Abstract
In the 1996–1997 cholera epidemic in Guinea-Bissau, surveillance for antimicrobial resistance showed the emergence of a multidrug-resistant strain of Vibrio cholerae O1 during the course of the epidemic. The strain was resistant to ampicillin, erythromycin, tetracycline, furazolidone, aminoglycosides, trimethoprim, and sulfamethoxazole. Concomitant with the emergence of this strain, we observed a resurgence in the number of registered cholera cases as well as an increase in the case fatality rate from 1.0% before the emergence of the multiple-drug-resistant strain to 5.3% after the emergence of the strain. Our study shows that the strain contained a 150-kb conjugative multiple-antibiotic resistance plasmid with class 1 integron-borne gene cassettes encoding resistance to trimethoprim (dhfrXII) and aminoglycosides [ant(3")-1a]). The finding of transferable resistance to almost all of the antibiotics commonly used to treat cholera is of great public health concern. Studies should be carried out to determine to what extent the strain or its resistance genes have been spread to other areas where cholera is endemic.
Following its arrival on the African continent in 1970, the seventh cholera pandemic, caused by Vibrio cholerae O1 biotype El Tor, spread throughout West Africa with devastating consequences (35). However, Guinea-Bissau, situated along the West African coast north of Guinea, first reported outbreaks in 1987 (42), which were followed by an epidemic in 1994 that extended into early 1995 (8). We previously determined by phenotypic and genotypic analyses how the epidemics in 1987 and 1994–1995 were caused by different O1 strains, with the 1994–1995 epidemic strain likely to have been introduced by fishermen or travelers from Guinea (8).
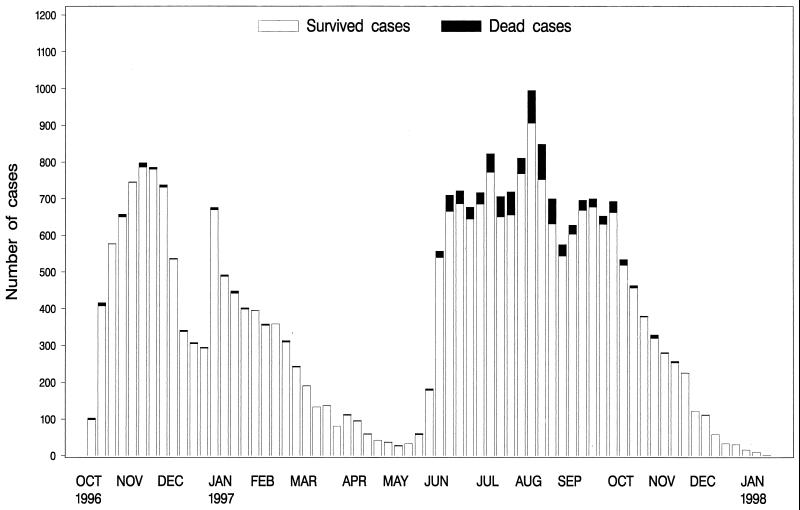
The most recent epidemic in Guinea-Bissau began in October 1996, extended throughout 1997, and included a total of 26,967 reported cases. The epidemic curve showed a bimodal pattern, with the first peak occurring in November 1996 and another peak extending over the summer of 1997 (Fig. 1). During the epidemic, monitoring of antimicrobial resistance was carried out by the National Public Health Laboratory in Bissau, Guinea-Bissau. This surveillance showed the emergence of a multidrug-resistant strain during 1997, concomitant with the second peak of the epidemic. At the same time, an increase in the case fatality rate was observed.
FIG. 1.
Number of cases of cholera and number of deaths reported during the cholera epidemic in Guinea-Bissau in 1996–1997.
Reports of toxigenic V. cholerae strains resistant to the antibiotics commonly used for treatment are appearing with increasing frequency, although the genetic mechanisms for the resistance are often not determined (2, 9, 13). Of particular interest and concern are the acquisition and transfer of resistance genes through mobile genetic elements, as such elements may spread among bacteria of different genera. Horizontal transferable plasmid-encoded resistance has been found on several occasions in V. cholerae O1 (4, 12, 13). Recently, Waldor et al. (41) showed that resistance genes among V. cholerae O1 and O139 strains in India were carried on a 62-kb self-transmissible, chromosomally integrating genetic element with properties similar to those of a conjugative transposon. A relatively new group of gene expression elements named integrons has been described as vehicles for the acquisition of antibiotic resistance genes (reviews have been provided by Hall and Stokes [16], Recchia and Hall [32], and Sundström [34]) (15, 33). So far three classes of integrons have been identified, and class 1 integrons have been found to be prevalent among clinical isolates. Class 1 elements were originally defined to be composed of two conserved segments, the 5′ conserved segment (5′-CS) and the 3′ conserved segment (3′-CS), and an internal variable region which contains gene cassettes encoding antibiotic resistance determinants (15, 33). The 5′-CS segment contains the intI1 gene, which encodes the type 1 integrase and which is responsible for site-specific insertion and excision of gene cassettes (3). Furthermore, the 5′-CS segment harbors the attI1 site, which is responsible for recombination. The 3′-CS segment contains the qacEΔ1 and sul1 genes, which encode resistance to quaternary ammonium compounds and sulfonamides, respectively. It should be noted that class 1 integrons may not always contain the entire 3′-CS segment (32, 34).
The objective of the present study was to determine the genetic mechanisms of antibiotic resistance and the genotypes of V. cholerae O1 strains isolated during the cholera outbreak in Guinea-Bissau in 1996–1997.
MATERIALS AND METHODS
Surveillance of the cholera epidemic and characterization of bacterial strains.
Information about the number of cholera cases and the number of deaths was collected and reported to the Ministry of Health in Bissau for the period from October 1996 to February 1998. Ten V. cholerae O1, biotype El Tor, serotype Ogawa strains recovered during the cholera outbreak in Guinea-Bissau in 1996–1997 were examined. Four and three O1 strains isolated during cholera outbreaks in 1987 (8) and 1994, respectively, were included for comparison purposes (V. cholerae O1 strains from the 1994 outbreak were kindly provided by Joy G. Wells and Fred Angulo, Centers for Disease Control and Prevention, Atlanta, Ga.) (Table 1). Each stool specimen was enriched in alkaline peptone-water (pH 8.6) for 6 h, after which a loopful of the surface pellicle was streaked onto thiosulfate-citrate-bile salt-sucrose agar (Difco, Detroit, Mich.). Suspected V. cholerae isolates were tested by agglutination tests with polyvalent O1-specific and monospecific Ogawa and Inaba antisera (Difco). V. cholerae isolates and appropriate controls were tested for DNA sequences encoding cholera toxin (CT) in hybridization studies with an alkaline phosphatase-labeled 23-base oligonucleotide probe (44).
TABLE 1.
V. cholerae O1 strains isolated in Guinea-Bissau during cholera outbreaks in 1987, 1994, and 1996–1997
| Strain no. | Place/date (mo and yr) of isolation | Antibiograma | Plasmid size (kb) | Plasmid RFLP typeb | Ribotypec | CT typec | Retrieval of amplicon by PCR with primers qacEΔ1-F and sul1-Bd | Size (bp) of amplicons obtained by PCR with primers in-F and in-B |
|---|---|---|---|---|---|---|---|---|
| 1407 | Bissau/Oct 1987 | COL | —e | Ia | A | −f | ||
| 1445 | Bissau/Oct 1987 | COL | — | Ia | A | − | ||
| 1452 | Bissau/Oct 1987 | COL | — | Ia | A | − | ||
| 1482 | Bissau/Oct 1987 | COL | — | Ia | A | − | ||
| F2107 | Bissau/Aug 1994 | COL, FUR, O129, STR, SUL, SXT | II | B | − | |||
| F2196 | Biombo/Dec 1994 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | F | II | B | − | |
| F2189 | —g/1994 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | E | II | B | − | |
| 9861 | Bissau/Dec 1996 | COL, FUR, O129, STR, SUL, SXT | — | II | B | − | ||
| 9862 | Bissau/Dec 1996 | AMP, COL, FUR, GEN, KANI, O129, STR, SUL, SXT | 150 | A | II | B | − | |
| 9863 | Bissau/Dec 1996 | COL, FUR, O129, STR, SUL, SXT | — | II | B | − | ||
| 9868 | Bissau/Dec 1996 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | D | II | B | + | 1,800 |
| C230 | São Domingo/June 1997 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | D | II | B | + | 1,800 |
| C231 | Ingóré/June 1997 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | D | II | B | + | 1,800 |
| C233 | Quinera/June 1997 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | D | II | B | + | 1,800 |
| C235 | —/June 1997 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | D | II | B | + | 1,800 |
| C236 | —/June 1997 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | D | II | B | + | 1,800 |
| C240 | Biombo/Nov 1997 | AMP, COL, FUR, GEN, KAN, O129, STR, SUL, SXT, TET | 150 | D | II | B | + | 1,800 |
AMP, ampicillin; COL, colistin; FUR, furazolidone; GEN, gentamicin; KAN, kanamycin; O129, vibriostatic agent (2,4-diamino-6,7-diisopropylpteridine phosphate); STR, streptomycin; SUL, sulfamethoxazole; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; superscript I, intermediate resistance.
RFLP analysis was done with the enzyme EcoRI.
The ribotype and CT genotype were established with the enzyme BglI.
The presence of class 1 integrons was determined with the PCR primers qacEΔ1-F and sul1-B.
—, no plasmids were found.
−, no amplicons were obtained by PCR.
—, location unknown.
Antibiotic susceptibility testing and transfer of resistance.
Each isolate was tested for susceptibility to 14 antibacterial agents representing different antimicrobial classes by the disk diffusion method with Mueller-Hinton agar (Difco) as described by the National Committee for Clinical Laboratory Standards (28). A nalidixic acid-resistant mutant strain of Escherichia coli K-12, strain J53-1 (lac+ pro met Nalr) (37), and a streptomycin- and rifampin-resistant mutant strain of V. cholerae O1, strain 1407, isolated during the epidemic in Guinea-Bissau in 1987 (8) were used as recipients in conjugation experiments. Following mating on nonselective L agar (Difco) incubated at 37°C for 4 to 6 h of growth, exconjugants were harvested and appropriate dilutions were streaked onto plates of Fluorocult E. coli O157:H7 agar (Merck, Virum, Denmark) supplemented with 50 μg of nalidixic acid per ml and adequate concentrations of selected drugs. Phenotypic appearance was used to differentiate possible spontaneous nalidixic acid-resistant donors from the exconjugants.
Plasmid analysis.
Plasmid extraction was carried out by the method of Kado and Liu (22), modified by incubating the cells at elevated pH (12.54) for 30 min at 56°C during the lysis step. V. cholerae O1 strain 1075/25 carrying a 150-kb plasmid was used as a positive control (36). The enzyme EcoRI was used in restriction fragment length polymorphism (RFLP) analysis. Electrophoresis and visualization of plasmids were carried out essentially as described previously (29).
Ribotyping and CT genotyping.
The genotype of V. cholerae O1 was established by ribotyping with the restriction enzyme BglI to digest chromosomal DNA (31). Ribotyping was performed by the procedure described by Dalsgaard et al. (5) with digoxigenin-labeled 16S and 23S rRNA probes. RFLP analysis of DNA sequences encoding CT genes (CT genotyping) was performed by hybridization of nylon membranes with BglI-digested DNA with a digoxigenin-labeled oligonucleotide CT probe (44). A 1-kb DNA molecular size standard (GIBCO BRL, Gaithersburg, Md.) was used as a size marker in ribotyping and CT genotyping.
PCR amplification and sequencing of class 1 integrons.
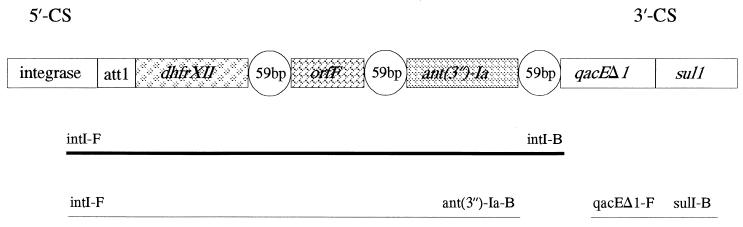
First, we selected a specific class 1 integron primer set, qacEΔ1 and sul1, directed at the 3′-CS conserved segment of class 1 integrons (30, 32). As few class 1 integrons are known to not contain the qacEΔ1 and sul1 genes, these would not be detected with the selected primers. Figure 2 demonstrates the general structure of class 1 integrons together with the amplicons sequenced and gene cassettes found. PCR was carried out as described by Dalsgaard et al. (7). DNAs from strains yielding a PCR product with the class 1 primers were subsequently amplified with the integron primers in-F and in-B (F is forward and B is backward) which amplify the region between the 5′-CS and 3′-CS conserved segments, generating products of variable sizes depending on the numbers and lengths of the inserted gene cassettes (Fig. 1 and Table 1). The in-B primer anneals at the 3′-CS conserved segment. Primers in-F and ant(3")-Ia-B (addA-B) were used to assess if the integron contained a gene cassette encoding resistance to streptomycin and spectinomycin (Fig. 2) (25). Appropriate control strains and molecular size markers were included, as described previously (7). Amplified DNA was purified before sequencing by using Microspin S-400HR columns purchased from Pharmacia Biotech (Hillerød, Denmark), and the nucleotide sequence was determined by cycle sequencing with the AmplitaqFS dye terminator kit and a Pharmacia Biotech ALF automated DNA sequencing apparatus. The identities of the sequences determined were analyzed by comparison of the sequences with the gene sequences in databases by using the BLAST software (1).
FIG. 2.
Integron structure and resistance gene cassettes found in V. cholerae O1. The PCR primers used and the amplicons obtained are shown below the integron structure, with the bold line representing the amplicon sequenced. att1 is a site which is responsible for recombination, and qacEΔ1 and sul1 encode resistance to disinfectants and sulphonamides, respectively. The individual gene cassettes are shown together with their recombination sites (59-bp element).
Statistical methods.
To adjust for regional differences in mortality, we used multivariate logistic regression analysis to compare the case fatality rates in the first and the second parts of the epidemic.
RESULTS
Epidemic surveillance data.
The epidemic in Guinea-Bissau, which started on October 6, 1996, and which ended on January 12, 1998, included a total of 26,967 reported cases (national attack rate, 24 cases per 1,000 inhabitants). The epidemic curve had a bimodal pattern (Fig. 1). During the period from October 1996 through March 1997, 10,386 of 10,574 cases (98%) were from the capital, Bissau, and the surrounding region, Biombo. During the period from April 1997 through January 1998, 11,020 (67%) of 16,393 cases were from the capital area and Biombo. Throughout the epidemic, the attack rate was highest in the capital area (68 cases per 1,000), but after April 1997, the epidemic spread to the interior regions of Cacheu, Oio, Bafatá, Quinara, and Tombali. Also, the archipelago of Bijagos was affected.
The case fatality rate increased from 1.0% (102 deaths per 10,574 cases) during the period from October 1996 through March 1997 to 5.3% (860 deaths per 16,393 cases) during the rest of the epidemic. There was a significantly higher rate of mortality among the case patients from the interior regions of Guinea-Bissau (P < 0.0001). Furthermore, the multivariate analysis showed that, independent of geography, there was an increase in the case fatality rate in the second epidemic period (adjusted odds ratio, 1.8; 95% confidence interval, 1.4 to 2.3; P < 0.0001). Among hospitalized case patients from Bissau, the case fatality rate increased from 3.2% (69 per 2,178) in the first period to 5.8% (155 per 2,680) in the second period (risk ratio, 1.9; P < 0.001).
Antibiotic susceptibility testing and plasmid analysis.
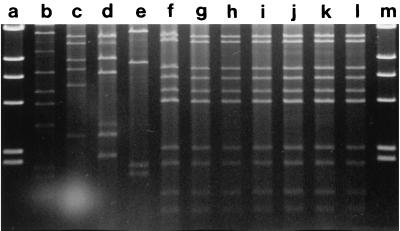
Table 1 shows the characteristics of the V. cholerae O1 strains isolated in Guinea-Bissau during the cholera outbreaks in 1987, 1994, and 1996–1997. Each of the strains hybridized with the CT probe. Seven strains, including all six strains recovered in 1997, isolated at different locations and different times during the epidemic in 1996–1997 had identical antibiograms that showed multiple-drug resistance, including resistance to furazolidone, sulphonamides, trimethoprim, and tetracycline (Table 1). In addition, each of these strains contained a 150-kb plasmid which was found to be identical in each case by RFLP analysis (profile D) with the restriction enzyme EcoRI (Fig. 3 and Table 1). Interestingly, three strains isolated when the epidemic began in December 1996 showed different resistance patterns, and one of these strains contained a 150-kb plasmid that showed a different RFLP profile (profile A). Strains isolated in 1987 were resistant to colistin only, did not contain plasmids, and showed a unique ribotype and CT genotype (8) (ribotypes are shown in Fig. 4).
FIG. 3.
RFLP analysis of the 150-kb plasmid isolated from V. cholerae O1 strains isolated during cholera outbreaks in Guinea-Bissau. The explanations of the lanes include the strain designation and RFLP type unless stated otherwise. Lanes: a, HindIII-digested phage lambda DNA (weight marker); b, reference strain 1075/25; c, F2189, type E; d, F2196, type F; e, 9862, type A; f, 9868, type D; g, C230, type D; h, C231, type D; i, C233, type D; j, C235, type D; k, C236, type D; 1, C240, type D; m, HindIII-digested phage lambda DNA (weight marker).
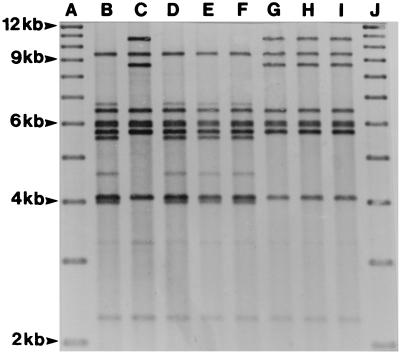
FIG. 4.
Examples of BglI ribotypes of V. cholerae O1 strains associated with cholera outbreaks in Guinea-Bissau. Unless stated otherwise, the explanations of the lanes include the strain designation, ribotype, and year of isolation. Lanes: A, 1-kb molecular weight standard; B, 1407, ribotype Ia, 1987; C, C235, type II, 1997; D, 1445, type Ia, 1992; E, 1452, type Ia, 1992; F, 1482, type Ia, 1992; G, F2107, type II, 1994; H, F2196, type II, 1994; I, F2189, type II, 1994; J, 1-kb molecular weight standard.
Ribotyping and CT genotyping.
Ribotyping showed that the 1996–1997 outbreak strains all belonged to the same type, type II, which was also shown for strains isolated in 1994 but which was clearly different from type Ia, as demonstrated with strains isolated in 1987 (Fig. 4). These findings were supported by CT genotyping, in which all strains from 1994 and 1996–1997 showed two fragments of 10.0 and 12.8 kb, whereas strains from 1987 demonstrated two fragments of 6.8 and 11.2 kb (Table 1).
Resistance transfer: PCR amplification and sequencing of class 1 integrons.
As transferable plasmid-encoded multiple-drug resistance has been reported for V. cholerae O1, strain C230 was selected as a representative strain for transfer experiments into E. coli K-12. One type of transconjugant that showed susceptibility patterns identical to that of the donor strain was found, except that the transconjugants were sensitive to colistin and furazolidone. Antibiotic susceptibility testing and plasmid analysis of the transconjugants, including RFLP analysis, revealed that the 150-kb plasmid was transferred and that the antibiotic resistance, except for resistance to colistin and furazolidone, was plasmid encoded. The 150-kb plasmid was subsequently transferred from the transconjugants back to strain 1407 and was further directly transferred from strain C230 to strain 1407. Again, the transfer of resistance was found as described above, with apparent higher frequencies in the direct transfer from V. cholerae strain C230 into strain 1407 than in transfer studies with E. coli K-12 (transfer frequencies were not calculated but were evaluated only by visual inspection of the agar plates on which mating occurred).
As the vast majority of strains containing class 1 integrons were resistant to sulphonamides, we investigated if the sulphonamide resistance was associated with the presence of class 1 integrons (30, 32). An approximately 800-bp 3′-CS PCR product was obtained from strain 9868 isolated in December 1996 and each of the six O1 strains isolated in 1997 with the primers qacEΔ1-F and sulI1-B (Fig. 2). None of the remaining strains yielded an amplicon with these primers and therefore did not seem to contain class 1 integrons (Table 1). The use of the in-F and in-B primers yielded a PCR product of 1,800 bp. DNA sequencing analysis of strain 9868 revealed that the 1,800-bp amplicon contained the dhfrXII (dfrA12) gene cassette encoding resistance to trimethoprim, an open reading frame, orfF, with an unknown function, and the gene cassette ant(3")-1a (aadA2), which confers resistance to streptomycin and spectinomycin (Fig. 2). The sequences of the dhfrXII gene cassette and orfF have been published in GenBank under accession number Z21672 (18), while the sequence for ant(3")-1a has the accession number D43625 (25). The sequences of the three cassettes were identical to those that have been published previously. PCR with in-F and ant(3")-Ia-B primers also yielded an approximately 1,800-bp amplicon and therefore confirmed the presence and location of the aminoglycoside resistance gene cassette ant(3")-Ia (Fig. 2) (25). Furthermore, PCR of the cell lysates of the transconjugants E. coli K-12 and V. cholerae O1 strain 1407 with qacEΔ1-F and sul1-B and with in-F and in-B primers yielded 800- and 1,800-bp amplicons, respectively, showing that the class 1 integron and gene cassettes are located and transferred on the 150-kb plasmid.
DISCUSSION
The 1996–1997 cholera epidemic in Guinea-Bissau, which involved a reported 26,967 cases, was caused by a V. cholerae O1 that had a genotype identical to that of the 1994–1995 epidemic strain (8) but that contained a 150-kb conjugative multiple-antibiotic resistance plasmid with class 1 integron-borne gene cassettes encoding resistance to trimethoprim (dhfrXII) and aminoglycosides [ant(3")-1a].
Studies indicate that class 1 integrons may be widespread in multiple-drug-resistant clinical isolates of bacterial enteropathogens (17, 21, 24). However, few studies have investigated their presence and importance in V. cholerae. Dalsgaard et al. (7) reported that V. cholerae O1 strains isolated in Vietnam after 1990 were resistant to sulphonamides and streptomycin and harbored chromosomally located class 1 integrons containing the aminoglycoside gene cassette ant(3")-1a. Furthermore, an extensive study of the distribution and content of class 1 integrons in different V. cholerae O serotypes strains isolated in Thailand revealed that O1 and non-O1 serotype strains mainly of clinical origin but not O139 serotypes contained antibiotic resistance genes located on class 1 integrons (6). Finally, class 1 integrons containing the aadA1 gene cassette, which encodes resistance to spectinomycin, were described among strains from a minor cholera outbreak in Albania and Italy in 1994 (10).
The 1996–1997 cholera epidemic was associated with a high case fatality rate, in particular during the second epidemic period. Cholera patients were managed according to standard guidelines, with emphasis on rehydration therapy. Antimicrobial treatment was, in principle, reserved for the patients with severe cases, but in practice, the therapeutic and prophylactic use of tetracycline was widespread. The increased mortality rate during the second period was partly due to the spread of the epidemic to the interior, where access to primary health care is more difficult and limited than in the capital area (14). However, the increase in the case fatality rate could not be attributed solely to the spread of the epidemic. Thus, an increased case fatality rate was observed among hospitalized patients from the capital and in the multivariate analysis by adjusting for regional differences. We suggest that the finding of transferable resistance to all of the antimicrobials most commonly used to treat cholera could contribute to the increased fatality rate. Although the primary objective of case management is the treatment and prevention of dehydration, it has been shown that appropriate antimicrobial therapy can reduce by half the volume of stool purged during illness, as well as shorten the duration of excretion of V. cholerae by the same amount (26). In addition, the presence of resistant strains may cause an increased risk of secondary transmission due to prolonged excretion. The widespread use of antimicrobials for any reason in the community may indeed favor the transmission of the resistant strain and may be one of the explanations for the second wave in the 1996–1997 epidemic.
In contrast to the strains from the earlier epidemics, the 1996–1997 strain was resistant not only to low-dose and high-dose tetracycline but also to other commonly used antimicrobials such as furazolidone and trimethoprim. It is likely that the multidrug-resistant outbreak strain in the 1996–1997 epidemic was derived from the same clone responsible for the 1994–1995 epidemic but had subsequently acquired the 150-kb resistance plasmid from an unknown source. The plasmid-containing strain may have come to dominate through the selective pressure caused by the prophylactic or therapeutic use of antimicrobials. This hypothesis is corroborated by the fact that some strains isolated early in the epidemic were less resistant and did not contain plasmids.
Although genes for resistance to eight antibiotics were shown to be located on the 150-kb plasmid, only a minor part of the resistance, namely, resistance to streptomycin, sulphonamides, and trimethoprim, were contained in gene cassettes of the class 1 integron. This corroborates the findings of the content of class 1 integrons in V. cholerae strains from Thailand (6) and in members of the family Enterobactericeae and Pseudomonas spp., in which gene cassettes often encode resistance to aminoglycosides, trimethoprim, and β-lactamases. The dhfrXII gene cassette product confers a level of resistance to trimethoprim which is about 10 times stronger than that conferred by dhfrI (18), the most widespread gene coding trimethoprim resistance (38, 39). It is believed that the spread of trimethoprim resistance genes among clinical isolates is mostly by the integron mechanism (20).
The cluster of three gene cassettes described in this paper has been found previously in five Escherichia coli isolates from patients with urinary tract infections and geriatric patients and in four patients with Shigella enteritis in Finland (18). The integrons were borne on a Tn21-like transposon located on a 70-kb plasmid (18), whereas our integron was located on a 150-kb self-transmissible plasmid. We did not investigate if the integron was inserted in a transposon, but this may be an explanation for the presence of the apparent identical integron on different plasmids. Although it is believed that the integrons themselves are mobile elements, this possibility remains to be verified experimentally. The presence of the same arrangement of gene cassettes in different bacterial species and on different plasmids suggests that the integron-associated mechanism of gene capture is a widespread and an important mechanism by which bacteria acquire novel resistance genes.
Surprisingly and in contrast to our earlier report of the cholera epidemic in 1994–1995, in which strains were found to be resistant to colistin, trimethoprim, and the vibriostatic agent O129, the three O1 strains isolated in 1994 showed increased antibiotic resistance, with strains F2196 and F2189 carrying a 150-kb plasmid (Table 1) (19). Two strains isolated early in the 1996–1997 epidemic did not carry plasmids, whereas another strain carried a 150-kb plasmid without class 1 integrons and was clearly different from the 1996–1997 epidemic strains, as determined by RFLP analysis. These findings suggest that several large plasmids are present in V. cholerae and that the 150-kb plasmid containing the integron may have been received by strains in the early stages of the epidemic.
Since the 1996–1997 epidemic in Guinea-Bissau, the eastern and southern parts of Africa have been hit severely by several cholera epidemics, with mortality rates as high as 20% being reported (43). Of the total of 147,425 cases of cholera reported to the World Health Organization in 1997, 80% occurred in Africa (43). Despite the tremendous negative impact on human health and economy caused by the cholera epidemics, little information has been published about the transmission patterns and characteristics of the V. cholerae O1 strains involved. Also, research suggests that V. cholerae is undergoing genetic changes at higher rates than previously thought. Manning et al. (27) described a new mega-integron genetic structure named VCR (V. cholerae repeat) which was proposed to represent a genetic hot spot in the capture of genes, thereby enabling more rapid adaptation and evolution in V. cholerae. Also, the recent evidence that the structural genes for CT and the toxin-coregulated pilus antigen are encoded by filamentous bacteriophages, which may integrate chromosomally or replicate as a plasmid following horizontal gene transfer, have provided new and intriguing information about the emergence of toxigenic V. cholerae (11, 23, 40). Thus, it is of utmost clinical importance that studies be carried out to characterize the current epidemic strains, including determination of the levels and mechanisms of antibiotic resistance and the extent which the 1996–1997 strain and its resistance genes have spread to other parts of Africa.
ACKNOWLEDGMENTS
We thank Joy G. Wells and Fred Angulo of the Centers for Disease Control and Prevention for providing the strains from the cholera epidemic in 1994. We are grateful for the technical assistance provided by Anne-Mette Petersen.
Kåre Mølbak and Anders Dalsgaard were supported by the Danish Council for Development Research (DANIDA grants 104.Dan.8/1114 and 90928, respectively).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bag P K, Maiti S, Sharma C, Ghosh A, Basu A, Mitra R, Bhattacharya S K, Nakamura S, Yamasaki S, Takeda Y, Blair B G. Rapid spread of the new clone of Vibrio cholerae O1 biotype El Tor in cholera endemic areas in India. Epidemiol Infect. 1998;121:245–251. doi: 10.1017/s0950268898001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collis C M, Grammaticopoulow G, Briton J, Stokes H W, Hall R M. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 4.Coppo A, Colombo M, Pazzani C, Bruni R, Mohamud K A, Omar K H, Mastrandrea S, Salvia A M, Rotigliano G, Maimone F. Vibrio cholerae in the horn of Africa: epidemiology, plasmids, tetracycline resistance gene amplification, and comparison between O1 and non-O1 strains. Am J Trop Med Hyg. 1995;53:351–359. doi: 10.4269/ajtmh.1995.53.351. [DOI] [PubMed] [Google Scholar]
- 5.Dalsgaard A, Echeverria P, Larsen J L, Siebeling R, Serichantalergs O, Huss H H. Application of ribotyping for differentiating Vibrio cholerae non-O1 isolates from shrimp farms in Thailand. Appl Environ Microbiol. 1995;61:245–251. doi: 10.1128/aem.61.1.245-251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalsgaard A, Forslund A, Serichantalergs O, Sandvang D. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotypes strains isolated in Thailand. Antimicrob Agents Chemother. 2000;44:1315–1321. doi: 10.1128/aac.44.5.1315-1321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalsgaard A, Forslund A, Tam N, Vinh D X, Cam P D. Cholera in Vietnam: changes in genotypes and emergence of class I integrons containing aminoglycoside resistance gene cassettes in Vibrio cholerae O1 strains isolated from 1979 to 1996. J Clin Microbiol. 1999;37:734–741. doi: 10.1128/jcm.37.3.734-741.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalsgaard A, Mortensen H F, Mølbak K, Dias F, Serichantalergs O, Echeverria P. Molecular characterization of Vibrio cholerae O1 strains isolated during cholera outbreaks in Guinea-Bissau. J Clin Microbiol. 1996;34:1189–1192. doi: 10.1128/jcm.34.5.1189-1192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubon J M, Palmer C J, Ager A L, Shor-Posner G, Baum M K. Emergence of multiple drug-resistant Vibrio cholerae O1 in San Pedro Sula, Honduras. Lancet. 1997;349:924. doi: 10.1016/s0140-6736(05)62699-2. [DOI] [PubMed] [Google Scholar]
- 10.Falbo V, Carattoli A, Tosini F, Pezzella C, Dionisi A M, Luzzi I. Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob Agents Chemother. 1999;43:693–696. doi: 10.1128/aac.43.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque S, Asadulghani M, Saha M N, Abdul Alim A R M, Albert M J, Nasirul Islam K M, Mekalanos J J. Analysis of clinical and environmental strains of nontoxogenic Vibrio cholerae for susceptibility to CTXφ: molecular basis for origination of new strains with epidemic potential. Infect Immun. 1998;66:5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finch M J, Morris J G, Jr, Kaviti J, Kagwanja W, Levine M M. Epidemiology of antimicrobial resistant cholera in Kenya and East Africa. Am J Trop Med Hyg. 1988;39:484–490. doi: 10.4269/ajtmh.1988.39.484. [DOI] [PubMed] [Google Scholar]
- 13.Glass R I, Huq I, Alim A R M A, Yunus M. Emergence of multiply antibiotic-resistant Vibrio cholerae in Bangladesh. J Infect Dis. 1980;142:939–943. doi: 10.1093/infdis/142.6.939. [DOI] [PubMed] [Google Scholar]
- 14.Gunnlaugsson G, Angulo F J, Einarsdottir J, Passa A, Tauxe R V. Epidemic cholera in Guinea-Bissau: the challenge of preventing deaths in rural West Africa. Int J Infect Dis. 2000;4:8–13. doi: 10.1016/s1201-9712(00)90059-6. [DOI] [PubMed] [Google Scholar]
- 15.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 16.Hall R M, Stokes H W. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90:115–132. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 17.Hannecart-Pokorni E, Depuydt F, Wit L D, Bossuyt E V, Content J, Vanhoof R. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Il associated with a sulI-type integron. Antimicrob Agents Chemother. 1997;41:314–318. doi: 10.1128/aac.41.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heikkila E, Skurnik M, Sundström L, Huovinen P. A novel dihydrofolate reductase cassette inserted in an integron borne on a Tn21-like element. Antimicrob Agents Chemother. 1993;37:1297–1304. doi: 10.1128/aac.37.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmberg S D, Day D E, Parker R D R, Rao N U, Harris J R, Hargrett N T, Kansou N, Blake P A. Foodborne transmission of cholera in Micronesian households. Lancet. 1984;11:325–328. doi: 10.1016/s0140-6736(84)90370-2. [DOI] [PubMed] [Google Scholar]
- 20.Huovinen P, Sundström L, Swedberg G, Sköld O. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother. 1995;39:279–289. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones M E, Peters E, Weersink A-M, Fluit A, Verhoef J. Widespread occurrence of integrons causing multiple antibiotic resistance in bacteria. Lancet. 1997;349:1742–1743. doi: 10.1016/S0140-6736(05)62954-6. [DOI] [PubMed] [Google Scholar]
- 22.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karaolis D K R, Somara S, Maneval D R J, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 24.Kazama H, Hamashima H, Sasatsu M, Arai T. Distribution of the antiseptic resistance genes qacE and qacE1 in gram-negative bacteria. FEMS Microbiol Lett. 1998;159:173–178. doi: 10.1111/j.1574-6968.1998.tb12857.x. [DOI] [PubMed] [Google Scholar]
- 25.Kazama H, Kizu K, Iwasaki M, Hamashima H, Sasatsu M, Arai T. Isolation and structure of a new integron that includes a streptomycin resistance gene from the R plasmid of Pseudomonas aeroginosa. FEMS Microbiol Lett. 1995;134:137–141. doi: 10.1111/j.1574-6968.1995.tb07927.x. [DOI] [PubMed] [Google Scholar]
- 26.Mahalanabis D, Molla A M, Sack D A. Clinical management of cholera. In: Barua D, Greenough W B I, editors. Cholera. New York, N.Y: Plenum Medical Book Company; 1992. pp. 253–283. [Google Scholar]
- 27.Manning P A, Clark C A, Focareta T. Gene capture in Vibrio cholerae. Trends Microbiol. 1999;7:93–95. doi: 10.1016/s0966-842x(99)01464-x. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S8. Vol. 18. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 29.Olsen J E, Larsen J L. Restriction fragment length polymorphism of the Vibrio anguillarum serovar O1 virulence plasmid. Appl Environ Microbiol. 1990;56:3130–3132. doi: 10.1128/aem.56.10.3130-3132.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paulsen I T, Littlejoh T G, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popovic T, Bopp C A, Olsvik Ö, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 33.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 34.Sundström L. The potential of integrons and connected programmed rearrangements for mediating horizontal gene transfer. APMIS. 1998;84:37–42. doi: 10.1111/j.1600-0463.1998.tb05646.x. [DOI] [PubMed] [Google Scholar]
- 35.Swerdlow D L, Isaäcson M. The epidemiology of cholera in Africa. In: Wachsmuth I K, Blake P A, Olsvik Ö, editors. Vibrio cholerae and cholera. Washington, D.C.: ASM Press; 1994. pp. 297–307. [Google Scholar]
- 36.Tabtieng R, Wattanasri S, Echeverria P, Seriwatana J, Bodhidatta L, Chatkaeomorakot A, Rowe B. An epidemic of V. cholerae El Tor Inaba resistant to several antibiotics with a conjugative group C plasmid coding for type II dihydrofolate reductase in Thailand. Am J Trop Med Hyg. 1989;41:680–686. doi: 10.4269/ajtmh.1989.41.680. [DOI] [PubMed] [Google Scholar]
- 37.Taylor D E, Levine J G, Bradley D E. In vivo formation of a plasmid cointegrate expressing two incompatibility phenotypes. Plasmid. 1981;5:233–244. doi: 10.1016/0147-619x(81)90001-9. [DOI] [PubMed] [Google Scholar]
- 38.Towner K J, Brennan A, Zhang Y, Holtham C A, Brough J L, Carter G I. Genetic structures associated with spread of the type Ia trimethoprim-resistant dihydrofolate reductase gene amongst Escherichia coli strains isolated in the Nottingham area of the United Kingdom. J Antimicrob Chemother. 1994;33:25–32. doi: 10.1093/jac/33.1.25. [DOI] [PubMed] [Google Scholar]
- 39.Tsakris A, Johnson A P, Legakis N J, Tzouvelekis L S. Prevalence of the type I and type II DHFR genes in trimethoprim-resistant urinary isolates of Escherichia coli from Greece. J Antimicrob Chemother. 1993;31:665–671. doi: 10.1093/jac/31.5.665. [DOI] [PubMed] [Google Scholar]
- 40.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 41.Waldor M K, Tschäpe H, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. Cholera in 1986. Weekly Epidemiol Rec. 1987;62:141–142. [Google Scholar]
- 43.World Health Organization. Cholera in 1997. Weekly Epidemiol Rec. 1998;73:201–208. [PubMed] [Google Scholar]
- 44.Wright A C, Guo Y, Johnson J A, Nataro J P, Morris J G., Jr Development and testing of a non-radioactive DNA oligonucleotide probe that is specific for Vibrio cholerae cholera toxin. J Clin Microbiol. 1992;30:2302–2306. doi: 10.1128/jcm.30.9.2302-2306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]