Abstract
In order to ascertain the microbiological quality of stored semen specimens processed for artificial insemination by a donor (AID), we developed a PCR assay targeting the chlamydial plasmid to detect Chlamydia trachomatis in semen. The lower limit of detection of this assay corresponded to 2.5 to 5 elementary bodies per μl of semen. A total of 669 cryopreserved ejaculates from 97 asymptomatic donors were tested for C. trachomatis infection. Twelve ejaculates, originating from four donors, were found to be positive, indicating a 4% prevalence of C. trachomatis infection among the donor population studied. Cross-contamination between the cryopreserved specimens in the storage container was studied by typing using sequence analysis of PCR-amplified omp1 genes of the strains. Two donors were infected with serovar E, one was infected with serovar F, and one was infected with serovar K. For two donors, the duration of C. trachomatis positivity could be assessed. One donor donated C. trachomatis-positive semen for at least 4 successive months, and the other did so for at least 16 months. The occurrence of C. trachomatis infection in cryopreserved donor semen indicates that ejaculates from donors not tested for a C. trachomatis infection just prior to donation should be tested for infection by a direct test such as the PCR described here. Direct testing of semen specimens will detect not only donors with an active infection but also C. trachomatis-infected ejaculates already stored and will thus improve the microbiological quality of AID, since discrepancies in the presence of C. trachomatis in urine and semen specimens have been reported.
Chlamydia trachomatis infections are the most prevalent bacterial sexually transmitted diseases worldwide (11). It has been estimated that there are approximately 4 million new C. trachomatis infections each year in the United States (7). A C. trachomatis infection, if not treated in an early stage, can lead to severe sequelae, such as pelvic inflammatory disease, ectopic pregnancy, and tubal infertility (16). However, 50 to 80% of infected men and women are asymptomatic (9, 16, 32). This high number of unrecognized infected individuals provides the reservoir for spreading the infection to men and women via sexual contact. In addition, women may also contract C. trachomatis infection after artificial insemination by a donor (AID) (20, 26). Considering the high prevalence of C. trachomatis infections worldwide, guidelines have been formulated to prevent the transmission of C. trachomatis infections through AID (2, 3). According to the recommendations of the American Society for Reproductive Medicine (2) and the European Society for Human Reproduction and Embryology (3), semen donors should be screened by testing of either urethral, urine, or semen specimens for C. trachomatis.
A variety of DNA amplification methods to detect C. trachomatis in urethral, urine, and cervical specimens are currently available and have appeared to be far more sensitive than cell culturing or antigen detection (4). These methods are now widely used in diagnostic laboratories for screening high-risk populations for C. trachomatis infection and donors providing semen for AID (4).
However, it has been reported that C. trachomatis was not detected in urinary or urethral specimens despite its presence in testicular specimens (10, 14). In addition, our semen bank contains large numbers of cryopreserved ejaculates from donors for whom no corresponding urethral or urine specimens have been tested.
As part of the quality requirements for human tissues processed for therapeutic purposes, we developed a PCR assay to detect C. trachomatis in semen specimens. This PCR was used to detect and to type C. trachomatis in stored semen specimens.
(The results of this study were presented in part at the 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, Calif., September 1999 [abstr. no. 1739, 1999], and at the annual meeting of the American Society for Reproductive Medicine, Toronto, Ontario, Canada, September 1999 [abstr. no. 227, 1999].)
MATERIALS AND METHODS
Semen samples.
Semen samples collected between 1984 and 1999 are stored in the Center for Reproductive Medicine, Academic Medical Center. In total, 669 ejaculates, donated by 97 men between 20 and 45 years old, were divided into aliquots in 0.25-ml straws with egg yolk, human sperm preservation medium (15), or SpermFreeze (Fertipro NV, Breemeng, Belgium). Straws were cryopreserved in liquid nitrogen and stored in a single container.
C. trachomatis EB preparation.
C. trachomatis biovar Lymphogranuloma venereum L2/434/Bu was obtained from P. B. Wyrick, East Tennessee State University, Johnson City (originally provided by C. C. Kuo and S. P. Wang, University of Washington, Seattle). Bacteria were propagated in McCoy cells (CRL 1696; American Type Culture Collection) according to standard procedures (21), and elementary bodies (EBs) were released from the cells by sonication in a water bath at 60 W for 5 min (Branson 2200; Bransonic, Shelton, Conn.). Host cell debris was removed by centrifugation at 500 × g for 10 min, and EBs were concentrated by centrifugation at 12,000 × g for 30 min. Pellets were washed twice with 0.02 M phosphate buffer (pH 7.2) containing 0.2 M sucrose and 0.73 mg of glutamine per ml (SPG), resuspended in SPG, divided into aliquots, and stored at −70°C. The number of EBs was quantified using direct immunofluorescence (Microtrak; Syva, San Jose, Calif.).
Purification of DNA from semen specimens.
Duplicate semen samples of 50 μl, derived from one straw, were processed. Prior to DNA extraction, 250 EBs (strain L2/434/Bu; Advanced Biotechnologies Inc., Columbia, Md.) (5 EBs per μl of semen) were added to one of the two samples as a positive control for lysis, DNA extraction efficiency, and inhibition of the PCR. Then, both samples were subjected to DNA extraction by the silica-guanidinium thiocyanate (GuSCN) procedure using buffer L6 and 20 μl of size-fractionated silica particles (6). DNA was eluted in 100 μl of sterile water. For comparison, DNA of some semen samples was also extracted in the presence of buffer L7A (5).
PCR to assess C. trachomatis infection in semen (C-PCR).
Primers, purified by high-pressure liquid chromatography, were obtained from Perkin-Elmer (PE; Nieuwekerk a/d IJssel, The Netherlands). The primers used for amplification of a 201-bp fragment of the cryptic chlamydial plasmid were CTP1 and CTP2 (Table 1); this primer pair has been described previously (12). The final reaction mixture (50 μl) contained 20 μl of eluate (the equivalent of 10 μl of semen); 100 ng of each primer; 1.25 U of Taq DNA polymerase (Promega); 0.5 U of Amperase (PE); 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 2.0 mM MgCl2; dATP, dGTP, and dCTP each at a concentration of 200 μM; and 400 μM dUTP (PE). PCRs were performed with a Trio thermoblock (Biometra, Göttingen, Germany) as follows: 5 min at 50°C; 5 min at 95°C; 40 cycles each consisting of 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C; and a final elongation step of 10 min at 72°C. Semen samples previously testing negative were simultaneously processed with other samples and served as a negative control for the extraction procedure. Water and chlamydial DNA from strain L2/434/Bu were used as negative and positive controls, respectively, for the reaction mixture. All controls were used every 10 samples.
TABLE 1.
Oligonucleotides used in this study
| Primer | Purpose | 5′→3′ Sequence (reference)a | Positions |
|---|---|---|---|
| CTP1 | C-PCR | TAGTAACTGCCACTTCATCA (12) | 2939–2958b |
| CTP2 | C-PCR | TTCCCCTTGTAATTCGTTGC (12) | 3139–3120b |
| CTP5 | Probe | ATCTCATTACCATGCATTAGCAGCTATCCA (12) | 3077–3106b |
| MOMP108 | M-PCR | GGCCATTAATTGCTACAGGACATCTTGTC (23) | 6867–6839c |
| RVS1163 | M-PCR | CGGAATTGTGCATTTACGTGA (23) | 5585–5605c |
| MOMP87 | M-PCR | TGAACCAAGCCTTATGATCGACGGA (23) | 6673–6649c |
| RVS1095 | M-PCR | GCAATACCGCAAGATTTTCTAGATTTCATC (23) | 5681–5710c |
| OMP334 | Sequencing | CAYATGCARGAYGCWGA (this study) | 6426–6410c |
| ROMP388 | Sequencing | ACATCAAARCGATCCCA (this study) | 6356–6372c |
| OMP625 | Sequencing | TTAGGSGCTTCWTTC (this study) | 6135–6121c |
| ROMP715 | Sequencing | CCWACATAYCCTTTMGG (this study) | 6029–6045c |
| ROMP856 | Sequencing | CCYTACATTGGAGTTA (this study) | 5904–5889c |
Detection of PCR products.
PCR was carried out using the same reagent mixture for nonspiked as well as for spiked samples, and amplicons were loaded on the same gel and detected simultaneously by Southern hybridization. Twenty microliters of the PCR product, the equivalent of 4 μl of semen, was analyzed by 1% agarose gel electrophoresis and Southern blot hybridization. The DNA was transferred to a Zeta probe membrane (Bio-Rad) by vacuum blotting. The blot was saturated with a 2× SSC solution (1× SSC is 0.315 M NaCl plus 0.015 M sodium citrate) in 0.2 M Tris-HCl (pH 7.5). DNA was cross-linked to the membrane by exposure to UV light for 5 min. Hybridizations were performed using the digoxigenin (DIG) detection system (Roche Diagnostics, Almere, The Netherlands). The C. trachomatis plasmid PCR products were probed with a specific internal probe, CTP5 (12) (Table 1), end labeled with DIG. After hybridization, the blot was washed and processed further with the DIG detection system, and chemiluminescent signals were detected on Royal X-Omat film (Eastman Kodak Co., Rochester, N.Y.) using intensifying screens.
Assessment of the detection limit of the C-PCR.
In order to determine the detection limit of the C-PCR, 10 semen samples from 10 donors, found negative for C. trachomatis by the C-PCR, were pooled, and 100 μl was spiked with 1,000 EBs. Then, serial twofold dilutions were made using the C. trachomatis-negative pooled semen. For each dilution, DNA was extracted from 50 μl of semen and subjected to PCR, and amplicons were detected by Southern hybridization.
PCR targeting the C. trachomatis omp1 gene (M-PCR).
Two semen samples from a positive ejaculate were processed separately for the M-PCR. DNA was extracted from 100 μl of the C. trachomatis-positive semen samples by the silica-GuSCN procedure (6). The omp1 gene was amplified by nested PCR using primers described previously (23) (Table 1). The primer pair MOMP108-RVS1163 was used in the first PCR. This primer pair amplifies a 1.2-kb fragment of the omp1 gene. The reaction mixture (50 μl) contained 20 μl of eluate; 100 ng of each primer; 1.25 U of Taq DNA polymerase; 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 2.0 mM MgCl2; and dATP, dGTP, dCTP, and dTTP (PE) each at a concentration of 200 μM. PCRs were performed with a thermocycler as follows: 5 min at 95°C; 40 cycles each consisting of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C; and a final elongation step of 10 min at 72°C. The nested PCR product was generated by using primer pair MOMP87-RVS1095 (23) (Table 1) and 1 or 10 μl from the first PCR product as a template. PCR conditions were essentially as described above. The omp1 gene fragment so obtained from approximately 1.1 kb contains the four variable domains on which the typing of C. trachomatis is based (31).
omp1 amplicon cloning.
The amplicons obtained from the C. trachomatis-positive semen samples were loaded on a 1% agarose gel, gel purified (Qiaex; Qiagen Inc., Chatsworth, Calif.), cloned into the TA vector pCR2.1, and transformed into Escherichia coli Top10F′ cells (Invitrogen, Groningen, The Netherlands). Recombinants were selected by blue-white screening and ampicillin resistance. Insert sizes were checked by performing a standard colony PCR with the universal −21M13 and M13Reverse primers (PE). In brief, a portion of one colony was resuspended in 100 μl of distilled water, boiled for 10 min, and centrifuged for 1 min at maximum speed in an Eppendorf table centrifuge; 2.5 μl of the supernatant was used as a template in a 25-μl PCR mixture. Two independent recombinant clones from each transformation were subjected to sequence analysis.
Fluorescence-based sequencing.
The recombinant clones containing an almost full-size omp1 gene were used as templates for sequencing with either fluorescence dye-labeled universal M13 primers or dye terminators and internal omp1 primers (Table 1). Analysis was performed on an automatic sequencer (model 373), according to the instructions supplied by Applied Biosystems Incorporated (Foster City, Calif.). Multiple sequence alignments and database similarity searches were performed using CLUSTAL W (25) and the BLASTN algorithms (1), respectively.
Nucleotide sequence accession numbers.
The nucleotide sequence data have been deposited in the EMBL/GenBank/DDBJ nucleotide sequence databases under the accession numbers AF265237, AF265238, AF265239, and AF265240.
RESULTS
Detection limit of the C-PCR.
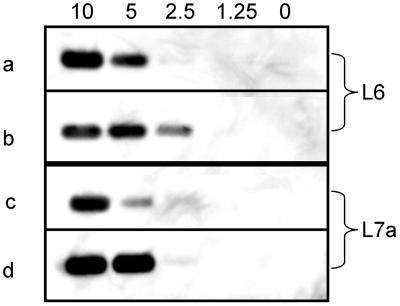
Using the silica-GuSCN procedure with buffer L6 for lysis and DNA extraction, it was possible to detect the equivalent of 10 to 20 EBs on the blot (Fig. 1). Since the equivalent of 4 μl of semen was loaded on the gel, a detection limit of 2.5 to 5 EBs per μl of semen was achieved. Some variation in the strength of the signals obtained between experiments was observed (Fig. 1). Using buffer L7A instead of buffer L6 in the silica-GuSCN procedure (5) did not improve the detection limit of the C-PCR (Fig. 1).
FIG. 1.
Lower limit of detection of C. trachomatis EBs. With buffer L6 (rows a and b) or buffer L7A (rows c and d), DNA was purified from 50 μl of semen containing 500, 250, 125, 62.5, or 0 EB particles. One-fifth of the extracted DNA was subjected to PCR, and the equivalent of 4 μl of semen was loaded on an agarose gel, blotted, and analyzed by Southern hybridization. Numbers above the lanes represent the numbers of EBs added per microliter of semen. Results from two representative experiments carried out in duplicate (a and b; c and d) are shown.
Assessment of C. trachomatis in semen samples present in the semen bank by the C-PCR.
A total of 669 semen samples, derived from 97 donors, were blindly assessed for the presence of C. trachomatis by the C-PCR. Duplicate semen samples, to one of which EBs were added as a positive control, were processed. The semen samples supplied with EBs yielded a positive signal on the blot, indicating successful DNA extraction and amplification. Again, some variation in the strength of the signals obtained was observed (data not shown).
Twelve (1.8%) of the 669 nonspiked semen samples yielded a positive signal on the blot, indicating the presence of C. trachomatis DNA. At least two or more additional semen samples from these 12 C-PCR-positive ejaculates were also positive. Decoding of the C. trachomatis-positive ejaculate specimens revealed that the C. trachomatis-positive samples were obtained from four donors (encoded K23, H38, V12, and B38).
Duration of C. trachomatis infection.
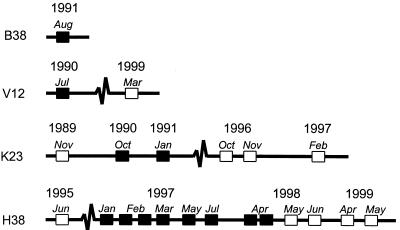
The results of C-PCR testing of all of the ejaculates obtained from the four positive donors over time were used to study the persistence of infection. Donor K23 donated between November 1989 and February 1997 six ejaculates. Only two ejaculates, donated in October 1990 and January 1991, were C-PCR positive, demonstrating that donor K23 carried C. trachomatis for at least four successive months (Fig. 2). Donor H38 provided 13 ejaculates between June 1995 and May 1999, of which 8, donated between January 1997 and April 1998, were found C. trachomatis positive. Therefore, the duration of the infection was at least 16 months (Fig. 2). From donor V12, only two ejaculates were left in the semen bank. The first ejaculate, donated in July 1990, represented the positive ejaculate. The last ejaculate, donated in March 1999, was negative. From donor B38, only one ejaculate was still present; it was found C. trachomatis positive by the C-PCR (Fig. 2).
FIG. 2.
Persistence and clearance of asymptomatic C. trachomatis infection in semen samples, as demonstrated by the C-PCR. Semen samples from all ejaculates (obtained from donors B38, V12, K23, and H38) previously testing positive were analyzed in retrospect by the C-PCR. C-PCR-positive ejaculates are indicated by filled squares; negative ejaculates are indicated by open squares.
Sequence analysis of the chlamydial omp1 gene.
C. trachomatis can be discriminated into several serovars on the basis of reactions with monoclonal antibodies against epitopes of Omp1. These serovars can also be identified by sequencing of omp1 (31). To assess whether cross-contamination had occurred from C. trachomatis-positive semen samples during storage in liquid nitrogen, the serovars of C. trachomatis present in semen samples from donors B38 and V12, from whom only one C. trachomatis-positive ejaculate each was available, and the serovars present in the first and last C. trachomatis-positive semen samples from donors K23 and H38 were determined by sequencing of omp1. From each processed semen sample, two independent recombinant clones containing the omp1 insert were sequenced in order to study whether sequence errors had occurred. Alignment of the two independent sequences revealed that they were always identical, indicating that sequence errors had not occurred. The ejaculate from donor B38 and both ejaculates from donor H38 contained omp1 sequences corresponding to C. trachomatis serovar E. The omp1 sequence found in the positive ejaculate from donor V12 represented serovar F, and the two ejaculates from donor K23 both contained omp1 sequences representing serovar K. The omp1 sequence of C. trachomatis L2/434/Bu, the strain used as a positive control throughout the study, was not found in any of the positive ejaculates.
DISCUSSION
We developed and evaluated a PCR assay (C-PCR) to assess C. trachomatis infection in semen specimens processed for artificial insemination. A total of 669 semen samples, donated by 97 donors participating in an AID program, were examined for C. trachomatis infection by the C-PCR. Twelve positive ejaculates originating from four donors were identified, indicating a prevalence of 4% among the donor population studied.
By using semen samples spiked with a quantified amount of purified C. trachomatis EBs, the lower limit of detection of the assay was found to range between 2.5 and 5 EBs per μl of semen. The observed range of the detection limit and the variation in the strength of the signals obtained between experiments are most likely not due to variation in the DNA extraction efficiency. We used the silica-GuSCN procedure (6) to extract DNA from semen specimens. This procedure has been acknowledged for its potency in the removal of inhibitory substances in a variety of clinical specimens interfering with PCR (5, 17, 30). All spiked samples were clearly positive, indicating that the silica-GuSCN procedure (6) can also be successfully used to extract C. trachomatis DNA from semen specimens. Spiking with purified EBs may have caused the variation in the strength of the signals obtained. It is well known that EBs have a strong tendency to form aggregates (21). Although stocks were subjected to sonication in order to reduce the number of aggregates, new aggregates could have been formed during the time span between sonication and addition to the semen samples, leading to an uneven distribution of EBs in the SPG stock solutions used. This problem could have easily been avoided by using naked plasmid DNA instead of EBs, but we chose to use EBs to control the efficiency of lysis of EB particles. With this approach, the detection limit obtained was similar to that previously reported by Van den Brule et al. (26). These investigators used cloned C. trachomatis plasmid DNA to spike semen samples and obtained a detection limit of between 10 and 100 copies of target plasmid DNA, corresponding to 1 to 10 EBs per μl of semen, assuming a plasmid copy number of 10 (24). Therefore, the GuSCN procedure used to lyse the EBs and to extract the DNA is very efficient for semen samples infected with C. trachomatis.
Samples from 12 ejaculates were found positive for C. trachomatis by the C-PCR. Since cracking of or spilling from straws with C. trachomatis-infected semen during processing and cryopreservation may occur, leading to contamination of the liquid nitrogen and cross-contamination of other straws (13), we determined the serovars of the C. trachomatis strains found in the positive ejaculates. Three different serovars were identified, indicating that the semen samples of at least three donors were C. trachomatis positive prior to storage. Serovar E was found to be present in two straws from the single ejaculate from donor B38 and in two straws from two ejaculates from donor H38. The second ejaculate from this donor was obtained 16 months after the first ejaculate. Taking into consideration that serovar E is one of the most prevalent C. trachomatis serovars in The Netherlands (18, 28), it is not surprising that strains of serovar E were present in the ejaculates from two of the four infected donors. In addition, the eight positive ejaculates from donor H38, donated in 16 consecutive months, as well as the three straws from the positive ejaculate from donor B38 were consistently C-PCR positive. Among the remaining 660 ejaculates in the same container, two ejaculates, from donor K23, were infected with serovar K, and one ejaculate, from donor V12, was infected with serovar F. The finding that the other 657 ejaculates were not infected indicates that cross-contamination of straws during processing and cryopreservation had not occurred.
The 4% prevalence of asymptomatic C. trachomatis infection among the donor population in this study is in accordance with the recent reported prevalences (2.3 to 4.7%) of C. trachomatis infection among men in The Netherlands in age groups similar to the age group of the donors tested here (27, 29). Transmission of C. trachomatis due to insemination with infected semen has been reported (20, 26). Whether such transmission also occurred in our recipient population was addressed, but no conclusive data were obtained.
The European Society for Human Reproduction and Embryology recommends testing of urine, urethral, or semen specimens from donors for C. trachomatis infection before they enter the AID program as well as at the end of the donation period (3). Of the 97 donors investigated in this study, 40 were tested in 1995 for C. trachomatis infection by the ligase chain reaction with urine specimens (LCx; Abbott Laboratories, North Chicago, Ill.). None of these 40 donors, who were actively participating in the AID program in 1995, tested positive then. Donors V12, B38, and K23 were not tested in 1995 because they either had stopped participating in the AID program or were not actively donating when urine testing was done. In 1995, donor H38 tested negative by the urine LCx. Results of C-PCR tests of semen donated in 1995 as well as after April 1998 were also negative, but semen samples obtained during this period were positive. This information indicates that screening at entry and at the end of the donation period does not guarantee a C. trachomatis-free interval. Whether screening at 6-month intervals, as suggested by the American Society for Reproductive Medicine (2), does exclude infected ejaculates is questionable, since molecular evidence of infection after antibiotic treatment is erased after 1 month (19). Therefore, we strongly recommend that stored ejaculates from donors not tested for C. trachomatis infection just prior to donation should be tested for C. trachomatis infection by a direct test such as the C-PCR. Testing of the specimens themselves will detect not only donors with active infection but also C. trachomatis-infected ejaculates already stored and will thus improve the microbiological quality of AID.
ACKNOWLEDGMENTS
We thank René Boom for scientific support and Nadia Binnekamp and Remco de Vries for technical assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Society for Reproductive Medicine. Guidelines for gamete and embryo donation. Fertil Steril. 1998;4(Suppl. 3):1S–13S. [PubMed] [Google Scholar]
- 3.Barratt, C., Y. Englert, C. Gottlieb, and P. Jouannet. 1998. Gamete donation guidelines. The Corsendonk consensus document for the European Union. Hum. Reprod. 13(Suppl. 2):vii–ix. [PubMed]
- 4.Black C M. Current methods of laboratory diagnosis of Chlamydia trachomatis infections. Clin Microbiol Rev. 1997;10:160–184. doi: 10.1128/cmr.10.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boom R, Sol C J A, Beld M, Weel J F L, Goudsmit J, Wertheim-van Dillen P M E. Improved silica-guanidinium thiocyanate DNA isolation procedure based on selective binding of bovine alpha-casein to silica particles. J Clin Microbiol. 1999;37:615–619. doi: 10.1128/jcm.37.3.615-619.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control. Chlamydia trachomatis infections: policy guidelines for prevention and control. Morb Mortal Wkly Rep. 1995;34:53S–74S. [PubMed] [Google Scholar]
- 8.Comanducci M, Ricci S, Cevenini R, Ratti G. Diversity of the Chlamydia trachomatis common plasmid in biovars with different pathogenicity. Plasmid. 1990;23:149–154. doi: 10.1016/0147-619x(90)90034-a. [DOI] [PubMed] [Google Scholar]
- 9.Fish A N, Fairweather D V, Oriel J D, Ridgway G L. Chlamydia trachomatis infection in a gynaecology clinic population: identification of high-risk groups and the value of contact tracing. Eur J Obstet Gynecol Reprod Biol. 1989;31:67–74. doi: 10.1016/0028-2243(89)90027-0. [DOI] [PubMed] [Google Scholar]
- 10.Fujisawa M, Nakano Y, Matsui T, Okada H, Arakawa S, Kamidono S. Chlamydia trachomatis detected by ligase chain reaction in the semen of asymptomatic patients without pyospermia or pyuria. Arch Androl. 1999;42:41–44. doi: 10.1080/014850199263039. [DOI] [PubMed] [Google Scholar]
- 11.Gerbase A C, Rowley J T, Heymann D H, Berkley S F, Piot P. Global prevalence and incidence estimates of selected curable STDs. Sex Transm Infect. 1998;74(Suppl. 1):S12–S16. [PubMed] [Google Scholar]
- 12.Griffais R, Thibon M. Detection of Chlamydia trachomatis by the polymerase chain reaction. Res Microbiol. 1989;140:139–141. doi: 10.1016/0923-2508(89)90047-8. [DOI] [PubMed] [Google Scholar]
- 13.Hawkins A E, Zuckerman M A, Briggs M, Gilson R J, Goldstone A H, Brink N S, Tedder R S. Hepatitis B nucleotide sequence analysis: linking an outbreak of acute hepatitis B to contamination of a cryopreservation tank. J Virol Methods. 1996;60:81–88. doi: 10.1016/0166-0934(96)02048-4. [DOI] [PubMed] [Google Scholar]
- 14.Krieger J N, Riley D E, Roberts M C, Berger R E. Prokaryotic DNA sequences in patients with chronic idiopathic prostatitis. J Clin Microbiol. 1996;34:3120–3128. doi: 10.1128/jcm.34.12.3120-3128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahadevan M, Trounson A O. Effect of cryoprotective media and dilution methods on the preservation of human spermatozoa. Andrologia. 1983;15:355–366. doi: 10.1111/j.1439-0272.1983.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 16.Marrazzo J M, Stamm W E. New approaches to the diagnosis, treatment, and prevention of chlamydial infection. Curr Top Clin Infect Dis. 1998;18:37–59. [PubMed] [Google Scholar]
- 17.Meijer A, van der Vliet J A, Schouls L M, de Vries A, Roholl P J M, Ossewaarde J M. Detection of microorganisms in vessel wall specimens of the abdominal aorta: development of a PCR assay in the absence of a gold standard. Res Microbiol. 1998;149:577–583. doi: 10.1016/s0923-2508(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 18.Morre S A, Moes R, Van Valkengoed I, Boeke J P, van Eijk J T, Meijer C J L M, Van den Brule A J C. Genotyping of Chlamydia trachomatis in urine specimens will facilitate large epidemiological studies. J Clin Microbiol. 1998;36:3077–3078. doi: 10.1128/jcm.36.10.3077-3078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morre S A, Sillekens P T G, Jacobs M V, De Blok S, Ossewaarde J M, Van Aarle P, Van Gemen B, Walboomers J M M, Meijer C J L M, Van den Brule A J C. Monitoring of Chlamydia trachomatis infections after antibiotic treatment using RNA detection by nucleic acid sequence-based amplification. J Clin Pathol. 1998;51:149–154. doi: 10.1136/mp.51.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagel T C, Tagatz G E, Campbell B F. Transmission of Chlamydia trachomatis by artificial insemination. Fertil Steril. 1986;46:959–960. doi: 10.1016/s0015-0282(16)49842-0. [DOI] [PubMed] [Google Scholar]
- 21.Schachter J, Wyrick P B. Culture and isolation of Chlamydia trachomatis. Methods Enzymol. 1994;236:377–390. doi: 10.1016/0076-6879(94)36028-6. [DOI] [PubMed] [Google Scholar]
- 22.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 23.Stothard D R, Boguslawski G, Jones R B. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect Immun. 1998;66:3618–3625. doi: 10.1128/iai.66.8.3618-3625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam J E, Davis C H, Thresher R J, Wyrick P B. Location of the origin of replication for the 7.5-kb Chlamydia trachomatis plasmid. Plasmid. 1992;27:231–236. doi: 10.1016/0147-619x(92)90025-6. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van den Brule A J C, Hemrika D J, Walboomers J M M, Raaphorst P, van Amstel N, Bleker O P, Meijer C J L M. Detection of Chlamydia trachomatis in semen of artificial insemination donors by the polymerase chain reaction. Fertil Steril. 1993;59:1098–1104. doi: 10.1016/s0015-0282(16)55935-4. [DOI] [PubMed] [Google Scholar]
- 27.Van den Hoek J A R, Mulder-Folkerts D K F, Coutinho R A, Dukers N H T M, Buimer M, van Doornum G J J. Opportunistic screening for genital infections with Chlamydia trachomatis among the sexually active population of Amsterdam. I. Over 90% participation and almost 5% prevalence. Ned Tijdschr Geneeskd. 1999;143:668–672. [PubMed] [Google Scholar]
- 28.van Duynhoven Y T H P, Ossewaarde J M, Derksen-Nawrocki R P, van der Meijden W I, van de Laar M J W. Chlamydia trachomatis genotypes: correlation with clinical manifestations of infection and patients' characteristics. Clin Infect Dis. 1998;26:314–322. doi: 10.1086/516291. [DOI] [PubMed] [Google Scholar]
- 29.van Valkengoed I G M, Boeke A J P, van den Brule A J C, Morre S A, Dekker J H, Meijer C J L M, van Eijk J T M. Systematic home screening for Chlamydia trachomatis infections of asymptomatic men and women in family practice by means of mail-in urine samples. Ned Tijdschr Geneeskd. 1999;143:672–676. [PubMed] [Google Scholar]
- 30.Vernazza P L, Dyer J R, Fiscus S A, Eron J J, Cohen M S. HIV-1 viral load in blood, semen and saliva. AIDS. 1997;11:1058–1059. [PubMed] [Google Scholar]
- 31.Yang C L, Maclean I, Brunham R C. DNA sequence polymorphism of the Chlamydia trachomatis omp1 gene. J Infect Dis. 1993;168:1225–1230. doi: 10.1093/infdis/168.5.1225. [DOI] [PubMed] [Google Scholar]
- 32.Zimmerman H L, Potterat J J, Dukes R L, Muth J B, Zimmerman H P, Fogle J S, Pratts C I. Epidemiologic differences between chlamydia and gonorrhea. Am J Public Health. 1990;80:1338–1342. doi: 10.2105/ajph.80.11.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]