Abstract
Due to its peculiar morphological characteristics, there is dispute as to whether the genus of Annamocarya sinensis, a species of Juglandaceae, is Annamocarya or Carya. Most morphologists believe it should be distinguished from the Carya genus while genomicists suggest that A. sinensis belongs to the Carya genus. To explore the taxonomic status of A. sinensis using chloroplast genes, we collected chloroplast genomes of 16 plant species and assembled chloroplast genomes of 10 unpublished Carya species. We analyzed all 26 species’ chloroplast genomes through two analytical approaches (concatenation and coalescence), using the entire and unique chloroplast coding sequence (CDS) and entire and protein sequences. Our results indicate that the analysis of the CDS and protein sequences or unique CDS and unique protein sequence of chloroplast genomes shows that A. sinensis indeed belongs to the Carya genus. In addition, our analysis shows that, compared to single chloroplast genes, the phylogeny trees constructed using numerous genes showed higher consistency. Moreover, the phylogenetic analysis calculated with the coalescence method and unique gene sequences was more robust than that done with the concatenation method, particularly for analyzing phylogenetically controversial species. Through the analysis, our results concluded that A. sinensis should be called C. sinensis.
Keywords: Carya sinensis, Annamocarya sinensis, unique gene, concatenation, coalescence, phylogeny
1. Introduction
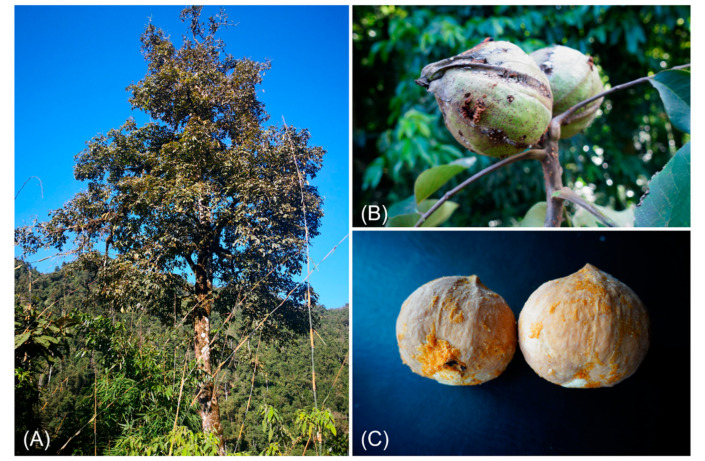
Annamocarya sinensis, controversially called Carya sinensis, is widely distributed in southern China and northern Vietnam. As a deciduous tree, it is generally about 30 m tall, has a 125-cm diameter trunk, and has grayish bark with alternate leaves (Figure 1) [1]. As the individual number of A. sinensis has reduced and its distribution narrowed, it is now a Class II endangered species in China [2]. It belongs to the Juglandaceae family, but its genus is still controversial; while most plant scientists classify it as Annamocarya, some classify it as Carya. This dispute has been caused by different taxonomic approaches, especially in morphology and evolutionary genomics.
Figure 1.
Annamocarya sinensis (Carya sinensis). (A) Tree; (B) Fruits in tree; (C) Fruits without husk (Photos provided by Weibang Sun, Kunming Institute of Botany, China).
Dode (1912) first described A. sinensis as C. sinensis based on limited herbarium samples, seed specimens without leaves, and flower samples [3]. Hence, his classification was controversial [4]. Chevalier (1941) [5] described the morphological characteristics of the leaves and nuts and concluded that it belonged to a new, single genus, Annamocarya. Nevertheless, Manning and Hjelmqvist (1951) [4] questioned whether, given that Carya and it shared plenty of similar morphological characteristics, it was necessary to describe this uncertain species as a single genus. Most morphologists believe that it is distinguished from Carya based on several morphological characteristics. For example, the leaves of A. sinensis are smooth, while Carya has serrated leaves; A. sinensis has five to eight catkins per bundle, whereas Carya has only three. The staminate bract of A. sinensis have three or more vascular strands, but only one in Carya [6]. And Leroy (1955) [7] proposed that the characters of the shell might be an important reason to distinguish it from Carya. Grauke et al. (1991) [3] compared the differences between Carya and Annamocarya in terms of tree shape, leaves, flowers, fruit shape, and other samples collected from Cuc Phuong National Forest in Vietnam. They concluded that the separate genus Annamocarya was necessary. As more morphological pieces of evidence became available, the genus Annamocarya was adopted over a long time.
In the 21st century, with the rapid development of sequencing technology, researchers have increasingly used gene sequences to study the evolutionary history of species with a phylogenomic approach. Manos and Stone (2001) [6] analyzed the phylogeny of Juglandaceae by using the internal transcribed spacer (ITS) and a few chloroplast genes. Their results suggest that A. sinensis is closely related to the Carya genus. In contrast, Li et al. (2004) [8] separated A. sinensis and Carya on different branches of infrafamilial relationships of the order Fagales based on six genes (atpB, matK, matR, rbcL, trnL-F and 18S rDNA). Manos et al. (2007) [9] investigated the evolution of Juglandaceae by integrating fossils and nucleic sequence evidence. Several results in their study suggest that A. sinensis should still be grouped within the Carya genus. Zhang et al. (2013) [10] used six plastid fragments (matK, rbcL-atpB, rpoC1, rps16, trnH-psbA, and trnL-F) and nuclear markers (ITS and phyA) in their study; the results suggest that A. sinensis should be clustered within the Carya genus. It seems, based on these examples, that sequence analysis is likely to classify A. sinensis in the Carya genus.
Recently, nuclear and organelle genes have commonly been used to analyze the phylogeny of species. Among them, chloroplast genome data has been utilized due to its low evolution rate and stable in variation [11]. It has been widely used to study phylogeny, chloroplast inheritance, domestication history, and adaptative evolution [12,13]. For example, Wu et al. (2020) [14] constructed a phylogenetic analysis of Chrysosplenium based on the whole chloroplast genome. They conclude that Chrysosplenium could be divided into two subgroups: those having alternate leaves or opposite leaves. Summarizing empirical studies that studied the phylogeny of Carya based on chloroplast genes, the similarity among them is that only a few chloroplast genes or fragments were utilized [6,9,10]. Researchers have found that phylogeny trees built from individual genes may show discordance on the species tree [15,16]. Furthermore, as the number of gene trees increases, they may converge in probability to the true species tree [17]. Hence, phylogenetic trees constructed entirely of chloroplast genome genes may be reliable.
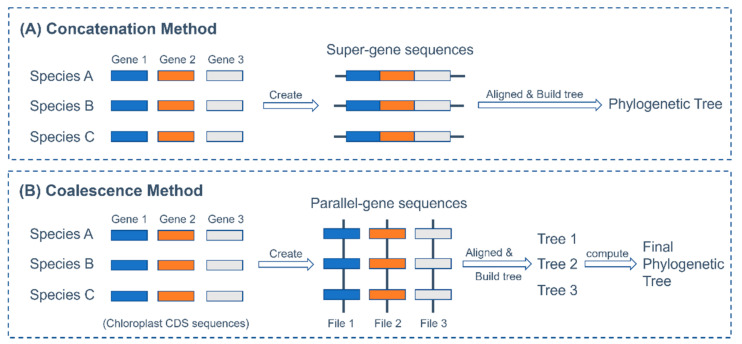
Since the development of next-generation sequencing and computational phylogenomics, the reconstruction of angiosperm phylogenies from multiple genes has relied upon concatenation methods [18]. The concatenation method concatenates multiple gene sequences of each species and treats them as one alignment to generate a phylogenetic tree (Figure 2A). As the number of subsampled nuclear genes increases, the result of concatenation analyses becomes dependable [18]. However, a simulation study by Kubatko and Degnan (2007) [19] shows that such approaches could lead to misleading phylogenetic calculations. They demonstrated that using concatenated data, species tree estimation performed deficiently and was statistically inconsistent even under stable population size without selection or population stratification. In addition to concatenation method, recent studies have also taken advantage of coalescence method for phylogenetic analysis [20,21].This method first computes gene trees by the individual gene sequences and then aggregates all gene trees to obtain the final species tree (Figure 2B) [15]. Goncalves and colleagues [22] conducted a phylogenetic analysis of 78 plastid genes, and their results showed that the phylogenetic tree inferred by the coalescence method was reliable. Xi et al. (2013) [23] propose that the coalescence method might reduce the potential deleterious effect of elevated substitution rates in phylogenomic analyses. To test this, Xi et al. (2014) [18] used the two methods to analyze the phylogeny of Amborella within 45 seed plants and found that fast-evolving sites likely disrupt the concatenation method, while the coalescence method appears more robust in response to elevated substitution rates.
Figure 2.
Methods of concatenation and coalescence. (A) Concatenation method joins all gene sequences of each species into each super-gene sequence. These super-gene sequences are then used in aligning and building phylogeny trees; (B) Coalescence method extracts the same gene sequences from all species and merges them into each single parallel-gene sequence file. Then all parallel-gene sequence files are aligned separately to build trees. Finally, all those trees are computed to get the final tree.
In this work, we first assembled 10 chloroplast genomes of the Carya species for observing the phylogenetic relationship between A. sinensis and Carya. We determined the true phylogenetic location of A. sinensis by comparing and evaluating the concatenation and coalescence methods with the chloroplast genome of 26 species. Our results indicate that A. sinensis was clustered within the Carya genus. Hence, we conclude that Annamocarya sinesis should be referred to as Carya sinensis. Moreover, the phylogeny results constructed by the coalescence method are more robust than those by the concatenation method.
2. Results
We analyze the phylogenetic position of A. sinensis using the genes of the chloroplast genome. A total of 14 species belonging to the Carya genus of Eastern Asia and North America were included in this work (four species of Eastern Asia include C. cathayensis, C. dabieshanensis, C. hunanensis, and C. kweichowensis; 10 species of North America include C. aquatica, C. cordiformis, C. glabra, C. illinoinensis, C. laciniosa, C. myristiciformis, C. ovata, C. palmeri, C. texana, and C. tomentosa) [10]. The CDS and protein sequences of all 26 species of six genera (Betula, Carya, Cyclocarya, Juglans, Platycarya, and Pterocarya) were used for the analysis.
2.1. Assemblies and Annotations of Chloroplast Genomes
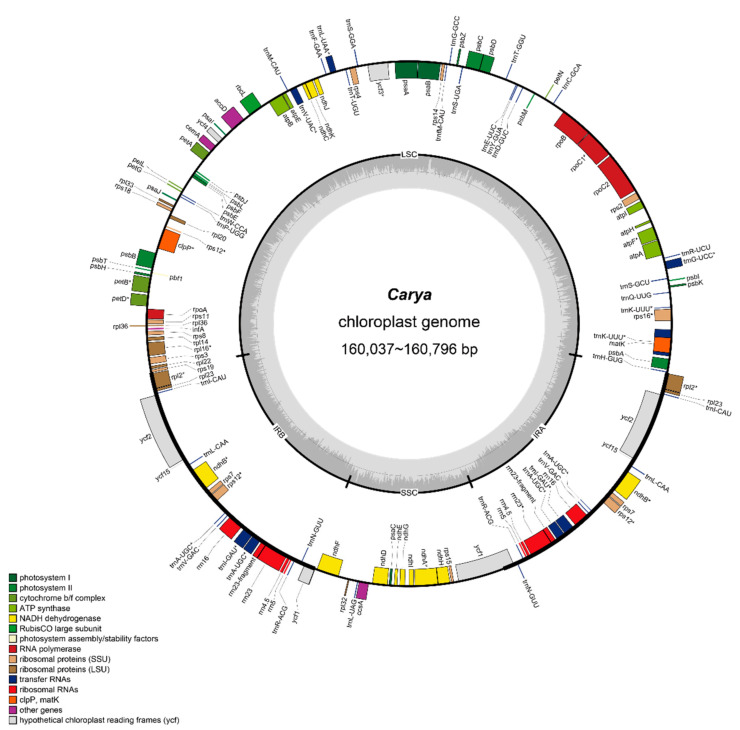
We first obtained the raw sequencing data of 10 species of the Carya genus (Table S1). Then, we assembled the 10 complete closed-loop chloroplast genomes of these species (Figure 3 and Figure S1) for the analysis below. From the assemblies, we learned that the average size of the 10 species was about 160 kb (Table 1). The chloroplast genome of C. cordiformis was the longest at 160,796 bp, and the shortest one was C. dabieshanensis at 160,037 bp; the difference between the two was 759 bp. The GC content of the assembled 10 chloroplast genomes was similar, around 36%.
Figure 3.
The circular chloroplast genome map of Carya (take C. cordiformis as reference graph). The genes shown inside and outside of the circle are transcribed in clockwise and counterclockwise directions, respectively. Genes from different functional groups are shown in different colors. The thick dark lines in inner circle show the extent of the Inverted repeats (IRA and IRB) separating the Large Single-Copy (LSC) and the Small Single-Copy (SSC) regions. The gray ring represents the GC content. The circular chloroplast genome maps of 10 species were show in Figure S1.
Table 1.
The assembled ten chloroplast genome structure of 10 Carya species.
| Species | LSC | SSC | IR | Total Length | GC Content(%) |
|---|---|---|---|---|---|
| Carya aquatica | 89,966 | 18,791 | 26,003 | 160,763 | 36.16 |
| Carya cordiformis | 89,992 | 18,798 | 26,003 | 160,796 | 36.15 |
| Carya dabieshanensis | 89,508 | 18,861 | 25,834 | 160,037 | 36.20 |
| Carya glabra | 89,888 | 18,786 | 25,989 | 160,652 | 36.18 |
| Carya laciniosa | 89,927 | 18,842 | 26,001 | 160,771 | 36.17 |
| Carya myristiciformis | 89,990 | 18,792 | 26,003 | 160,788 | 36.15 |
| Carya ovata | 89,930 | 18,809 | 25,994 | 160,727 | 36.17 |
| Carya palmeri | 89,818 | 18,778 | 26,004 | 160,604 | 36.18 |
| Carya texana | 89,964 | 18,793 | 25,994 | 160,745 | 36.17 |
| Carya tomentosa | 89,988 | 18,792 | 26,002 | 160,784 | 36.16 |
We also obtained the chloroplast genomes of 16 species of six genera from other database, used GeSeq [24] to reannotate the chloroplast genomes of all 26 species to standardize the annotations previously generated by different annotation tools. The chloroplast genomes of the Carya genus contained an average of 79 coding genes, suggesting that they were highly conserved (Table 2). The Cyclocarya and Pterocarya genera contained the most coding genes. The CDS GC content of all species was approximately 37.22%, with little difference. The uniformly reannotated chloroplast genomes were used for downstream analysis.
Table 2.
Chloroplast genome features of 26 species.
| Species | Genome Size (bp) | Coding Gene Number | tRNA Genes | rRNA Genes | CDS Total Length (bp) | CDS GC Content (%) |
|---|---|---|---|---|---|---|
| A. sinensis | 158,484 | 79 | 35 | 8 | 68,261 | 37.26 |
| B. platyphylla | 160,518 | 84 | 37 | 8 | 78,972 | 37.43 |
| C. aquatica | 160,763 | 79 | 37 | 8 | 68,673 | 37.24 |
| C. cathayensis | 160,666 | 80 | 36 | 8 | 69,595 | 37.22 |
| C. cordiformis | 160,796 | 79 | 37 | 8 | 68,673 | 37.24 |
| C. dabieshanensis | 160,037 | 80 | 37 | 8 | 69,250 | 37.24 |
| C. glabra | 160,652 | 79 | 37 | 8 | 68,674 | 37.25 |
| C. hunanensis | 160,397 | 80 | 36 | 8 | 69,449 | 37.21 |
| C. illinoinensis | 160,819 | 79 | 37 | 8 | 68,673 | 37.25 |
| C. kweichowensis | 175,313 | 79 | 38 | 8 | 68,314 | 37.27 |
| C. laciniosa | 160,771 | 79 | 37 | 8 | 68,672 | 37.26 |
| C. myristiciformis | 160,788 | 79 | 37 | 8 | 68,673 | 37.24 |
| C. ovata | 160,727 | 79 | 37 | 8 | 68,680 | 37.26 |
| C. palmeri | 160,604 | 79 | 37 | 8 | 68,673 | 37.25 |
| C. texana | 160,745 | 79 | 37 | 8 | 68,674 | 37.25 |
| C. tomentosa | 160,784 | 79 | 37 | 8 | 68,673 | 37.25 |
| Cy. paliurus | 160,562 | 89 | 40 | 8 | 81,015 | 37.19 |
| J. cathayensis | 159,730 | 87 | 40 | 8 | 80,331 | 37.26 |
| J. cinerea | 160,288 | 84 | 37 | 8 | 78,333 | 37.26 |
| J. hopeiensis | 159,714 | 86 | 40 | 8 | 80,259 | 37.27 |
| J. major | 160,276 | 83 | 37 | 8 | 77,970 | 37.2 |
| J. nigra | 160,274 | 83 | 37 | 8 | 77,958 | 37.21 |
| J. regia | 160,370 | 83 | 37 | 8 | 77,979 | 37.2 |
| P. strobilacea | 160,994 | 85 | 36 | 8 | 78,915 | 37.18 |
| Pt. hupehensis | 159,770 | 89 | 40 | 8 | 81,315 | 37.2 |
| Pt. stenoptera | 160,202 | 89 | 40 | 8 | 81,021 | 37.23 |
2.2. Unique Genes of the Chloroplast Genomes
After uniformly reannotating the chloroplast genomes, we used Orthofinder to identify 50 unique genes (Table 3) based on these chloroplast protein sequences. All 50 unique genes were single-copy genes in the chloroplast genome of all 26 species. Most of them were related to photoreaction and ribosome function. And according to these unique genes, we then extracted 50 unique CDS sequences and 50 unique protein sequences for downstream analysis.
Table 3.
A total of 50 unique genes existed in 26 chloroplast genomes.
| Fuctional Groups | Name of Genes |
|---|---|
| Ribosomal protein small subunit | rps2, rps3, rps4, rps8, rps11, rps14, rps18 |
| Ribosomal protein large subunit | rpl14, rpl20, rpl22, rpl32, rpl33 |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 |
| Photosystcm I | psaA, psaB, psaI, psaJ |
| Photosystem II | psbA, psbC, psbD, psbE, psbH, psbJ, psbK, psbL, psbM, psbT |
| Cythochrome b/f complex | petA, petG, petL, petN |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI |
| NADH-dehydrogenase | ndhC, ndhF, ndhJ, ndhK |
| Large subunit Rubisco | rbcL |
| Acetyl-CoA carboxylase | accD |
| Maturase | matk |
| Inner membrane protein | cemA |
| Conserved open reading frames | ycf3, ycf4 |
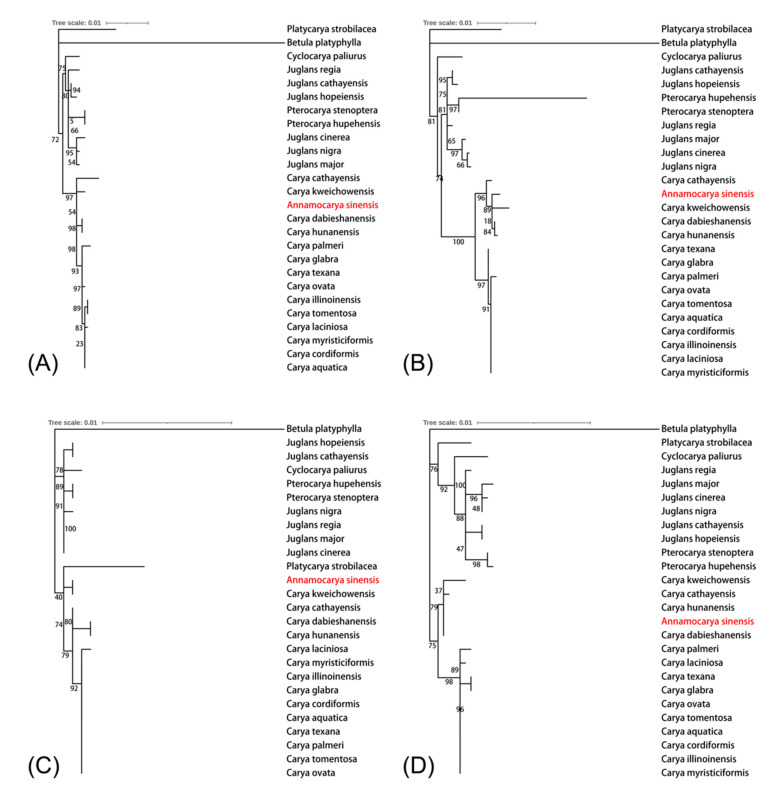
After extracting 50 unique genes, we constructed the phylogenetic trees using each individual gene. However, most of the 50 chloroplast gene trees (Figure S2) showed chaotic phylogenetic relationships. For example, matK, ndhF, rbcL and rpoC1 genes (Figure 4), which are commonly used to construct phylogenetic trees, indicated that species of the genus Juglans failed to group together. And even if we rerooted Betula platyphylla as an outgroup, it cannot be separated from Platycarya strobilaceaat some point (Figure 4A,B). The rpoC1 gene phylogenetic tree was the only tree correctly classified all genera and clustered pecans into East Asia and North America. Most phylogenetic trees showed the analyzed species were confusingly classified into the wrong genera. The evolutionary relationship of these results was unstable and inconsistent. Hence, we utilize the following two methods to achieve a certain degree of reliability.
Figure 4.
Phylogenetic trees constructed by unique chloroplast gene. (A) matK; (B) ndhF; (C) rbcL; (D) rpoC1.
2.3. Phylogenetic Analysis Based on the Concatenation Method
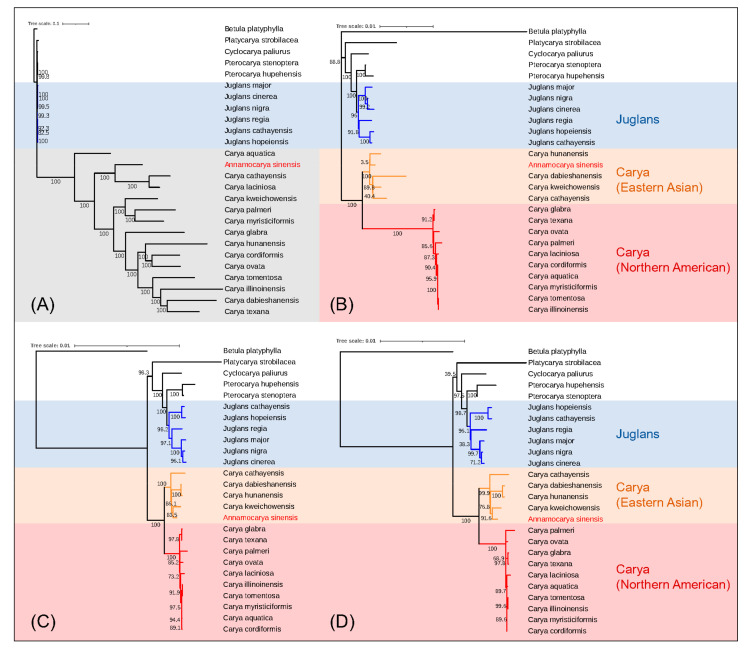
We collected whole CDS sequences and protein sequences (79–89 genes in each) from the reannotated chloroplast genomes of all 26 species. Then, we extracted the unique CDS sequences and unique protein sequences (50 genes in each) according to the analysis result of Orthofinder. The four phylogenetic trees of entire CDS sequences, entire protein sequences, unique CDS sequences, and unique protein sequences constructed by the concatenation method are shown in Figure 5A–D. Impressively, we noticed that the out-of-group branches (genera of Betula, Cyclocarya, Juglans, Platycarya, and Ptercocarya) in these four trees were clustered in the same clades. Species in genera such as Juglans and Pterocarya were always clustered together. In all the results, as we hypothesized, A. sinensis was classified into the Carya genus branch. The results (Figure 5B–D) clustered all Carya into Eastern Asian and North American subgroups well except for the phylogenetic tree using the entire CDS sequences (Figure 5A), which showed low bootstrap credibility. Interestingly, compared to the results of entire gene sequences, the two results based on unique genes (Figure 5C,D) showed more similarity, indicating that the phylogeny analysis using unique genes was stable. Moreover, A. sinensis was close with C. kweichowensis in these two result trees (Figure 5C,D). The results are in accordance with the geographical distribution of these two species [2,25].
Figure 5.
Phylogeny analysis based on the concatenation method. Visualized using ITOL. (A) Analyzed with entire CDS sequences; (B) Analyzed with entire protein sequences; (C) Analyzed with unique CDS sequences; (D) Analyzed with unique protein sequences.
2.4. Phylogenetic Analysis Based on Coalescence Method
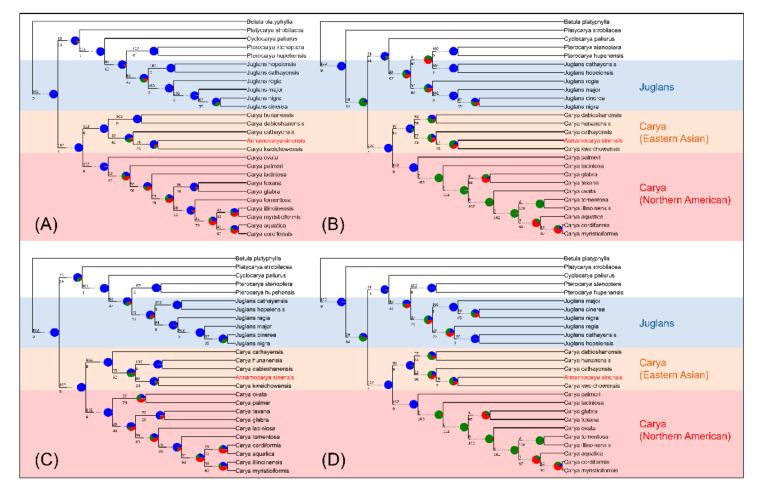
Subsequently, we again analyzed the phylogenetics of A. sinensis based on the coalescence method with entire CDS sequences, entire protein sequences, unique CDS sequences, and unique protein sequences. The resulting four phylogenetic trees are shown in Figure 6. It should be noted that these coalescence results were computed by ASTRAL, which used a quartet score to score species trees instead of bootstrap. The closer the score is to 1, the more credible the tree is. Scores of the four results were all greater than 0.8, denoting credibility. As was conjectured, the out-of-group branches (the five genera mentioned before) of the trees were clustered in correct phylogenetic positions with high confidence support (blue pie charts, Figure 6). Similarly, A. sinensis in all results was consistently classified into the Eastern Asian Carya genus branch, especially close with C. kweichowensis. The conflict analysis of CDSs (Figure 6A,C) showed more concordant with gene trees than protein sequences (Figure 6B,D). The clades of Northern American Carya, on the other hand, showed much common conflicts (green pie charts, Figure 6B,D), perhaps because of the degeneracy among these close species. Surprisingly, all leaves of phylogenetic tree constructed by the coalescence method, either using entire-gene or unique-gene sequences, were almost identical (Figure 6).
Figure 6.
Phylogeny analysis based on the coalescence method. (A) Analyzed with entire CDS sequences, final normalized quartet score: 0.954; (B) Analyzed with entire protein sequences, final normalized quartet score: 0.804; (C) Analyzed with unique CDS sequences, final normalized quartet score: 0.948; (D) Analyzed with unique protein sequences, final normalized quartet score: 0.807. The pie chart at each node showed the support of gene trees as the following: concordant with gene trees (blue), most common conflict bipartition (green), other conflicting bipartitions (red) and unsupported with gene trees (grey).
3. Discussion
The taxonomic status of A. sinensis has long been debated. Morphologists distinguished it from the Carya genus by morphological features such as leaves, bunches, bracts, and nut shells. However, the accuracy of sequencing technologies nowadays makes molecular phylogeny being more common and credible [26]. Following the release of raw sequencing data (Table S1) for some species of Carya [27], we have assembled and annotated the chloroplast genomes of 10 species of Carya that have not yet been published. Among them, nine North American species were assembled using C. illinoinensis (MH909600) as a reference chloroplast genome, and only C. dabieshanensis (Eastern Asian) was assembled using C. cathayensis (NC_046572) as a reference chloroplast genome (Table S1). The chloroplast genomes we assembled (Table 1) using GetOrganelle were of high quality; both the size of the LSC/SSC/IR region and the GC content were similar to the published chloroplast genomes of other Carya species [28]. All these characteristics of Juglandaceae chloroplast genomes show stability in variation and low evolutionary rate [11].
Juglans has been divided into a four-section classification (Cardiocaryon, Dioscaryon, Rhysocaryon, Trachycaryon) [29]. Six species of these four sections were selected as the reference taxa in our evolutionary trees. As with the eight phylogenetic trees mentioned in Figure 5 and Figure 6, we observed that all outgroup taxa were clustered into proper phylogenetic positions. In particular, the Carya genus were categorized correctly into Eastern Asia and North America in both methods, which means that the analytical strategies we employed were reliable. Our results show that A. sinensis does have a close phylogenetic relationship with the Carya species, rather than separating itself into an independent clade. Our results also show that A. sinensis close to C. kweichowensis in the Carya clade. Both of these two species are scattered in southwestern China with an endemic distribution area [2,25]. The phylogenetic relationship of our results was highly consistent with geographical distribution, indicating that A. sinensis is reliably clustered in the Carya genus, in the Eastern Asian branch, whether based on molecular or geographical evidence. We conclude that A. sinensis should belong in Carya rather than divided into a single genus. Hence, it should be referred to as Carya sinensis.
The phylogenetic trees created by a single gene indicate that most of these single-gene trees cannot classify all species into the correct genera. It reveals that the evolutionary tree created by this single chloroplast gene may be unreliable. We also analyzed the phylogeny of the entire chloroplast genome of 26 species (Figure S3). However, the resulting taxa did not conform to the geographical distribution of these species. Therefore, it is necessary to adopt appropriate methods to make the evolutionary results stable and reliable. The two main approaches we put used in this study were concatenation and coalescence. Compared with the traditional concatenation method, the coalescence method, a new computer method, was more effective and robust when applied to phylogeny analysis [20]. The phylogenetic trees created by ASTRAL includes the conflicts of each node (displayed as pie charts), indicating that coalescence results were concordant with highly supported gene trees (blue pie charts, Figure 6). However, some nodes still showed conflicts (green or red pie charts), indicating that the gene trees which were inconsonant with species trees were still not negligible, especially in the American Carya genus branch. Conflicts caused by individual genes trees may be related to several reasons. Some plastid genes are multiple in one species while haploid in another species or the inverted repeats of genes were confused when extracting them and constructing phylogenetic trees. The correction of such conflicts between genes trees and species trees probably should be investigated in further study. The phylogenetic relationship between Carya species computed with the coalescence method (Figure 6) showed high similarity. In contrast, these results analyzed by the concatenation method showed some heterogeneity (Figure 5). In addition to the entire chloroplast genes, we also combined unique genes for analysis. The utilization of combined unique genes may provide a complete selection of independent characters for plant phylogenetic analysis [30]. Our results showed that A. sinensis was always next to C. kweichowensis in the Carya clade when using the combined unique gene sequences for the analysis (Figure 5C,D and Figure 6C,D), indicating that the combined unique gene sequences were stable and consistent for the phylogenetic analysis. The very low non-conservative percentage of the sequence alignment (Figure S4) of the chloroplast genes of A. sinensis and C. kweichowensis suggested that this approach is reasonable. All these results conclude that the coalescence method using unique genes is recommended when computing phylogenetic species trees, especially for the analysis of phylogenetic controversial or close related species.
Even if molecular analysis evidence seems more accurate in classification, we cannot ignore the practicality and reliability of morphological evidence. Compared with molecular analysis, morphological analysis is serviceable to identify plants in field studies. With permitted conditions, by comparing the molecular and morphological analysis of closely related species, it may be possible to find out the reasons for the differences between them and perhaps prove the correct classification of these species better.
4. Materials and Methods
4.1. Data Sets
In this work, a total of 26 chloroplast genomes in five genera of Juglandaceae (Carya, Cyclocarya, Juglans, Platycarya, and Pterocarya) and Betula platyphylla were used for phylogeny analysis. Among them, 15 complete chloroplast genomes of Juglandaceae and Betula platyphylla were obtained from National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/, accessed on 1 March 2021; Table S2). The raw sequencing data of another 10 species of Carya (Table S1) [27] whose complete chloroplast genome had not been disclosed were downloaded and assembled. The assembled chloroplast genome of these 10 species was then uploaded to NCBI (Table S3). The chloroplast genome of C. illinoinensis (cultivar Pawnee) was sequenced and assembled by our lab and can be downloaded by the accession GWHBGBH00000000 at National Genomics Data Center (https://ngdc.cncb.ac.cn/, accessed on 1 November 2021) [31,32].
4.2. Chloroplast Genomes Assembly and Gene Annotation
The quality of raw sequencing data (raw reads) mentioned above was assessed using fastp v0.20.1 [33] with default parameters. After the quality assessment, the reads were mapped against the chloroplast genome of C. illinoinensis cultivar 87MX3-2.11 (MH909600) using BWA-MEM 0.7.17 [34] to obtain clean chloroplast reads. The SAM mapping results were then transformed to BAM format by SAMtools v1.9 [35,36] for assembly. The complete closed-loop chloroplast genome of each species was assembled using GetOrganelle v1.7.137 [37] with particular reference chloroplast genomes (Table S1). These assembled chloroplast genomes were annotated by the GeSeq tool [24] with C. illinoinensis (MH909600) as the reference genome.
4.3. Analysis of Unique Chloroplast Genes
We first used GeSeq to uniformly reannotate the chloroplast genomes of all 26 species to standardize the annotations previously generated by different annotation tools. Then the genomes’ protein sequences were subjected to Orthofinder [38,39] analysis with default parameters to obtain the single-copy genes (unique genes). Then both the protein and nucleotide sequences of these unique genes were extracted and prepared for subsequent phylogeny analysis.
The alignment of unique protein and nucleotide sequence files was created with L-INS-I parameters using MAFFT v7.271 [40,41]. Next, these alignment results were then used to construct maximum likelihood phylogenetic trees using IQtree [42] with K3Pu+F as the best fit model and 500 bootstraps.
4.4. Phylogeny Analysis Based on Concatenation Method
The entire chloroplast coding sequence (CDS) and protein sequences (entire-genes) of each species (Table 2) were concatenated to make super sequences (Figure 2A) using in-house Linux scripts. The super-entire-CDSs and super-entire-protein sequences of all the chloroplast genomes were acquired. Then, the unique CDS and protein sequences (unique-genes) of all species were concatenated into super-unique-CDSs and super-unique-protein sequences. The multiple alignment of all entire-genes and unique-genes of the super sequences were individually constructed using MAFFT v7.271 with FFT-NS-2 parameters. The phylogenetic tree of the concatenation method was obtained using FastTree [43,44] with the Jukes-Cantor model. Finally, these phylogenetic trees were visualized using ITOL [45].
4.5. Phylogeny Analysis Based on Coalescence Method
All coding gene sequences of chloroplast genomes of 26 species were extracted using Seqkit [46]. After gathering common coding gene sequences into common files, a total of 73 specific gene sequence files were acquired (Table S4). These 73 CDS and protein sequences were used to contract the parallel-entire-genes phylogeny trees (Figure 2B). Similarly, the alignment of each specific sequence file was also created using MAFFT v7.271 and constructed phylogenetic trees using IQtree with the same parameters as described in unique chloroplast genes above. All parallel-genes phylogeny trees generated by IQtree were first merged into one file using ASTRAL with the “-b” parameter. And the output (102 trees in default) was then computed to the final species trees using ASTRAL again. The conflicts of nodes of species trees were analyzed using PhyParts [47]. The visualization of trees with pie charts were drawn using PhypartsPieChart (https://github.com/mossmatters/phyloscripts/tree/master/phypartspiecharts, accessed on 7 November 2021).
5. Conclusions
The phylogenetic trees created using a single unique gene are unreliable. The concatenation and coalescence methods possibly are appropriate approaches to study the phylogeny of close species. Compared to the concatenation method, the phylogenetic trees constructed using the coalescence method shows stability and reliability. We collected 16 chloroplast genomes and assembled 10 chloroplast genomes of Carya species to analyze the phylogeny of A. sinensis. Our analysis of the chloroplast genomes with concatenation and coalescence methods showed that A. sinensis is clustered into the Carya genus. Our results concluded that A. sinensis, therefore, should be called C. sinensis.
Acknowledgments
We acknowledge Sun-Li Chong (ZAFU) for her reading and comment on the manuscript. We thank Weibang Sun of Kunmming Institute of Botany, Chinese Academy of Sciences, for kindly providing the photos of Carya sinensis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/2223-7747/11/1/52/s1, Figure S1: The circular chloroplast genome map of 10 assembled chloroplast genome; Figure S2: 50 phylogenetic results constructed by unique chloroplast genes; Figure S3: Phylogeny analysis of the entire chloroplast genome of 26 species; Figure S4: The alignment of chloroplast genomes of A. sinensis, C. cathayensis and C. kweichowensis; Table S1: Assembled chloroplast genome of 10 species; Table S2: Complete chloroplast genome of 15 species from NCBI; Table S3: NCBI accession number of assembled chloroplast genomes; Table S4: Mutual coding genes within all 26 chloroplast genomes (73 in total).
Author Contributions
Conceptualization, J.C., J.L.; methodology, J.C., J.L.; analyzation, J.L.; validation, W.G.; data curation, J.L., W.G.; writing—original draft preparation, J.L.; writing—review and editing, K.-J.L., J.L.; visualization, J.L.; funding acquisition, Z.W., K.-J.L., Z.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the National Natural Science Foundation of China (32001335), Key Research and Development Project of Zhejiang Province (2021C02054), Zhejiang Agriculture New Variety Breeding Major Science and Technology Special (2021C02066-12), National Key Research and Development Project (2018YFD1000604).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 1 March 2021) and National Genomics Data Center (https://ngdc.cncb.ac.cn/, accessed on 1 November 2021), accession number GWHBGBH00000000.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Long X. Annamocarya Sinensis, a Rare Species of Guizhou, China. For. By-Prod. Spec. China. 2002;2:63. [Google Scholar]
- 2.Zhang Z.Y., Pang X.M., Han J.W., Wang Y., Li Y.Y. Conservation Genetics of Annamocarya Sinensis (Dode) Leroy, an Endangered Endemic Species. Genet. Mol. Res. 2013;12:3965–3974. doi: 10.4238/2013.March.11.9. [DOI] [PubMed] [Google Scholar]
- 3.Grauke L.J., Wood B., Payne J. Genetic Resources of Carya in Vietnam and China. Ann. Report Northern Nut Growers Assoc. 1991;82:80–87. [Google Scholar]
- 4.Manning W.E., Hjelmqvist H. Annamocarya, Rhamphocarya, and Carya Sinensis. Botaniska Notiser. 1951;4:319–330. [Google Scholar]
- 5.Chevalier A. Variabilité Et Hybridité Chez Les Noyers. Notes Sur Des Juglans Peu Connus, Sur L’annamocarya Et Un Carya D’indochine. Rev. Bot. Appl. d’Agric. Coloniale. 1941;21:241. doi: 10.3406/jatba.1941.1646. [DOI] [Google Scholar]
- 6.Manos P.S., Stone D.E. Evolution, Phylogeny, and Systematics of the Juglandaceae. Ann. Mo. Bot. Gard. 2001;88:231–269. doi: 10.2307/2666226. [DOI] [Google Scholar]
- 7.Leroy J.F. Etude sur les Juglandaceae: A la Recherche d’une Concepcion Morphologique de la Fleur Femelle et du Fruit. Éditions du Muséum; Paris, France: 1955. [Google Scholar]
- 8.Li R., Chen Z., Lu A., Soltis D.E., Soltis P.S., Manoss P.S. Phylogenetic Relationships in Fagales Based on DNA Sequences from Three Genomes. Int. J. Plant Sci. 2004;165:311–324. doi: 10.1086/381920. [DOI] [Google Scholar]
- 9.Manos P.S., Soltis P.S., Soltis D.E., Manchester S.R., Oh S.-H., Bell C.D., Dilcher D.L., Stone D.E. Phylogeny of Extant and Fossil Juglandaceae Inferred from the Integration of Molecular and Morphological Data Sets. Syst. Biol. 2007;56:412–430. doi: 10.1080/10635150701408523. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J.-B., Li R.-Q., Xiang X.-G., Manchester S.R., Lin L., Wang W., Wen J., Chen Z.-D. Integrated Fossil and Molecular Data Reveal the Biogeographic Diversification of the Eastern Asian-Eastern North American Disjunct Hickory Genus (Carya Nutt.) PLoS ONE. 2013;8:e70449. doi: 10.1371/journal.pone.0070449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leebens-Mack J., Raubeson L.A., Cui L., Kuehl J.V., Fourcade M.H., Chumley T.W., Boore J.L., Jansen R.K., Claude W., DePamphilis C.W. Identifying the Basal Angiosperm Node in Chloroplast Genome Phylogenies: Sampling One’s Way out of the Felsenstein Zone. Mol. Biol. Evol. 2005;22:1948–1963. doi: 10.1093/molbev/msi191. [DOI] [PubMed] [Google Scholar]
- 12.Alzahrani D.A., Yaradua S.S., Albokhari E.J., Abba A. Complete chloroplast genome sequence of Barleria prionitis, comparative chloroplast genomics and phylogenetic relationships among Acanthoideae. BMC Genom. 2020;21:393. doi: 10.1186/s12864-020-06798-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z., Gui S., Quan Z., Pan L., Wang S., Ke W., Liang D., Ding Y. A precise chloroplast genome of Nelumbo nucifera (Nelumbonaceae) evaluated with Sanger, Illumina MiSeq, and PacBio RS II sequencing platforms: Insight into the plastid evolution of basal eudicots. BMC Plant Biol. 2014;14:289. doi: 10.1186/s12870-014-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z., Liao R., Yang T., Dong X., Lan D., Qin R., Liu H. Analysis of six chloroplast genomes provides insight into the evolution of Chrysosplenium (Saxifragaceae) BMC Genom. 2020;21:621. doi: 10.1186/s12864-020-07045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabiee M., Sayyari E., Mirarab S. Multi-Allele Species Reconstruction Using Astral. Mol. Phylogenet. Evol. 2019;130:286–296. doi: 10.1016/j.ympev.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 16.Roch S., Steel M. Likelihood-based tree reconstruction on a concatenation of aligned sequence data sets can be statistically inconsistent. Theor. Popul. Biol. 2015;100:56–62. doi: 10.1016/j.tpb.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Sayyari E., Mirarab S. Fast Coalescent-Based Computation of Local Branch Support from Quartet Frequencies. Mol. Biol. Evol. 2016;33:1654–1668. doi: 10.1093/molbev/msw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xi Z., Liu L., Rest J.S., Davis C.C. Coalescent Versus Concatenation Methods and the Placement of Amborella as Sister to Water Lilies. Syst. Biol. 2014;63:919–932. doi: 10.1093/sysbio/syu055. [DOI] [PubMed] [Google Scholar]
- 19.Kubatko L.S., Degnan J.H. Inconsistency of Phylogenetic Estimates from Concatenated Data under Coalescence. Syst. Biol. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- 20.Simmons M.P., Gatesy J. Coalescence Vs. Concatenation: Sophisticated Analyses Vs. First Principles Applied to Rooting the Angiosperms. Mol. Phylogenet. Evol. 2015;91:98–122. doi: 10.1016/j.ympev.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Edwards S.V., Xi Z., Janke A., Faircloth B., McCormack J.E., Glenn T.C., Zhong B., Wu S., Lemmon E.M., Lemmon A.R., et al. Implementing and testing the multispecies coalescent model: A valuable paradigm for phylogenomics. Mol. Phylogenet. Evol. 2016;94:447–462. doi: 10.1016/j.ympev.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 22.Gonçalves D.J., Simpson B.B., Ortiz E.M., Shimizu G.H., Jansen R.K. Incongruence between gene trees and species trees and phylogenetic signal variation in plastid genes. Mol. Phylogenet. Evol. 2019;138:219–232. doi: 10.1016/j.ympev.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Xi Z., Rest J.S., Davis C.C. Phylogenomics and Coalescent Analyses Resolve Extant Seed Plant Relationships. PLoS ONE. 2013;8:e80870. doi: 10.1371/journal.pone.0080870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye L., Fu C., Wang Y., Liu J., Gao L.-M. Characterization of the complete plastid genome of a Chinese endemic species Carya kweichowensis. Mitochondrial DNA Part B. 2018;3:492–493. doi: 10.1080/23802359.2018.1464414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Favre F., Jourda C., Besse P., Charron C. Molecular Plant Taxonomy. Humana; New York, NY, USA: 2020. Genotyping-by-Sequencing Technology in Plant Taxonomy and Phylogeny; pp. 167–178. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y., Xiao L., Zhang Z., Zhang R., Wang Z., Huang C., Huang R., Luan Y., Fan T., Wang J., et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. GigaScience. 2019;8:giz036. doi: 10.1093/gigascience/giz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Rhein H.S., Jenkins J., Schmutz J., Grimwood J., Grauke L.J., Randall J.J. Chloroplast Genome Sequences of Carya Illinoinensis from Two Distinct Geographic Populations. Tree Genet. Genomes. 2020;16:48. doi: 10.1007/s11295-020-01436-0. [DOI] [Google Scholar]
- 29.Mu X.-Y., Tong L., Sun M., Zhu Y.-X., Wen J., Lin Q.-W., Liu B. Phylogeny and divergence time estimation of the walnut family (Juglandaceae) based on nuclear RAD-Seq and chloroplast genome data. Mol. Phylogenet. Evol. 2020;147:106802. doi: 10.1016/j.ympev.2020.106802. [DOI] [PubMed] [Google Scholar]
- 30.Salas-Leiva D.E., Meerow A.W., Calonje M., Griffith M.P., Francisco-Ortega J., Nakamura K., Stevenson D.W., Lewis C.E., Namoff S. Phylogeny of the Cycads Based on Multiple Single-Copy Nuclear Genes: Congruence of Concatenated Parsimony, Likelihood and Species Tree Inference Methods. Ann. Bot. 2013;112:1263–1278. doi: 10.1093/aob/mct192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CNCB-NGDC Members and Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen M., Ma Y., Wu S., Zheng X., Kang H., Sang J., Xu X., Hao L., Li Z., Gong Z., et al. Genome Warehouse: A Public Repository Housing Genome-scale Data. Genom. Proteom. Bioinform. 2021 doi: 10.1016/j.gpb.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012;28:1838–1844. doi: 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin J.-J., Yu W.-B., Yang J.-B., Song Y., Depamphilis C.W., Yi T.-S., Li D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emms D.M., Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emms D.M., Kelly S. Orthofinder: Solving Fundamental Biases in Whole Genome Comparisons Dramatically Improves Orthogroup Inference Accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K., Asimenos G., Toh H. Multiple Alignment of DNA Sequences with Mafft. Methods Mol. Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 41.Katoh K., Standley D.M. Mafft Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. Iq-Tree 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price M.N., Dehal P.S., Arkin A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price M.N., Dehal P.S., Arkin A.P. Fasttree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen W., Le S., Li Y., Hu F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE. 2016;11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith S.A., Moore M.J., Brown J.W., Yang Y. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evol. Biol. 2015;15:150. doi: 10.1186/s12862-015-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available upon request from NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 1 March 2021) and National Genomics Data Center (https://ngdc.cncb.ac.cn/, accessed on 1 November 2021), accession number GWHBGBH00000000.