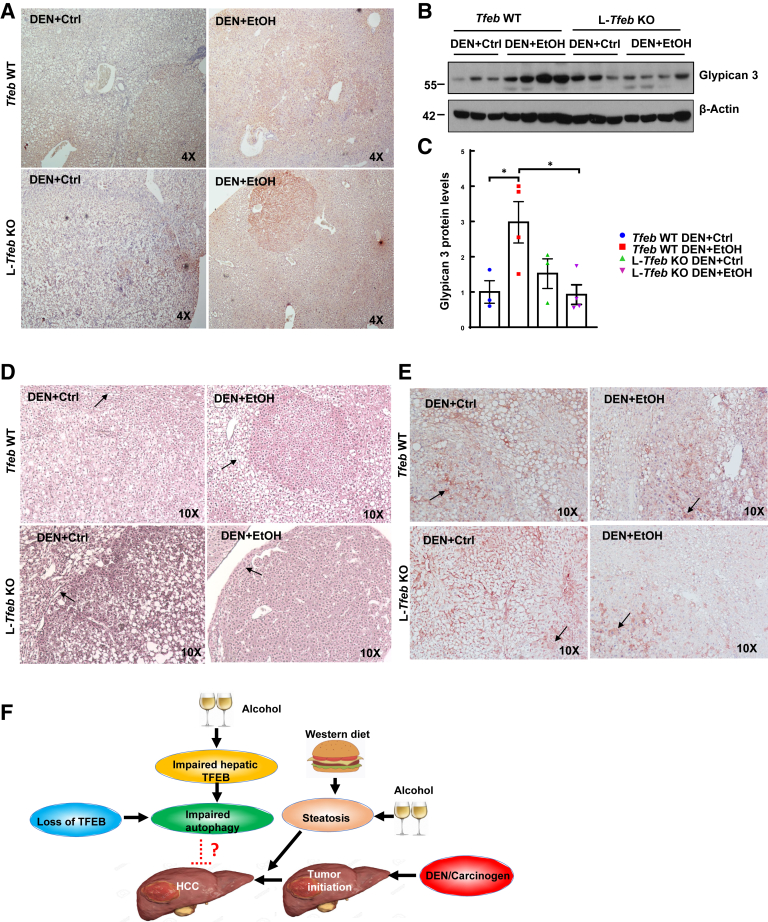
Loss of Hepatic TFEB Reduces Malignant Characteristics of DEN and Chronic Ethanol Feeding–Induced HCC in Mice
Glypican 3 is a cell-surface glycoprotein in which heparan sulfate glycosaminoglycan chains are covalently linked to a protein core. Glypican 3 is specifically expressed in HCC tissues and presents as soluble glypican 3 in peripheral blood of HCC patients. The expression of glypican 3 is generally not detected in healthy adult liver tissues, and not in fatty liver and cirrhotic liver.47 Therefore, glypican 3 has been widely used as a reliable specific marker for HCC. Immunostaining and immunoblotting analysis revealed that the protein levels of glypican 3 significantly increased in DEN-treated WT mouse livers fed ethanol compared with the mice fed control diet (Figure 9, A–C), suggesting ethanol may enhance tumor malignancy in DEN-treated mice. Unlike the WT mice, the levels of glypican 3 did not change in DEN-treated L-Tfeb KO mice after ethanol feeding (Figure 9, A–C). The normal liver and benign hepatocellular adenoma generally have abundant reticulin staining, which is markedly decreased or absent in DEN-treated mice regardless of control or ethanol diet feeding and Tfeb genotypes of mice (Figure 9D). Cytosolic and nuclear accumulation of β-catenin is highly associated with human HCC. Although membrane staining of β-catenin was observed in normal hepatocytes of all the four experimental groups, cytosolic and nuclear staining of β-catenin was also found in all four experimental groups regardless of ethanol feeding and Tfeb genotypes (Figure 9E). Taken together, the data suggest that loss of TFEB may reduce certain but not all malignant HCC markers.
Figure 9.
Loss of hepatic transcription factor EB (TFEB) reduces certain malignant tumor markers of hepatocellular carcinoma (HCC) induced by diethylnitrosamine (DEN) and chronic ethanol (EtOH) feeding in mice. Male Tfeb wild-type (WT) and L-Tfeb knockout (KO) mice were subjected to the DEN-alcohol–associated HCC model. A: Representative images of glypican 3 immunohistochemistry staining are shown. B: Total lysates from mouse livers were subjected to Western blot analysis. C: Densitometry analysis of B. D: Representative images of reticulin staining (arrows indicate tubular reticular staining). E: Representative images of β-catenin immunohistochemistry staining (arrows indicate cytosolic and nuclear β-catenin staining). F: A proposed working model for the role of TFEB in alcohol- and nonalcoholic fatty liver disease–associated HCC. Chronic alcohol consumption and Western diet increase hepatic steatosis, which promotes DEN-initiated liver carcinogenesis to HCC. Alcohol consumption also impairs hepatic TFEB, and genetic loss of Tfeb in mouse livers may slightly decrease hepatic lysosomal and autophagy activity, which may reduce some malignant HCC markers and attenuate the progression of HCC. Future studies are needed to get rid of the potential compensatory effects of loss of hepatic Tfeb to better understand the role of lysosomal biogenesis and autophagy in liver carcinogenesis and HCC pathogenesis. Data are presented as means ± SEM (C). n = 3 to 4 (C). ∗P < 0.05 (one-way analysis of variance with Bonferroni post hoc test). Original magnifications, ×4 (A); ×10 (D and E). Ctrl, control.
Discussion
The present study used chronic ethanol feeding together with hepatic carcinogen DEN injection to establish an alcohol-associated HCC mouse model. The role of TFEB in DEN-induced liver tumorigenesis was studied in WT and L-Tfeb KO mice fed an ethanol diet or pair-fed control liquid diet. Hepatic TFEB deficiency tended to attenuate DEN-initiated and alcohol-associated liver tumorigenesis in mice. Additionally, the role of TFEB in NAFLD-associated HCC was evaluated, and loss of hepatic TFEB was found to have more striking protection against DEN-induced and Western diet–associated liver tumorigenesis compared with that from the alcohol-associated liver tumorigenesis in mice. Collectively, these data suggest that TFEB may have a role in promoting the progression of both ALD- and NAFLD-associated HCC (Figure 9F).
Autophagy plays distinctive roles in liver tumor initiation and progression, in which autophagy inhibits early tumor initiation while promoting progression and the malignancy of already formed liver tumors.13 Genetic deletion of Atg5 or Atg7 in mouse livers leads to spontaneous benign adenoma,45,48 supporting the tumor suppressor role of autophagy at the initiation stage. Interestingly, DEN induces HCC in WT mice but only benign tumors in L-Atg5 KO mice,49 suggesting autophagy may be required for the malignant progression of tumors from the benign stage. As a master regulator of lysosome biogenesis and autophagy, TFEB may also be required for the liver tumor progression in alcohol-associated HCC. The present study found that both the number and size of tumors increased in DEN-treated mice fed ethanol compared with mice fed control diet, supporting the notion that alcohol drinking may promote progression of HCC. However, loss of hepatic TFEB had no effect on the total number of tumors but only decreased the number of large-sized tumors after DEN treatment and alcohol feeding. Moreover, the tumor glypican 3 levels were also lower in alcohol-fed L-Tfeb KO mice, suggesting that TFEB may also play more important roles in tumor progression than in tumor initiation in alcohol-associated HCC. In contrast, in Western diet–fed mice treated with DEN, L-Tfeb KO mice not only had a decreased number of total tumors, but also decreased number of large-sized tumors, suggesting that TFEB may promote both tumor initiation and progression in the NAFLD-associated HCC. In our DEN-alcohol–associated HCC model, the mice fed the liquid control diet already developed steatosis, likely due to the high fat content in the liquid diet. It is likely that steatosis may facilitate TFEB's effects on the tumor initiation, which may explain why TFEB does not affect the tumor initiation in alcohol-fed mice but potentiates tumor initiation in Western diet–fed mice. The mechanism of how steatosis coordinates with TFEB for the liver tumor initiation needs to be studied further.
Although alcohol consumption is a known risk factor for HCC, animal models to recapitulate the pathogenesis of alcohol-associated HCC are limited. Alcohol enhances HCC formation in mice that received a single dose of DEN for 16 or 40 weeks and further fed alcohol (10%/20% v/v) in drinking water for 8 weeks.50 However, feeding alcohol in the drinking water can only lead to mild steatosis with little liver injury and inflammation,51 which cannot mimic the human ALD and alcohol-associated HCC. This study showed that mice fed ethanol Lieber-DeCarli diet for 24 weeks had markedly increased steatosis with relatively high serum ALT values, significantly increased the number of middle-sized tumors (1 to 5 mm), and had a trend towards increasing the tumor burden in DEN-treated mice. However, one of the limitations of this model is that pair-fed mice with the liquid diet for 24 weeks also develop severe hepatic steatosis, which may diminish the effects of ethanol on DEN-initiated tumorigenesis when comparing the ethanol feeding group with the control diet group.
In addition to regulating autophagy, TFEB has been shown to have nonautophagy functions in regulating cancer cell behavior through modulating cell cycle metabolism in cancer cells in response to a high metabolic flexibility to adapt themselves to cell-extrinsic and cell-intrinsic cues.41,42,52, 53, 54, 55, 56, 57, 58 TFEB also plays an important role in choreographing tumor-microenvironment interaction. Tumor microenvironment encompasses a heterogeneous population of differentiated cells and progenitor cells, which are critical for tumor fate regulation. In addition to hepatocytes, hepatic nonparenchymal cells, such as hepatic stellate cells and macrophages, also play critical roles in HCC. TFEB has a principal role in regulating innate immunity,56 through both regulating the biogenesis of lysosomes and their role in phagocytosis, and activating both M1- or M2-like macrophages. Although the number of Kupffer/hepatic macrophages did not change in L-Tfeb KO mice compared with that in WT mice in the DEN-alcohol-HCC model, future experiments are needed to further determine the plasticity of these macrophages. Moreover, the roles of TFEB in liver nonparenchymal cells, such as macrophages and hepatic stellate cells, in HCC, also need to be investigated by using liver nonparenchymal cell-specific TFEB deletion mice. Although mechanistic target of rapamycin complex 1 negatively regulates TFEB activation by directly phosphorylating TFEB at serine 142 and serine 211, resulting in TFEB cytosolic retention and degradation, TFEB also forms a feedback loop to activate mechanistic target of rapamycin complex 1 to promote cancer cell growth by transcription up-regulation of RagD.59 Therefore, it is also likely that TFEB may promote liver cancer progression via this feedback mechanistic target of rapamycin complex 1 activation. In the future, it will be interesting to determine whether pharmacologic inhibition of mechanistic target of rapamycin complex 1 would be beneficial in alcohol-associated HCC.
Although the overall tumor-related parameters, including the tumor number and size, tended to be improved in DEN-treated L-Tfeb KO mice compared with DEN-treated WT mice fed either ethanol or control diet, these differences are generally not striking. As the MITF family has four redundant members, it is likely that loss of TFEB in the mouse livers could be compensated by other MITF family member proteins. Indeed, TFE3 has been shown to compensate for the loss of TFEB in mouse livers.39 Moreover, most autophagy-related and lysosomal proteins were not significantly altered in TFEB KO mice compared with WT mice after DEN treatment, regardless of ethanol or control diet feeding, further supporting the potential compensatory effects by other MITF family proteins. Future work using Tfeb/Tfe3 double-knockout mice in this DEN-alcohol–associated HCC model may help to overcome the possible compensatory effects in L-Tfeb KO mice.
In summary, we successfully established an alcohol-associated HCC mouse model by using hepatic carcinogen DEN injection with chronic alcohol feeding in mice. These findings indicate that chronic alcohol consumption promotes liver tumor development in DEN-injected mice, and loss of hepatic TFEB attenuates alcohol-associated liver tumorigenesis.
Footnotes
Supported in part by NIH grants R37 AA020518 (W.X.D.), R01 DK102142 (W.X.D.), U01 AA024733 (W.X.D.), R01 AG072895 (W.X.D.), and R01-NS078072; a pilot project that was supported by the National Cancer Institute Cancer Center Support grant P30 CA168524 (W.X.D.); Telethon Foundation, MIUR FIRB RBAP11Z3YA, European Research Council H2020 AdG LYSOSOMICS 694282, U.S. National Institutes of HealthR01-NS078072, Huffington Foundation, Associazione Italiana per la Ricerca sul Cancro A.I.R.C. IG-22103 and 5x1000-21051 (A.B.).
Disclosures: A.B. is cofounder of CASMA Therapeutics and advisory board member of Next Generation Diagnostics and Avilar Therapeutics.
Author Contributions
W.X.D. conceived and supervised the project; H.M.N. and W.X.D. conceived and designed the experiments; X.C., H.M.N., S.W., and M.H. performed the experiments and analyzed the data; A.B. provided key reagents and discussed the manuscript; X.C. and W.X.D. analyzed data and wrote the manuscript.
References
- 1.Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parzych K.R., Klionsky D.J. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin X.M., Ding W.X., Gao W. Autophagy in the liver. Hepatology. 2008;47:1773–1785. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- 5.Rautou P.E., Mansouri A., Lebrec D., Durand F., Valla D., Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Ding W.X. Role of autophagy in liver physiology and pathophysiology. World J Biol Chem. 2010;1:3–12. doi: 10.4331/wjbc.v1.i1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams J.A., Manley S., Ding W.X. New advances in molecular mechanisms and emerging therapeutic targets in alcoholic liver diseases. World J Gastroenterol. 2014;20:12908–12933. doi: 10.3748/wjg.v20.i36.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Rodriguez A., Mayoral R., Agra N., Valdecantos M.P., Pardo V., Miquilena-Colina M.E., Vargas-Castrillon J., Lo Iacono O., Corazzari M., Fimia G.M., Piacentini M., Muntane J., Bosca L., Garcia-Monzon C., Martin-Sanz P., Valverde A.M. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. doi: 10.1038/cddis.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka S., Hikita H., Tatsumi T., Sakamori R., Nozaki Y., Sakane S., Shiode Y., Nakabori T., Saito Y., Hiramatsu N., Tabata K., Kawabata T., Hamasaki M., Eguchi H., Nagano H., Yoshimori T., Takehara T. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology. 2016;64:1994–2014. doi: 10.1002/hep.28820. [DOI] [PubMed] [Google Scholar]
- 10.Fallowfield J.A. Therapeutic targets in liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G709–G715. doi: 10.1152/ajpgi.00451.2010. [DOI] [PubMed] [Google Scholar]
- 11.Wang L., Ou J.H. Hepatitis C virus and autophagy. Biol Chem. 2015;396:1215–1222. doi: 10.1515/hsz-2015-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sir D., Liang C., Chen W.L., Jung J.U., Ou J.H. Perturbation of autophagic pathway by hepatitis C virus. Autophagy. 2008;4:830–831. doi: 10.4161/auto.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao X., Qian H., Wang S., Fulte S., Ding W.-X. Autophagy and liver cancer. Clin Mol Hepatol. 2020;26:606–617. doi: 10.3350/cmh.2020.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 15.Xu J. Trends in liver cancer mortality among adults aged 25 and over in the United States, 2000-2016. NCHS Data Brief. 2018;313:1–8. [PubMed] [Google Scholar]
- 16.Nagy L.E., Ding W.X., Cresci G., Saikia P., Shah V.H. Linking pathogenic mechanisms of alcoholic liver disease with clinical phenotypes. Gastroenterology. 2016;150:1756–1768. doi: 10.1053/j.gastro.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao B., Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141:1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson D.E., Jarman D.W., Rehm J., Greenfield T.K., Rey G., Kerr W.C., Miller P., Shield K.D., Ye Y., Naimi T.S. Alcohol-attributable cancer deaths and years of potential life lost in the United States. Am J Public Health. 2013;103:641–648. doi: 10.2105/AJPH.2012.301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seitz H.K., Stickel F. Risk factors and mechanisms of hepatocarcinogenesis with special emphasis on alcohol and oxidative stress. Biol Chem. 2006;387:349–360. doi: 10.1515/BC.2006.047. [DOI] [PubMed] [Google Scholar]
- 20.Hemesath T.J., Steingrimsson E., McGill G., Hansen M.J., Vaught J., Hodgkinson C.A., Arnheiter H., Copeland N.G., Jenkins N.A., Fisher D.E. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 21.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., Banfi S., Parenti G., Cattaneo E., Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 22.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D.C., Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napolitano G., Ballabio A. TFEB at a glance. J Cell Sci. 2016;129:2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis I.J., Hsi B.L., Arroyo J.D., Vargas S.O., Yeh Y.A., Motyckova G., Valencia P., Perez-Atayde A.R., Argani P., Ladanyi M., Fletcher J.A., Fisher D.E. Cloning of an alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A. 2003;100:6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kauffman E.C., Ricketts C.J., Rais-Bahrami S., Yang Y., Merino M.J., Bottaro D.P., Srinivasan R., Linehan W.M. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol. 2014;11:465–475. doi: 10.1038/nrurol.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka M., Homme M., Yamazaki Y., Shimizu R., Takazawa Y., Nakamura T. Modeling alveolar soft part sarcoma unveils novel mechanisms of metastasis. Cancer Res. 2017;77:897–907. doi: 10.1158/0008-5472.CAN-16-2486. [DOI] [PubMed] [Google Scholar]
- 27.Kundu S.T., Grzeskowiak C.L., Fradette J.J., Gibson L.A., Rodriguez L.B., Creighton C.J., Scott K.L., Gibbons D.L. TMEM106B drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Nat Commun. 2018;9:2731. doi: 10.1038/s41467-018-05013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giatromanolaki A., Kalamida D., Sivridis E., Karagounis I.V., Gatter K.C., Harris A.L., Koukourakis M.I. Increased expression of transcription factor EB (TFEB) is associated with autophagy, migratory phenotype and poor prognosis in non-small cell lung cancer. Lung Cancer. 2015;90:98–105. doi: 10.1016/j.lungcan.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Ploper D., Taelman V.F., Robert L., Perez B.S., Titz B., Chen H.W., Graeber T.G., von Euw E., Ribas A., De Robertis E.M. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci U S A. 2015;112:E420–E429. doi: 10.1073/pnas.1424576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perera R.M., Stoykova S., Nicolay B.N., Ross K.N., Fitamant J., Boukhali M., Lengrand J., Deshpande V., Selig M.K., Ferrone C.R., Settleman J., Stephanopoulos G., Dyson N.J., Zoncu R., Ramaswamy S., Haas W., Bardeesy N. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J.H., Lee J., Cho Y.R., Lee S.Y., Sung G.J., Shin D.M., Choi K.C., Son J. TFEB supports pancreatic cancer growth through the transcriptional regulation of glutaminase. Cancers (Basel) 2021;13:483. doi: 10.3390/cancers13030483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He R., Wang M., Zhao C., Shen M., Yu Y., He L., Zhao Y., Chen H., Shi X., Zhou M., Pan S., Liu Y., Guo X., Li X., Qin R. TFEB-driven autophagy potentiates TGF-beta induced migration in pancreatic cancer cells. J Exp Clin Cancer Res. 2019;38:340. doi: 10.1186/s13046-019-1343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu X., Zhuo Y., Wu S., Chen Y., Ye J., Deng Y., Feng Y., Liu R., Cai S., Zou Z., Wang B., Wu C.L., Zeng G., Zhong W. TFEB promotes prostate cancer progression via regulating ABCA2-dependent lysosomal biogenesis. Front Oncol. 2021;11:632524. doi: 10.3389/fonc.2021.632524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang J., Jia X., Wang K., Zhao N. High expression of TFEB is associated with aggressive clinical features in colorectal cancer. Onco Targets Ther. 2018;11:8089–8098. doi: 10.2147/OTT.S180112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S., Liu F., Xu L., Li C., Yang X., Guo B., Gu J., Wang L. Wnt/beta-catenin signaling axis is required for TFEB-mediated gastric cancer metastasis and epithelial-mesenchymal transition. Mol Cancer Res. 2020;18:1650–1659. doi: 10.1158/1541-7786.MCR-20-0180. [DOI] [PubMed] [Google Scholar]
- 36.Slade L., Biswas D., Ihionu F., El Hiani Y., Kienesberger P.C., Pulinilkunnil T. A lysosome independent role for TFEB in activating DNA repair and inhibiting apoptosis in breast cancer cells. Biochem J. 2020;477:137–160. doi: 10.1042/BCJ20190596. [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Hodge J., Liu Q., Wang J., Wang Y., Evans T.D., Altomare D., Yao Y., Murphy E.A., Razani B., Fan D. TFEB is a master regulator of tumor-associated macrophages in breast cancer. J Immunother Cancer. 2020;8:e000543. doi: 10.1136/jitc-2020-000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Astanina E., Bussolino F., Doronzo G. Multifaceted activities of transcription factor EB in cancer onset and progression. Mol Oncol. 2021;15:327–346. doi: 10.1002/1878-0261.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao X., Wang S., Zhao K., Li Y., Williams J.A., Li T., Chavan H., Krishnamurthy P., He X.C., Li L., Ballabio A., Ni H.M., Ding W.X. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. 2018;155:865–879.e12. doi: 10.1053/j.gastro.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao X., Ni H.M., Ding W.X. Insufficient autophagy: a novel autophagic flux scenario uncovered by impaired liver TFEB-mediated lysosomal biogenesis from chronic alcohol-drinking mice. Autophagy. 2018;14:1646–1648. doi: 10.1080/15548627.2018.1489170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pastore N., Huynh T., Herz N.J., Calcagni A., Klisch T.J., Brunetti L., Kim K.H., De Giorgi M., Hurley A., Carissimo A., Mutarelli M., Aleksieva N., D'Orsi L., Lagor W.R., Moore D.D., Settembre C., Finegold M.J., Forbes S.J., Ballabio A. TFEB regulates murine liver cell fate during development and regeneration. Nat Commun. 2020;11:2461. doi: 10.1038/s41467-020-16300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Settembre C., De Cegli R., Mansueto G., Saha P.K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T.J., Wollenberg A.C., Di Bernardo D., Chan L., Irazoqui J.E., Ballabio A. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni H.M., Bockus A., Boggess N., Jaeschke H., Ding W.X. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55:222–232. doi: 10.1002/hep.24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni H.M., Bhakta A., Wang S., Li Z., Manley S., Huang H., Copple B., Ding W.X. Role of hypoxia inducing factor-1beta in alcohol-induced autophagy, steatosis and liver injury in mice. PLoS One. 2014;9:e115849. doi: 10.1371/journal.pone.0115849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni H.M., Woolbright B.L., Williams J., Copple B., Cui W., Luyendyk J.P., Jaeschke H., Ding W.X. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18:223–238. doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo M., Zhang H., Zheng J., Liu Y. Glypican-3: a new target for diagnosis and treatment of hepatocellular carcinoma. J Cancer. 2020;11:2008–2021. doi: 10.7150/jca.39972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian Y., Kuo C.F., Sir D., Wang L., Govindarajan S., Petrovic L.M., Ou J.H. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 2015;22:1025–1034. doi: 10.1038/cdd.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandon-Warner E., Walling T.L., Schrum L.W., McKillop I.H. Chronic ethanol feeding accelerates hepatocellular carcinoma progression in a sex-dependent manner in a mouse model of hepatocarcinogenesis. Alcohol Clin Exp Res. 2012;36:641–653. doi: 10.1111/j.1530-0277.2011.01660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coleman R.A., Young B.M., Turner L.E., Cook R.T. A practical method of chronic ethanol administration in mice. Methods Mol Biol. 2008;447:49–59. doi: 10.1007/978-1-59745-242-7_4. [DOI] [PubMed] [Google Scholar]
- 52.Pisonero-Vaquero S., Soldati C., Cesana M., Ballabio A., Medina D.L. TFEB modulates p21/WAF1/CIP1 during the DNA damage response. Cells. 2020;9:1186. doi: 10.3390/cells9051186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doronzo G., Astanina E., Cora D., Chiabotto G., Comunanza V., Noghero A., Neri F., Puliafito A., Primo L., Spampanato C., Settembre C., Ballabio A., Camussi G., Oliviero S., Bussolino F. TFEB controls vascular development by regulating the proliferation of endothelial cells. EMBO J. 2019;38:e98250. doi: 10.15252/embj.201798250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brady O.A., Jeong E., Martina J.A., Pirooznia M., Tunc I., Puertollano R. The transcription factors TFE3 and TFEB amplify p53 dependent transcriptional programs in response to DNA damage. Elife. 2018;7:e40856. doi: 10.7554/eLife.40856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slade L., Pulinilkunnil T. The MiTF/TFE family of transcription factors: master regulators of organelle signaling, metabolism, and stress adaptation. Mol Cancer Res. 2017;15:1637–1643. doi: 10.1158/1541-7786.MCR-17-0320. [DOI] [PubMed] [Google Scholar]
- 56.Martina J.A., Puertollano R. TFEB and TFE3: the art of multi-tasking under stress conditions. Transcription. 2017;8:48–54. doi: 10.1080/21541264.2016.1264353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mansueto G., Armani A., Viscomi C., D'Orsi L., De Cegli R., Polishchuk E.V., Lamperti C., Di Meo I., Romanello V., Marchet S., Saha P.K., Zong H., Blaauw B., Solagna F., Tezze C., Grumati P., Bonaldo P., Pessin J.E., Zeviani M., Sandri M., Ballabio A. Transcription factor EB controls metabolic flexibility during exercise. Cell Metab. 2017;25:182–196. doi: 10.1016/j.cmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Settembre C., Fraldi A., Medina D.L., Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Malta C., Siciliano D., Calcagni A., Monfregola J., Punzi S., Pastore N., Eastes A.N., Davis O., De Cegli R., Zampelli A., Di Giovannantonio L.G., Nusco E., Platt N., Guida A., Ogmundsdottir M.H., Lanfrancone L., Perera R.M., Zoncu R., Pelicci P.G., Settembre C., Ballabio A. Transcriptional activation of RagD GTPase controls mTORC1 and promotes cancer growth. Science. 2017;356:1188–1192. doi: 10.1126/science.aag2553. [DOI] [PMC free article] [PubMed] [Google Scholar]