Abstract
Despite the global investment in One Health disease surveillance, it remains difficult and costly to identify and monitor the wildlife reservoirs of novel zoonotic viruses. Statistical models can guide sampling target prioritisation, but the predictions from any given model might be highly uncertain; moreover, systematic model validation is rare, and the drivers of model performance are consequently under-documented. Here, we use the bat hosts of betacoronaviruses as a case study for the data-driven process of comparing and validating predictive models of probable reservoir hosts. In early 2020, we generated an ensemble of eight statistical models that predicted host–virus associations and developed priority sampling recommendations for potential bat reservoirs of betacoronaviruses and bridge hosts for SARS-CoV-2. During a time frame of more than a year, we tracked the discovery of 47 new bat hosts of betacoronaviruses, validated the initial predictions, and dynamically updated our analytical pipeline. We found that ecological trait-based models performed well at predicting these novel hosts, whereas network methods consistently performed approximately as well or worse than expected at random. These findings illustrate the importance of ensemble modelling as a buffer against mixed-model quality and highlight the value of including host ecology in predictive models. Our revised models showed an improved performance compared with the initial ensemble, and predicted more than 400 bat species globally that could be undetected betacoronavirus hosts. We show, through systematic validation, that machine learning models can help to optimise wildlife sampling for undiscovered viruses and illustrates how such approaches are best implemented through a dynamic process of prediction, data collection, validation, and updating.
Introduction
Identifying the probable reservoirs of zoonotic pathogens is challenging.1 Sampling wildlife for the presence of an active or previous infection (ie, by testing for seropositivity) represents the first stage of a pipeline for the proper inference of a host species,2 but sampling is often limited on a phylogenetic, temporal, and spatial scale by logistical constraints.3 Given such restrictions, statistical models can play a crucial role by helping to identify which pathogen surveillance targets are a priority, by narrowing the set of plausible sampling targets by either ruling out clades of low-likelihood hosts4, 5 or predicting clades at a high risk of being hosts.6 For example, machine learning approaches have generated candidate lists of probable, but unsampled, primate reservoirs of Zika virus, bat reservoirs of filoviruses, and avian reservoirs of Borrelia burgdorferi.7, 8, 9
At the same time, host predictions are rarely validated empirically.10 Occasional case studies suggest both success and failures. For example, models predicted Eonycteris spelaea as an undetected bat host of filoviruses,7 which was later confirmed by field sampling in southeast Asia.11, 12 Similarly, models of mosquito–Zika virus interactions predicted Culex quinquefasciatus as a probable vector,13 which was rapidly validated by experimental competence trials.14, 15 A 2019 model of Nipah virus in India also predicted several bat species as undetected hosts.2 However, experimental infection of the predicted Rousettus aegyptiacus showed that this species could not support virus replication.16 Further, Nipah virus was found circulating in Pipistrellus pipistrellus in 2021, a species with a low predicted probability of being a host.17 More generally, predictions from most models are either untested or opportunistically validated, allowing for little insight into which approaches have greatest predictive accuracy. Systematically validating predictions would provide crucial insight into the broader usefulness (or inefficacy) of different models in zoonosis research. Moreover, these modelling approaches are generally developed in isolation; the implementation of multiple modelling approaches collaboratively and simultaneously, as part of a model-to-validation workflow, could reduce redundancy and apparent disagreement at the earliest stages of pathogen tracing at the same time as advancing predictive analytics by addressing inter-model reliability.
Coronaviruses are an ideal family of viruses with which to compare and validate predictive models of probable zoonotic reservoirs. Coronaviruses are positive-sense, single-stranded RNA viruses that have been detected in both mammals and birds.18 They have a broad host range, a high mutation rate, and the largest genomes of any RNA virus; but they have also evolved mechanisms for RNA proofreading and repair to mitigate the deleterious effects of a high recombination rate acting over a large genome.19 Consequently, coronaviruses fit the profile of viruses with a high potential for being a zoonotic disease. There are eight human coronaviruses (three in the genera alphacoronavirus and five in the genera betacoronavirus), of which three are highly pathogenic in humans: SARS-CoV, MERS-CoV, and SARS-CoV-2. These viruses are zoonotic and widely agreed to have evolutionary origins in bats.20, 21, 22, 23
The challenges caused by both SARS-CoV and MERS-CoV illustrate the difficulty of tracing the specific animal hosts of emerging viruses. During the 2002–03 severe acute respiratory syndrome (SARS) epidemic, SARS-CoV was traced to the masked palm civet (Paguma larvata),24 but the ultimate origin was unknown for several years. Horseshoe bats (family Rhinolophidae, genus Rhinolophus) were implicated as reservoir hosts in 2005, but their SARS-like coronaviruses were not identical to circulating human strains.21 Stronger data from 2017 placed the most likely evolutionary origin of SARS-CoV in Rhinolophus ferrumequinum or Rhinolophus sinicus.25 There is even less certainty about the origins of MERS-CoV, although spillover to humans often occurs through contact with dromedary camels (Camelus dromedarius). A virus with 100% nucleotide identity in an approximately 200 base pair region of the MERS-CoV polymerase gene was detected in Taphozous perforatus (family Emballonuridae) in Saudi Arabia;26 however, based on spike gene similarity, other sources treat the HKU4 virus from Tylonycteris pachypus (family Vespertilionidae) in China as the most closely related bat virus to MERS-CoV.27, 28 Several bat coronaviruses have shown close phylogenetic relationships with MERS-CoV, with a surprisingly broad geographical distribution from Mexico to China.29, 30, 31, 32
COVID-19 is caused by SARS-CoV-2, a novel virus with presumed evolutionary origins in bats. Although the earliest cases were linked to a wildlife market,23 contact tracing was low, and there has been no definitive identification of the wildlife contact that resulted in the spillover nor a true so-called index case. The divergence time between SARS-CoV-2 and two of the closest related bat viruses (RaTG13 from Rhinolophus affinis and RmYN02 from Rhinolophus malayanus) has been estimated to be 40–50 years,33 suggesting that the main host(s) involved in the spillover are unknown. A viral recombination in pangolins has been suggested but is unconfirmed.33 In 2020, SARS-like betacoronaviruses were isolated from Sunda pangolins (Manis javanica) traded in wildlife markets,34, 35 and these viruses have a high amino acid identity to SARS-CoV-2, but only show an approximately 90% similarity in nucleotide identity with SARS-CoV-2 or bat coronavirus RaTG13.36 None of these host species are universally accepted as the origin of SARS-CoV-2 nor are any of the viruses a clear SARS-CoV-2 progenitor, and a better fit wildlife reservoir could still be identified. However, substantial gaps in betacoronavirus sampling across wildlife reduce how much actionable inference can be made about plausible reservoir hosts and bridge hosts for SARS-CoV-2.37
Building a predictive ensemble
Here, we use betacoronaviruses in bats as a case study for the data-driven process of comparing and validating predictive models of probable reservoir hosts, with the aim of helping to identify which targets to prioritise for surveillance for known and future zoonotic viruses. We focused on betacoronaviruses rather than SARS-like coronaviruses (subgenus Sarbecovirus) specifically, because SARS-like coronaviruses are only characterised from a small number of bat species in publicly available data. This sparsity makes current modelling methods poorly suited to more precisely infer potential reservoir hosts of Sarbecoviruses specifically. Instead, we used predictive models to firstly identify bats (and other mammals) that might broadly host any betacoronavirus, and secondly to identify species with a high viral sharing probability with the two Rhinolophus species carrying the earliest known close viral relatives of SARS-CoV-2. In mid-2020, in the early stages of the COVID-19 pandemic, we developed a standardised dataset of mammal–virus associations by integrating a previously published edge list38 with a targeted scrape of all GenBank accessions for Coronaviridae and their associated hosts. Our final dataset spanned 710 host species and 359 virus genera, including 107 mammal hosts of betacoronaviruses as well as hundreds of other (non-coronavirus) association records. We integrated our host–virus data with a mammal phylogenetic supertree39 and more than 60 standardised ecological traits of bat species.7, 40, 41
We then used these data to generate an ensemble of predictive models and drew on two popular approaches, network-based and trait-based approaches, as well as a hybrid approach, to identify the candidate bat reservoir hosts of betacoronaviruses (table ). Network-based methods estimate a full set of true unobserved host–virus interactions on the basis of a recorded network of associations (here, pairs of host species and associated viral genera). These methods are increasingly popular to identify latent processes structuring ecological networks,42, 43, 44 but they are often confounded by sampling bias and often can only make predictions for species within the observed network (ie, those that have available virus data; in-sample prediction). In contrast, trait-based methods use observed relationships concerning host traits to identify species that fit the morphological, ecological, or phylogenetic profile of known host species of a given pathogen, and rank the suitability of unknown hosts on the basis of these trait characteristics.8, 45 These methods might be more likely to recapitulate patterns in observed host–pathogen association data (eg, geographical biases in sampling and phylogenetic similarity in host morphology), but they more easily correct for sampling bias and can predict host species without known viral associations (ie, out-of-sample prediction).
Table.
Scope and calibration of different predictive modelling approaches
| Prediction on hosts without known associations (out of sample) | Predictive extent and use of pseudoabsences | |
|---|---|---|
| Network 1: k-nearest neighbours | No | Only predicts link probabilities among species in the association data |
| Network 2: linear filter | No | Only predicts link probabilities among species in the association data |
| Network 3: plug and play | No | Uses pseudoabsences to predict over all mammals in association data, using latent approach |
| Network 4: scaled phylogeny | No | Only predicts link probabilities among species in the association data |
| Trait 1: boosted regression trees | Yes | Uses pseudoabsences for all bats in trait data to predict over all species, including those without known associations |
| Trait 2: Bayesian additive regression trees | Yes | Uses pseudoabsences for all bats in trait data to predict over all species, including those without known associations |
| Trait 3: neutral phylogeographic | Yes | Trains on a broader network, and predicts sharing probabilities among any mammals in phylogeny and International Union for Conservation of Nature range map data |
| Hybrid 1: two-step kernel ridge regression | Yes | Uses pseudoabsences for all bats in trait data to predict over all species, including those without known associations |
Some methods use pseudoabsences to expand the scale of prediction but still only analyse existing host–virus data, with no out-of-sample inference, whereas other methods can predict onto new data.
In total, we implemented eight different predictive models of host–virus associations, including four network-based approaches, three trait-based approaches, and one hybrid approach using trait and phylogenetic information to make network predictions. These efforts generated eight ranked lists of suspected bat hosts of betacoronaviruses. Each ranked list was then scaled proportionally and consolidated in an ensemble of recommendations for betacoronavirus sampling and broader ecological–evolutionary research. Next, approximately 1 year after our initial model ensemble, we reran our entire analytical pipeline with new bat betacoronavirus detections, taking advantage of the new proliferation of published research on bat coronaviruses. This process provided an unprecedented opportunity to rapidly compare model performance, provide up-to-date predictions of probable but unsampled bat hosts, and assess model accuracy in the context of ongoing sampling for bat coronaviruses.
Predicted bat reservoirs of betacoronaviruses
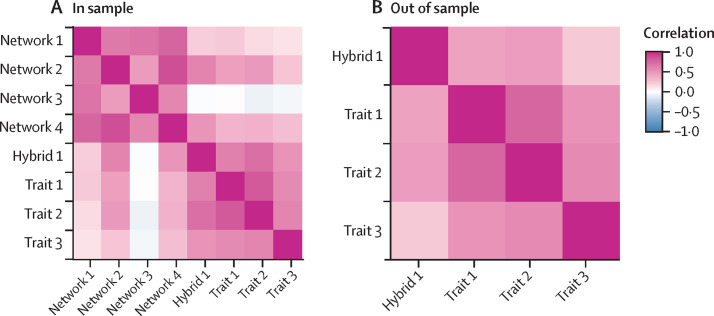
Our initial ensemble found a wide variation in model performance; individual models explained 0–69% of the variance in betacoronavirus positivity (with a mean of 25%), whereas the ensemble generally had improved predictive capacity (R 2=42%; appendix p 22). The predictions of bat betacoronavirus hosts derived from network-based and trait-based modelling approaches displayed strong inter-model agreement within each group but largely differed between groups (as measured by pairwise Spearman's rank correlations among predicted ranks; figure 1 ). Of the 1037 included bat species not known to be infected by betacoronaviruses during our initial analysis in 2020 (against 79 bat species known to be infected), our models identified between 7 and 723 potential hosts on the basis of a 10% omission threshold (90% sensitivity). Applying this same threshold to our ensemble predictions, our initial models identified 371 bat species as probable undetected hosts. Notably, only 48 suspect hosts were identified in sample, whereas we identified 323 suspect hosts out of sample, highlighting that most undiscovered hosts—and, in turn, undiscovered betacoronaviruses—should be in unsampled bat species.
Figure 1.
Agreement across an ensemble of predictive modelling approaches
Agreement across models identifying hosts with available virus data (in sample) (A) and without known viral associations (out of sample) (B). The pairwise Spearman's rank correlations between models' ranked species-level predictions were generally substantial and positive. Models were arranged in decreasing order of their mean correlation with other models. Models that used trait data made more similar predictions to each other than approaches using network methods with the same data. Network-based models that used some ecological data made more similar predictions than all other models (eg, network 4, which uses phylogeny, and hybrid 1, which uses both phylogeny and trait data). All models that could make out-of-sample predictions used trait data and showed strong agreement.
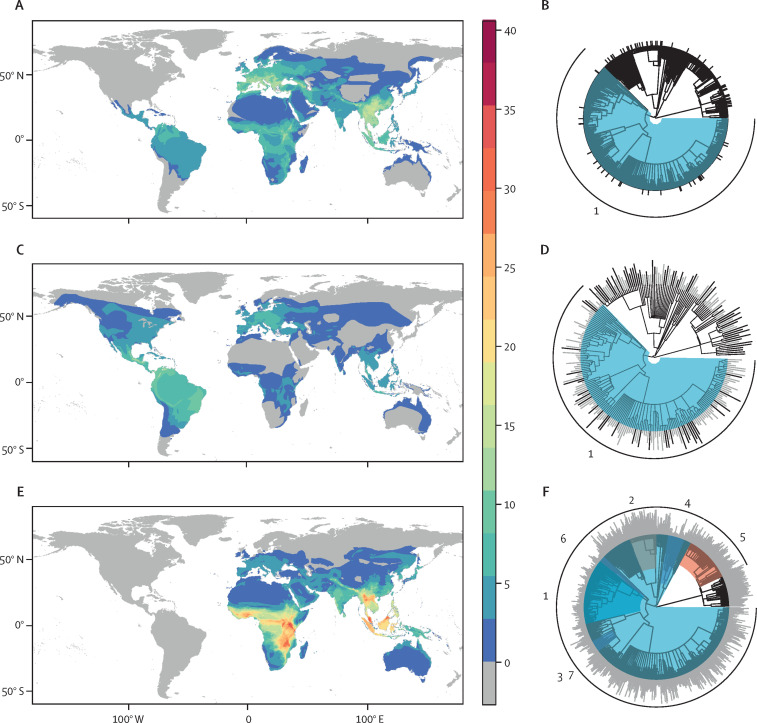
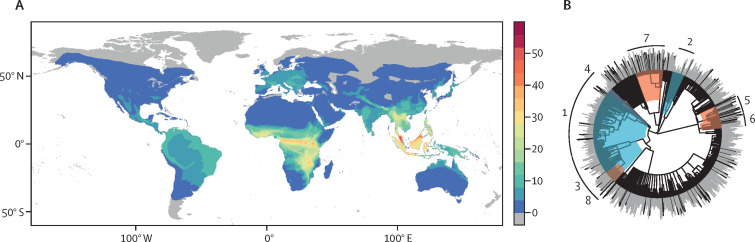
This multi-model ensemble predicted undiscovered betacoronavirus bat hosts with notable geographical and taxonomic patterning (figure 2 ). In-sample predicted hosts were globally distributed and recapitulated geographical patterns of known bat betacoronavirus hosts in Europe, the Neotropics, and southeast Asia; however, our models also predicted a high richness of probable bat reservoirs in North America. Applying a graph partitioning algorithm (phylogenetic factorisation) to the bat phylogeny,46 we similarly found that both betacoronavirus positivity and in-sample predictions were, on average, lowest for the superfamilies Noctilionoidea and Vespertilionoidea in the suborder Yangochiroptera. This finding makes intuitive sense, because these taxa do not include the groups known to harbour most of the betacoronaviruses detected in bats (eg, Rhinolophus and Hipposideridae). In contrast, our out-of-sample predicted hosts were more notably clustered in much of sub-Saharan Africa and southeast Asia (eg, Vietnam, Myanmar, and southern China), with no representation in the western hemisphere. Likewise, out-of-sample predictions were lower in Neotropical bat families (eg, Noctilionidae, Mormoopidae, and Phyllostomidae), most emballonurids, and primarily Neotropical molossids; whereas the Rhinolophus genus and most of the Old World subfamily Pteropodinae were predicted to be more likely to host betacoronaviruses (appendix p 18).
Figure 2.
Initial ensemble predictions of the geographical and evolutionary distribution of known and predicted bat hosts of betacoronaviruses
Known hosts of betacoronaviruses (A,B) are found worldwide, but particularly in southern Asia and southern Europe. Taxonomically, betacoronaviruses are less common in two superfamilies of the suborder Yangochiroptera, Noctilionoidea, and Vespertilionoidea (clade 1). The predicted in-sample bat hosts (ie, those with any viral association records; C,D) tend to recapitulate observed geographical patterns of known hosts but with a higher concentration in the Neotropics. Similarly, taxonomic patterns reflect those of known betacoronavirus hosts. In contrast, the out-of-sample bat host predictions based on phylogeny and ecological traits (E,F) are mostly clustered in Myanmar, Vietnam, and southern China, with none in the Neotropics, and North America. Predicted hosts are likewise more common in the Rhinolophidae (clade 2) and subfamilies of Old World bats (clade 5) and are rare in many Neotropical taxa (clades 1 and 7) and emballanurids (clades 3 and 4). In the phylogenies, bar height indicates betacoronavirus positivity (B) or predicted rank (D,F; higher values indicate lower proportional ranks). Colours indicate likelihood of clades to contain hosts identified through phylogenetic factorisation (red indicates clades more likely to contain hosts, blue indicates less likely hosts; appendix).
Because only trait-based models were capable of out-of-sample prediction, the differences in geographical and taxonomic patterns of our predictions probably reflect distinctions between the network-based and trait-based modelling approaches. We suggest that these should be considered as qualitatively different lines of evidence. Network approaches proportionally upweight species with a high observed viral diversity, recapitulating sampling biases largely unrelated to coronaviruses (eg, frequent screening for rabies lyssaviruses in the common vampire bat Desmodus rotundus, which has been sampled only a few times for coronaviruses31, 47, 48, 49). Highly ranked species might also have been previously sampled without evidence of a betacoronavirus presence; for example, Rhinolophus luctus from China and Macroglossus sobrinus from Thailand tested negative for betacoronaviruses, but the detection probability was limited by small sample sizes.50, 51, 52 In contrast, trait-based approaches are constrained by their reliance on phylogeny, ecological traits, and geographical covariates, all of which made the models more likely to recapitulate existing spatial (ie, clustering in southeast Asia) and taxonomic (ie, the Rhinolophus genus) patterns. However, out-of-sample predictions are, by definition, inclusive of unsampled bat hosts,53 which potentially offer a greater return on viral discovery investment.
Model validation
After this initial 2020 model ensemble, we used broad literature searches to systematically track betacoronavirus-positive bat species that were missed in our initial data compilation (eg, coronavirus sequences that were not annotated to genus on GenBank).52, 54 These searches also tracked the exponential increase in data on bat coronaviruses stemming from the emergence of SARS-CoV-2 that were published after our first model ensemble. This informal non-systematic Review used a combination of a Web of Science and PubMed searches (on Sept 24, 2020) and an ongoing Google Scholar search to update these results (from May 24, 2020, to Sept 30, 2021), including papers in English, with keywords such as “bat” and “betacoronavirus”; all specific search terms and species identified are given in the appendix (p 14). A year after our initial data compilation (in June, 2021), we also reran our initial scrape of GenBank to identify new betacoronavirus-positive bats, limiting our search to matches to betacoronavirus (taxid: 694002) and the order Chiroptera (taxid: 9397); however, this did not recover any additional host species positive for betacoronavirus not already recorded as positives in our updated data. We also mined publicly available metagenomic and transcriptomic datasets for evidence of a betacoronavirus infection.55, 56, 57 However, no published libraries contained evidence of betacoronaviruses (appendix p 15). Lastly, we analysed the wildlife testing data from the US Agency for International Development Emerging Pandemic Threats PREDICT programme, collected from 2009 to 2019, which was publicly released in June, 2021, and includes many betacoronaviruses that were discovered during the programme's run but have only published and identified down to the genus level in the full release.
In total, we uncovered 47 novel bat hosts of betacoronaviruses that were either absent from our original dataset or newly discovered after our initial analyses. This data update resulted in a total of 126 known bat species that host betacoronaviruses, and we continue to collate these records in a public online database. Of these 47 new hosts, the original ensemble correctly predicted only 36 (77% success rate), but some sub-models performed significantly better than others; for instance, three models (trait 3, network 1, and network 4) all correctly identified 100% of novel hosts in their predictive sample. The high performance of all these models, and their high performance on the training data (appendix p 22), suggest that both approaches contributed usefully to the initial ensemble.
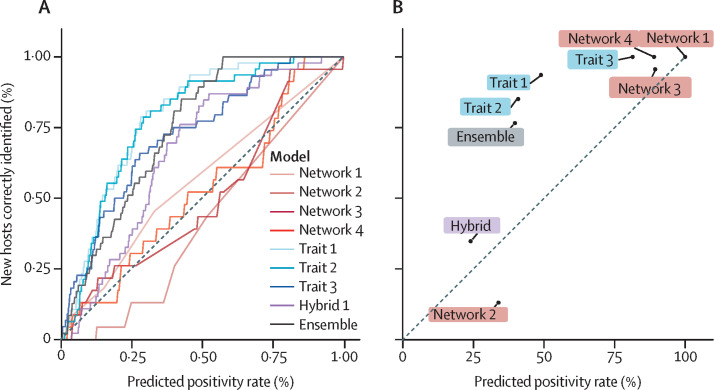
The 47 newly discovered hosts also enabled us to develop a new kind of performance metric for machine learning tasks with presence-only validation data (ie, new positives can be collected, whereas negatives are substantially more difficult to prove). If a model makes predictions at random, the predicted prevalence of positives in the training data should be roughly the same as the success rate with novel test data. For example, a so-called coin toss model will estimate that approximately 50% of species are betacoronavirus hosts and would likewise successfully identify approximately 50% of newly discovered hosts. A high-performing model, however, will identify a higher proportion of newly discovered hosts than expected at random. To evaluate how models perform in this regard, we developed a new diagnostic called the training prevalence-test sensitivity curve (TPTSC) that can be applied to modelling problems where the training data are composed of a mix of true positives, true negatives, and false negatives, but test data only include novel true positives (figure 3 ). The TPTSC plots the assumed prevalence in the training data against the sensitivity in the test data at every possible threshold from 0 to 1; these curves can be treated similarly to receiver–operator or precision-recall curves, where a higher area under the curve (AUC) indicates a better-than-random performance. Using the AUC-TPTSC scores, we found that trait-based and hybrid models consistently performed well (trait 1, AUC-TPTSC 0·79; trait 2, 0·78; trait 3, 0·73; hybrid 1, 0·67), whereas network methods performed at random or worse (network 1, 0·56; network 2, 0·42; network 3, 0·50; network 4, 0·52). Accordingly, the ensemble model performed similarly to the trait-based models (0·75).
Figure 3.
Measuring model performance with novel data
Performance is based on the comparison of total predicted prevalence (ie, what proportion of species are predicted hosts of betacoronaviruses) with the sensitivity measured from validation data (ie, how many of the 47 new species are correctly identified). The null expectation for a model with a random performance is that these should be equivalent, whereas a model with strong performance will be more than that null expectation (grey line). (A) The training prevalence–test sensitivity curve is a novel diagnostic that is conceptually similar to the receiver–operator curve, in that the model is evaluated at each possible scaled rank threshold between 0 and 1. (B) The same analysis as shown in (A), but only showing the point estimate of positivity created by each model's internally calibrated threshold. For model-guided sampling, the best model would be one that predicts a low-to-medium positivity rate and has a disproportionately high sensitivity (ie, in the upper left corner). Both (A) and (B) show that the trait-based models (including the hybrid model) perform well, whereas the network-only models perform roughly at-random or worse than random (ie, close to the line); the ensemble model, which includes all eight, performs similarly to the two best trait models and better than six of the eight component models.
These results have two key implications for future efforts in target sampling for putative reservoir hosts. First, ensemble modelling can be useful as a buffer against a variation in model quality, particularly in settings when the underlying drivers of model performance have yet to be identified. Second, and perhaps more importantly, models have been unable to have better-than-random performance without trait data that characterised bat ecology, even when they included phylogenetic data (eg, network 4). Part of this difference might also be attributable to the different scope of prediction: the response variable of trait-based models is betacoronavirus presence, whereas betacoronavirus-relevant predictions were extracted from a broader set of predictions made by the network models. However, this is contraindicated by the results of hybrid 1, which performed similarly to the other trait-based models. Therefore, we conclude that making meaningful predictions about probable zoonotic reservoirs is best accomplished by incorporating detailed information on the host ecology. The substantially greater performance of trait-based models compared with network-based models provides another compelling reason, in addition to other One Health and conservation rationales, to better understand the fundamental ecology and evolution of bats.
Dynamic prediction
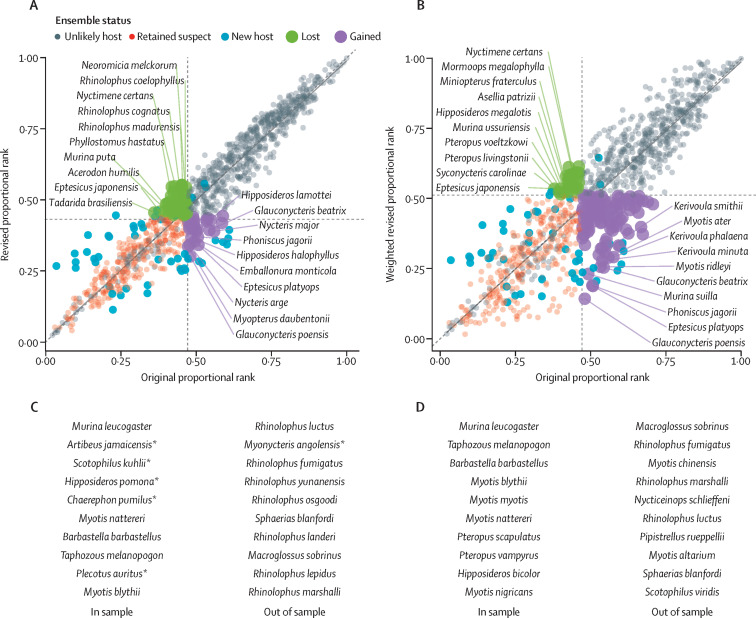
Inclusion of these 47 novel bat hosts substantially improved the performance of our predictive models. When revised with new data, our eight individual models explained 7–77% (mean, 33%) of the variance in betacoronavirus positivity, with the ensemble R 2 increasing to 62% (appendix p 23). Using our previously applied 90% sensitivity threshold, our revised ensemble identified a narrower set of 318 bat species as probable undetected hosts of betacoronaviruses. Predictions from the initial and revised ensembles were strongly correlated (ρ=0·97). However, after dynamically updating our models, our revised ensemble lost 46 suspected reservoirs and gained 29 new suspected reservoirs (figure 4A ). The predicted reservoir species that were lost from the initial ensemble were dominated by members of the family Vespertillionidae, whereas new suspect hosts were gained in the family Vespertillionidae, Hipposideridae, and Molossidae.
Figure 4.
Comparing bat betacoronavirus host prediction with dynamic model updates
Scatterplots show bat species predictions from our original ensemble in 2020 against the revised predictions after updating models with 47 new hosts (A), and the final predictions from the weighted revised ensemble (B). Species are coloured by their status in the respective revised ensemble: unlikely host, a retained suspected host, a new betacoronavirus-positive host (new host), lost as a suspected host (lost), or a novel suspected host (gained). Trendlines show a linear regression fit between the original and revised predictions against a 1:1 line, whereas dashed lines display the threshold cutoffs from each ensemble. The top ten in-sample and out-of-sample predictions from the original (C) and final (D) ensemble are also listed. *Five of the original top ten in-sample predictions, and one of the top ten out-of-sample predictions, have been empirically confirmed since the first iteration of our study.
Using the 47 newly discovered hosts, we were also able to tailor the updated ensemble responsively to model performance. To do so, we weighted the rank averaging across models based on their AUC-TPTSC score relative to the lowest performing model (network 2). In doing so, we effectively dropped network 2 from the ensemble, a choice supported by the fact the model's predictions were substantially poorer than expected at random. In the original ensemble, this correction would have created a marginal improvement in the model performance (unweighted ensemble: AUC-TPTSC, 0·746; weighted ensemble: AUC-TPTSC, 0·783). Therefore, we applied this weighting to the ensemble of updated predictions in the final copy released with this study.
This weighted, revised ensemble identified 412 suspect bat hosts, substantially expanding the scope of plausible candidates for future virus discovery compared with the two previous unweighted ensembles (figure 4B). Predictions from this final ensemble iteration were slightly less correlated with those from our initial ensemble (ρ=0·92) than those in the unweighted revised ensemble, and these final predictions retained most of the suspected hosts from the original ensemble. The top-ranked undiscovered hosts retained between our model updates included Murina leucogaster, Myotis nattereri, M blythii, Barbastella barbastellus, and Taphozous melanopogon in-sample, and the top out-of-sample hosts consistent between ensembles included Macroglossus sobrinus, Rhinolophus fumigatus, R marshalli, and Sphaerias blanfordi (figure 4C). Only 30 predicted hosts were lost, most of which were from the Pteropodidae and Vespertilionidae families, and the Rhinolophus genus. Of the 107 additional predicted hosts added to our final ensemble, most of these bat species were observed in the families Vespertilionidae (primarily the genus Myotis), Pteropodidae (primarily the genus Pteropus), Molossidae (primarily the genus Mops), and Hipposideridae (all in the genus Hipposideros), although we also identified several new predicted betacoronavirus hosts in the families Nycteridae, Emballonuridae, Rhinolophidae, and Phyllostomidae. The top-ranked novel predicted in-sample hosts included Myotis myotis, Molossus nigricans, Hipposideros bicolor, Pteropus scapulatus, and P vampyrus, and the most likely new out-of-sample hosts included Myotis chinensis, M altarium, Nycticeinops schlieffeni, Pipistrellus rueppellii, and Scotophilus viridis (figure 4D).
For Rhinolophus bats specifically, our final ensemble identified 44 suspected hosts relative to 21 known hosts, suggesting that more than two thirds of potential reservoirs in this genus could still be unidentified. Given the known roles of Rhinolophus bats as hosts of SARS-like coronaviruses,18, 21, 50 it is notable that our results suggest that the diversity of these viruses could be undescribed from approximately three quarters of Rhinolophus species not currently known to be hosts.
As in our initial ensemble, we lastly evaluated the geographical and taxonomic patterns in this finalised set of predicted betacoronavirus hosts. Spatially, undiscovered bat hosts were globally distributed (in not only the eastern but also western hemisphere), especially concentrated within a narrower band of equatorial sub-Saharan Africa, and more starkly in Malaysia and Borneo (figure 5A ). Notably, the geography of these predicted hosts contrasted with the distributions of both known bat hosts and probable hosts from our initial ensemble, each of which instead showed a stronger hotspot in southern China. We also identified distinct clades of bats highly predicted to be hosts by the weighted revised ensemble (figure 5B; appendix p 19). Both the Rhinolophus genus and subclades of the Pteropodidae family again had greater concentrations of predicted betacoronavirus hosts, although the phylogenetic factorisation now identified the Old World Molossidae family (ie, genus Mops and Chaerophon) as particularly likely to host these viruses, even though the Molossidae family as a whole had lower mean probabilities of having betacoronavirus hosts.
Figure 5.
Updated ensemble model predictions of geographical and evolutionary hotspots of bat betacoronavirus hosts
(A) Geographical map of the weighted revised ensemble predictions. Most predicted undiscovered betacoronavirus hosts were found in sub-Saharan Africa and southeast Asia, especially in Malaysia and Borneo (and less so in the high-elevation mainland hotspots where most reservoirs of severe acute respiratory syndrome coronavirus-like viruses are found). (B) Phylogeny of the weighted revised ensemble predictions. Predicted hosts from this final ensemble were also most likely in the Rhinolophus genus (clade 7), several subclades of the Pteropodidae (clades 5 and 6), and the Old World Molossidae (clade 8), even though the Molossidae family as a whole had less likely hosts (clade 3). Bar height in the phylogeny indicates predicted rank, and colours indicate clades identified through phylogenetic factorisation (red indicates clades more likely to contain hosts, blue indicates clades less likely to contain hosts; appendix p 19).
These geographical hotspots and clade-specific patterns of predictions could be particularly applicable for guiding future viral discovery and surveillance. On the one hand, betacoronavirus sampling in southeast Asian bat taxa (especially the genus Rhinolophus) might have a high success of viral detection (and isolation) of sarbecoviruses specifically, but might not substantially improve existing bat sampling gaps.5 On the other hand, the discovery of novel betacoronaviruses in pteropodid clades, Old World Molossidae, and bats in the Neotropics could substantially revise our understanding of the bat–virus association network relative to the coevolutionary distribution of bat betacoronaviruses.38 For example, predicted bat hosts in the Neotropics might be unlikely reservoirs of sarbecoviruses (given their known distribution in the eastern hemisphere) but would be expected to carry novel viruses from the subgenus merbecovirus. Such discoveries could be particularly important for global health security, given the surprising identification of MERS-like viruses within the merbecoviruses in Mexican and Belizean bats31, 58 and the likelihood that post-COVID research efforts will focus disproportionately on Asia, despite the near-global presence of bat betacoronavirus hosts.
Insight into SARS-CoV-2's emergence
Our work suggests that more than 400 species of bats might host undiscovered betacoronaviruses and that these species can be prioritised for sampling more efficiently via machine learning. Although our models do not target sarbecoviruses specifically, these efforts might help to find more SARS-like viruses in wildlife and might even uncover the direct progenitor of SARS-CoV-2, particularly given that 44 species of horseshoe bats are predicted to host undiscovered betacoronaviruses. However, our models provide otherwise limited insight into the origins of SARS-CoV-2, given the probable role of non-bat bridge hosts in spillover to humans.59, 60 We, thus, attempted a similar model ensemble in June, 2020, using five of our eight models to predict the broader mammal–virus network with a focus on potential betacoronavirus bridge hosts. At the time, only 30 non-bat hosts of betacoronaviruses were available in our data. Among the five models, we found a poor concordance in predictions (appendix p 24). The predictions were also heavily biased towards well studied and domesticated mammals (eg, Ovis aries, Vulpes vulpes, Capra hircus, Procyon lotor, and Rattus rattus), indicating that the sampling bias dwarfed biological signals. As such, we evaluated these models as having little value or consistency for an ensemble. This finding might be relevant given other studies have also modelled the susceptibility to SARS-CoV-2 across mammals; however, some have used more detailed trait data and thus probably make better predictions on this broader taxonomic scale.61, 62
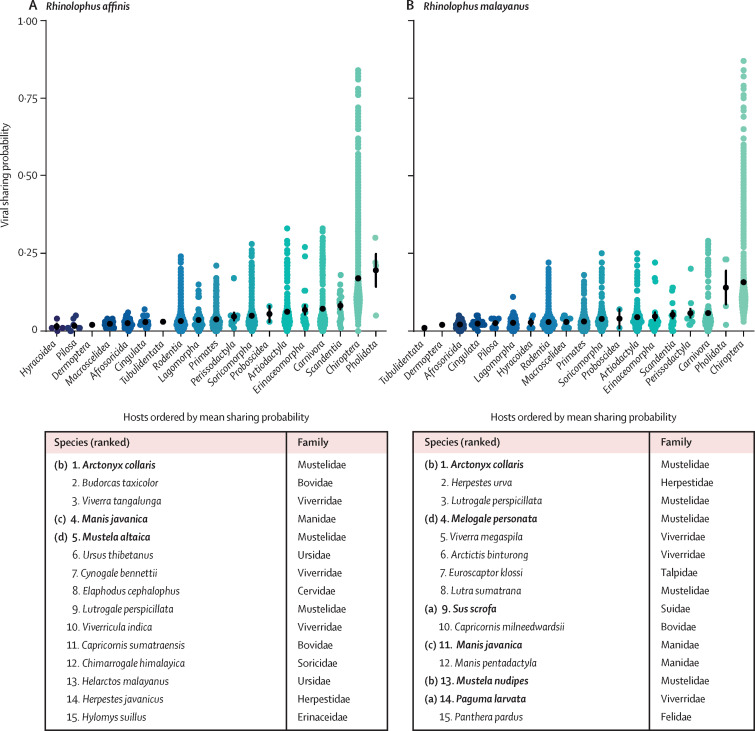
Instead of further calibrating this mammal-wide ensemble, we focused on the outputs of trait 3, which predicted how species should share viruses in nature based on their evolutionary history and geography. In June, 2020, we predicted the mammals expected to share viruses with R affinis and R malayanus, which hosted the two viruses (RaTG13 and RmYN02) most relevant to SARS-CoV-2's origins known at that time23, 63 (a closer related virus, RpYN06, has since been discovered in Rhinolophus pusillus).64 We predicted that these two bat species are disproportionately more likely to share viruses with pangolins (of the order Pholidota) and carnivores (of the order Carnivora), including civets (Viverridae family), mustelids (Mustelidae family), and cats (Felidae family; appendix p 25). These predictions have been broadly validated by the role of the masked palm civet (P larvata) in the original SARS-CoV outbreak,65, 66 the discovery of SARS-CoV-2-like viruses in the Sunda pangolin (Manis javanica),34 and extensive so-called spillback of SARS-CoV-2 into captive big cats, domestic cats, and both farmed and wild mink.67, 68 Notably, only the association between palm civets and SARS-CoV was present in the training data used for generating predictions from trait 3.
Given these successful predictions, we expect there might be potential insights into SARS-CoV-2's emergence when these predictions are paired with data on wildlife supply chains. Of the top 30 species (figure 6 ), two are known to have been traded in wildlife markets in Wuhan, China, immediately before the pandemic (the wild boar, Sus scrofa, and the palm civet, Paguma larvata), as were two species closely related to those in the top predictions (the Siberian weasel, Mustela siberica; the northern hog badger, Arctonyx albogularis).70 Another top species, the Burmese ferret badger (Melogale personata), was also reportedly of interest in WHO's origins investigation.71 Our models indicate that any of these species would be expected to regularly share viruses with relevant Rhinolophus bats in nature. Although bats use habitats differently than most of these probable bridge host species, opportunities for contact exist: one study from Gabon found cohabitation among pangolins, bats, and other mammals in burrows.72 Many species potentially implicated in the origins of SARS-CoV-2 could therefore have plausibly acquired a progenitor to SARS-CoV-2 in nature, at some point before contact with, and spillover into, humans.73 We suggest that this shortlist of species (figure 6) might, therefore, be useful for further investigations into the identity of potential bridge hosts, especially in combination with the experimental evaluation of susceptibility.
Figure 6.
Potential bridge hosts involved in SARS-CoV-2's emergence
Each dot represents predicted species-level sharing probabilities with Rhinolophus affinis (A) and R malayanus (B), estimated according to the phylogeographical viral sharing model trait-3.69 Each coloured point is a different mammal species. Black points and error bars denote the means and standard errors of viral sharing probability for each order; the mammal orders are arranged according to their mean sharing probability, ascending from left to right. The tables below report the top 15 predicted non-bat species for R affinis and R malayanus; several families are disproportionately represented, including pangolins (order, Pholidota; family, Manidae), mustelids (order, Carnivora; family, Mustelidae), and civets (order, Carnivora; family, Viverridae). Notable species are bolded (ordered based on immediate relevance to possible origins): (a) the wild boar S scrofa and palm civet P larvata were both traded in wildlife markets in Wuhan, China, before the pandemic; as were (b) close relatives of the greater hog badger, A collaris, (the northern hog badger, A albogularis), and of the mountain weasel, M altaica, and Malayan weasel, M nudipes (the Siberian weasel, M siberica). (c) SARS-CoV-2-like viruses have been found in traded Sunda pangolins (M javanica) outside of Wuhan, China, though the species was not reported in Wuhan. (d) The ferret badger (M personata) was also reportedly of interest in WHO's origins investigation, which explored the role of wildlife farm supply chains.
Conclusions
This Review is the first to show, by using predictive validation, that machine learning models could help to optimise wildlife sampling for undiscovered viruses. As such, the growing toolkit of models that predict host–pathogen interactions are likely to aid future efforts both to predict and prevent pandemics and to trace the origins of novel infections after they emerge. However, these tools will work best if they are implemented through a dynamic process of prediction, data collection, validation, and updating, as we have implemented here. Although some previous studies have incidentally tested specific hypotheses (eg, filovirus models and bat surveys,7, 11 henipavirus models and experimental infections,2, 16 and vector–virus models and competence trials13, 14, 15), predictions are almost never subject to systematic verification. More dialogue between modelers and empiricists is necessary to confront this research gap. This improved communication is particularly necessary when establishing a species' role as a viral reservoir rather than incidental hosts; susceptibility is only one aspect of host competence.1, 10 Future work, including the longitudinal tracking of viral shedding over space and time, the isolation of the live virus from wild animals, and the experimental confirmation of viral replication, can support more robust conclusions about whether predicted host species actually play a role in viral maintenance16 as well as inform related efforts to pinpoint and minimise risk factors for pathogen spillover.74, 75
This Review is also the first to benchmark the performance of a set of differently calibrated and designed statistical models all trained for one host prediction task. We found a range in model performances, even among a set of models that all performed well on training data. This finding underscores the need to incorporate long-term validation into similar studies and suggests there might be key lessons about viral ecology to be learned from this type of process. In our study, we found that network-based models performed mostly at random in validation against new bat hosts, whereas trait-based models were more successful in their predictions. There are two possible explanations for this difference in model performance. First, models that successfully predict the broader mammal–virus network are likely to vary in performance when subset to any given node. This likelihood might seem contradictory to the idea that understanding the broader so-called rules of life underpinning mammal–virus interactions will improve predictions in specific cases. However, there are ways to combine the strengths of both network-based and trait-based approaches. In a similar study, network-based predictions of zoonotic risk performed essentially at random, whereas a hybrid approach that embedded network predictions in targeted, trait-based models performed better than any approach in isolation.76 Future work should aim to develop and benchmark these types of hybrid model approaches more extensively and treat exclusively network-driven predictions with caution in the interim.
Second, models that integrate data on host ecology, evolution, and biogeography are likely to make more powerful predictions than those that mostly do not incorporate biology. This finding has many broader implications. Most notably, it suggests that filling gaps in the basic biology of bats is a key step towards zoonotic risk assessment and can benefit both pandemic prevention and bat conservation. High-quality host genomes are crucial to developing better predictive features, including genome composition bias metrics, improved host phylogenetic trees, and immunological traits.62, 77, 78, 79, 80 Whole-genome sequencing through initiatives such as the Bat1K project will expand the sparse available data on bat genomics and can facilitate other insights into the immune pathways used by bats to harbour virulent viruses.81, 82, 83 Targeted sequencing could also identify endogenised viral elements in bat genomes, shedding light on bat virus diversity and the evolution of bat immune systems.84, 85 Large-scale research networks, such as the Global Union of Bat Diversity Networks and its member networks, will further facilitate efficient sample sharing and ensure proper partnerships and equitable access and benefit sharing of knowledge across countries.86, 87 Additionally, museum specimens and historical collections offer opportunities to retrospectively screen samples for betacoronaviruses (thereby testing predictions), sequence tissue for an assembly of host genomes, and enhance understanding of complex host–virus interactions.88
Lastly, our iterative modelling of bat betacoronaviruses fits into a broader set of synergies in One Health research on bats, which can create win–win scenarios for conservation and outbreak prevention. For example, North American bats are threatened by an emerging disease, white-nose syndrome,89 which has documented synzootic interactions with other bat coronaviruses;90 at least seven North American bat species that can be infected by the fungal pathogen (Eptesicus fuscus, Myotis ciliolabrum, Myotis lucifugus, Myotis septentrionalis, Myotis velifer, Myotis volans, and Tadarida brasiliensis) are among the 412 bat species that we predicted could be undiscovered betacoronavirus hosts. Although our predictions do not imply bat susceptibility to SARS-CoV-2 specifically (and experimental infections of E fuscus have been unsuccessful91), efforts to minimise the risks of SARS-CoV-2 spillback into novel bat reservoirs,92, 93, 94, 95, 96 as well as to understand the dynamics of other bat coronaviruses, will both reduce zoonotic risk and help to understand and counteract disease-related population declines. Similarly, conservationists have expressed concern that the negative framing of bats as the source of SARS-CoV-2 has affected public and governmental attitudes toward bat conservation;97 this can fuel negative responses, including indiscriminate culling (ie, the reduction of populations by slaughter), which has already occurred in response to COVID-19 even outside of Asia (where a spillover probably occurred).98 Evidence shows that culling has many negative consequences, not only threatening population viability99 but also possibly increasing viral transmission within the very species that are targeted.100, 101 Bat conservation programmes and One Health practitioners should continue to work together to find sustainable solutions for humans to live safely alongside wildlife and to communicate with the public about the ecological importance of these highly vulnerable species and the science of pathogen spillover.
For more on the Viral Emergence Research Initiative consortium see https://viralemergence.org
This online publication has been corrected. The corrected version first appeared at thelancet.com/microbe on January 28, 2022
Data sharing
The standardised data on betacoronavirus associations, and all associated predictor data, are available from the Viral Emergence Research Initiative consortium's Github (github.com/viralemergence/virionette). All modelling teams contributed an individual repository with their methods, which are available in the organisational directory (github.com/viralemergence). Code for the 2020 analysis, and a working reproduction of each author's contributions, is available from the study repository (github.com/viralemergence/Fresnel), and code to produce the updated analyses from 2021 is available in a separate repository (github.com/viralemergence/Fresnel_Jun). A complete list of the 412 predicted betacoronavirus bat hosts from our final ensemble is provided in the appendix.
Declaration of interests
We declare no competing interests.
Acknowledgments
Acknowledgments
We thank Heather Wells for generously sharing thoughtful comments and code. The Viral Emergence Research Initiative consortium is supported by L'Institut de Valorisation de Données through the Université de Montreal and by US National Science Foundation BII 2021909. LMB was supported by the Wellcome Trust (217221/Z/19/Z). MS was supported by the Research Foundation – Flanders (FWO17/PDO/067) and the Flemish Government under the Onderzoeksprogramma Artificiële Intelligentie Vlaanderen programme. Lastly, we thank three anonymous reviewers for their constructive feedback.
Contributors
DJB, GFA, and CJC designed the study. ARS, TP, and BAH collected or contributed the initial data. GFA, TP, TAD, MJF, SG, MS, and CJC implemented the models. LMB, BC, LEC, ACF, ARS, and CJC collected secondary data. DJB, GFA, and CJC analysed the data. EAE, NBS, and ECT contributed to the data interpretation. DJB, GFA, and CJC wrote the manuscript with input from all coauthors.
Supplementary Material
References
- 1.Viana M, Mancy R, Biek R, et al. Assembling evidence for identifying reservoirs of infection. Trends Ecol Evol. 2014;29:270–279. doi: 10.1016/j.tree.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plowright RK, Becker DJ, Crowley DE, et al. Prioritizing surveillance of Nipah virus in India. PLoS Negl Trop Dis. 2019;13:e0007393. doi: 10.1371/journal.pntd.0007393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker DJ, Crowley DE, Washburne AD, Plowright RK. Temporal and spatial limitations in global surveillance for bat filoviruses and henipaviruses. Biol Lett. 2019;15:20190423. doi: 10.1098/rsbl.2019.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Washburne AD, Crowley DE, Becker DJ, et al. Taxonomic patterns in the zoonotic potential of mammalian viruses. PeerJ. 2018;6:e5979. doi: 10.7717/peerj.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowley D, Becker D, Washburne A, Plowright R. Identifying suspect bat reservoirs of emerging infections. Vaccines (Basel) 2020;8:228. doi: 10.3390/vaccines8020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker DJ, Washburne AD, Faust CL, Mordecai EA, Plowright RK. The problem of scale in the prediction and management of pathogen spillover. Philos Trans R Soc Lond B Biol Sci. 2019;374:20190224. doi: 10.1098/rstb.2019.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han BA, Schmidt JP, Alexander LW, Bowden SE, Hayman DTS, Drake JM. Undiscovered bat hosts of filoviruses. PLoS Negl Trop Dis. 2016;10:e0004815. doi: 10.1371/journal.pntd.0004815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han BA, Majumdar S, Calmon FP, et al. Confronting data sparsity to identify potential sources of Zika virus spillover infection among primates. Epidemics. 2019;27:59–65. doi: 10.1016/j.epidem.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Becker DJ, Han BA. The macroecology and evolution of avian competence for Borrelia burgdorferi. Glob Ecol Biogeogr. 2021;30:710–724. [Google Scholar]
- 10.Becker DJ, Seifert SN, Carlson CJ. Beyond infection: integrating competence into reservoir host prediction. Trends Ecol Evol. 2020;35:1062–1065. doi: 10.1016/j.tree.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X-L, Zhang Y-Z, Jiang R-D, et al. Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg Infect Dis. 2017;23:482–486. doi: 10.3201/eid2303.161119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laing ED, Mendenhall IH, Linster M, et al. Serologic evidence of fruit bat exposure to filoviruses, Singapore, 2011–2016. Emerg Infect Dis. 2018;24:114–117. doi: 10.3201/eid2401.170401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans MV, Dallas TA, Han BA, Murdock CC, Drake JM. Data-driven identification of potential Zika virus vectors. eLife. 2017;6:e22053. doi: 10.7554/eLife.22053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guedes DR, Paiva MH, Donato MM, et al. Zika virus replication in the mosquito Culex quinquefasciatus in Brazil. Emerg Microbes Infect. 2017;6:e69. doi: 10.1038/emi.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smartt CT, Shin D, Kang S, Tabachnick WJ. Culex quinquefasciatus (Diptera: Culicidae) from Florida transmitted Zika virus. Front Microbiol. 2018;9:768. doi: 10.3389/fmicb.2018.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seifert SN, Letko MC, Bushmaker T, et al. Rousettus aegyptiacus bats do not support productive Nipah virus replication. J Infect Dis. 2020;221(suppl 4):S407–S413. doi: 10.1093/infdis/jiz429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gokhale MD, Sreelekshmy M, Sudeep AB, et al. Detection of possible Nipah virus infection in Rousettus leschenaultii and Pipistrellus pipistrellus bats in Maharashtra, India. J Infect Public Health. 2021;14:1010–1012. doi: 10.1016/j.jiph.2021.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Anthony SJ, Johnson CK, Greig DJ, et al. Global patterns in coronavirus diversity. Virus Evol. 2017;3:vex012. doi: 10.1093/ve/vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren W, Li W, Yu M, et al. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J Gen Virol. 2006;87:3355–3359. doi: 10.1099/vir.0.82220-0. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 22.Yang X-L, Hu B, Wang B, et al. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol. 2015;90:3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 25.Hu B, Zeng L-P, Yang X-L, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Memish ZA, Mishra N, Olival KJ, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Qi J, Yuan Y, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Du L, Liu C, et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci USA. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu B, Ge X, Wang L-F, Shi Z. Bat origin of human coronaviruses. Virol J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anthony SJ, Gilardi K, Menachery VD, et al. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. MBio. 2017;8 doi: 10.1128/mBio.00373-17. e00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony SJ, Ojeda-Flores R, Rico-Chávez O, et al. Coronaviruses in bats from Mexico. J Gen Virol. 2013;94:1028–1038. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L, Wu Z, Ren X, et al. MERS-related betacoronavirus in Vespertilio superans bats, China. Emerg Infect Dis. 2014;20:1260–1262. doi: 10.3201/eid2007.140318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen R, Wang H, Pipes L. Synonymous mutations and the molecular evolution of SARS-Cov-2 origins. bioRxiv. 2020 doi: 10.1101/2020.04.20.052019. published online Oct 12. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam TT-Y, Jia N, Zhang Y-W, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 35.Xiao K, Zhai J, Feng Y, et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30:1578. doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fritz SA, Bininda-Emonds ORP, Purvis A. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol Lett. 2009;12:538–549. doi: 10.1111/j.1461-0248.2009.01307.x. [DOI] [PubMed] [Google Scholar]
- 40.Jones KE, Bielby J, Cardillo M, et al. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology. 2009;90:2648. [Google Scholar]
- 41.Wilman H, Belmaker J, Simpson J, de la Rosa C, Rivadeneira MM, Jetz W. EltonTraits 1·0: species-level foraging attributes of the world's birds and mammals. Ecology. 2014;95:2027. [Google Scholar]
- 42.Trifonova N, Kenny A, Maxwell D, Duplisea D, Fernandes J, Tucker A. Spatio-temporal Bayesian network models with latent variables for revealing trophic dynamics and functional networks in fisheries ecology. Ecol Inform. 2015;30:142–158. [Google Scholar]
- 43.Rohr RP, Scherer H, Kehrli P, Mazza C, Bersier L-F. Modeling food webs: exploring unexplained structure using latent traits. Am Nat. 2010;176:170–177. doi: 10.1086/653667. [DOI] [PubMed] [Google Scholar]
- 44.Dallas T, Park AW, Drake JM. Predicting cryptic links in host-parasite networks. PLoS Comput Biol. 2017;13:e1005557. doi: 10.1371/journal.pcbi.1005557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han BA, Schmidt JP, Bowden SE, Drake JM. Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci USA. 2015;112:7039–7044. doi: 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Washburne AD, Silverman JD, Morton JT, et al. Phylofactorization: a graph partitioning algorithm to identify phylogenetic scales of ecological data. Ecol Monogr. 2019;89:e01353. [Google Scholar]
- 47.Brandão PE, Scheffer K, Villarreal LY, et al. A coronavirus detected in the vampire bat Desmodus rotundus. Braz J Infect Dis. 2008;12:466–468. doi: 10.1590/s1413-86702008000600003. [DOI] [PubMed] [Google Scholar]
- 48.Corman VM, Rasche A, Diallo TD, et al. Highly diversified coronaviruses in neotropical bats. J Gen Virol. 2013;94:1984–1994. doi: 10.1099/vir.0.054841-0. [DOI] [PubMed] [Google Scholar]
- 49.Moreira-Soto A, Taylor-Castillo L, Vargas-Vargas N, Rodríguez-Herrera B, Jiménez C, Corrales-Aguilar E. Neotropical bats from Costa Rica harbour diverse coronaviruses. Zoonoses Public Health. 2015;62:501–505. doi: 10.1111/zph.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Fu S, Cao Y, et al. Discovery and genetic analysis of novel coronaviruses in least horseshoe bats in southwestern China. Emerg Microbes Infect. 2017;6:e14. doi: 10.1038/emi.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin X-D, Wang W, Hao Z-Y, et al. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wacharapluesadee S, Duengkae P, Rodpan A, et al. Diversity of coronavirus in bats from eastern Thailand. Virol J. 2015;12:57. doi: 10.1186/s12985-015-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guy C, Ratcliffe JM, Mideo N. The influence of bat ecology on viral diversity and reservoir status. Ecol Evol. 2020;10:5748–5758. doi: 10.1002/ece3.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wacharapluesadee S, Duengkae P, Chaiyes A, et al. Longitudinal study of age-specific pattern of coronavirus infection in Lyle's flying fox (Pteropus lylei) in Thailand. Virol J. 2018;15:38. doi: 10.1186/s12985-018-0950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bergner LM, Orton RJ, Broos A, et al. Diversification of mammalian deltaviruses by host shifting. bioRxiv. 2020 doi: 10.1101/2020.06.17.156745. published online Dec 22. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bergner LM, Orton RJ, da Silva Filipe A, et al. Using noninvasive metagenomics to characterize viral communities from wildlife. Mol Ecol Resour. 2019;19:128–143. doi: 10.1111/1755-0998.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergner LM, Orton RJ, Streicker DG. Complete genome sequence of an alphacoronavirus from common vampire bats in Peru. Microbiol Resour Announc. 2020;9:e00742–e00820. doi: 10.1128/MRA.00742-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neely BA, Janech MG, Brock Fenton M, Simmons NB, Bland AM, Becker DJ. Surveying the vampire bat (Desmodus rotundus) serum proteome: a resource for identifying immunological proteins and detecting pathogens. J Proteome Res. 2020;20:2547–2559. doi: 10.1021/acs.jproteome.0c00995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J, Cui W, Tian B-P. The potential intermediate hosts for SARS-CoV-2. Front Microbiol. 2020;11:580137. doi: 10.3389/fmicb.2020.580137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fischhoff IR, Castellanos AA, Rodrigues JPGLM, Varsani A, Han BA. Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2. Proc R Soc B. 2021;288:20211651. doi: 10.1098/rspb.2021.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wardeh M, Baylis M, Blagrove MSC. Predicting mammalian hosts in which novel coronaviruses can be generated. Nat Commun. 2021;12:780. doi: 10.1038/s41467-021-21034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou H, Chen X, Hu T, et al. A novel bat coronavirus reveals natural insertions at the S1/S2 cleavage site of the Spike protein and a possible recombinant origin of HCoV-19. bioRxiv. 2020 doi: 10.1101/2020.03.02.974139. published online March 11. (preprint). [DOI] [Google Scholar]
- 64.Zhou H, Ji J, Chen X, et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021;184:4380–4391. doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang M, Yan M, Xu H, et al. SARS-CoV infection in a restaurant from palm civet. Emerg Infect Dis. 2005;11:1860–1865. doi: 10.3201/eid1112.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song H-D, Tu C-C, Zhang G-W, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia P, Dai S, Wu T, Yang S. New approaches to anticipate the risk of reverse zoonosis. Trends Ecol Evol. 2021;36:580–590. doi: 10.1016/j.tree.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fagre AC, Cohen L, Eskew EA, et al. Spillback in the Anthropocene: the risk of human-to-wildlife pathogen transmission for conservation and public health. EcoEvoRxiv. 2021 doi: 10.32942/osf.io/sx6p8. published online April 11. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Albery GF, Eskew EA, Ross N, Olival KJ. Predicting the global mammalian viral sharing network using phylogeography. Nat Commun. 2020;11:2260. doi: 10.1038/s41467-020-16153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao X, Newman C, Buesching CD, Macdonald DW, Zhou Z-M. Animal sales from Wuhan wet markets immediately prior to the COVID-19 pandemic. Sci Rep. 2021;11:11898. doi: 10.1038/s41598-021-91470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McKay B, Page J, Hinshaw D. In hunt for Covid-19 origin, WHO team focuses on two animal types in China. Feb 18, 2021. The Wall Street Journal. https://www.wsj.com/articles/in-hunt-for-covid-19-origin-who-team-focuses-on-two-animal-types-in-china-11613665015
- 72.Lehmann D, Halbwax ML, Makaga L, et al. Pangolins and bats living together in underground burrows in Lopé National Park, Gabon. Afr J Ecol. 2020;58:540–542. doi: 10.1111/aje.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee J, Hughes T, Lee M-H, et al. No evidence of coronaviruses or other potentially zoonotic viruses in Sunda pangolins (Manis javanica) entering the wildlife trade via Malaysia. EcoHealth. 2020;17:406–418. doi: 10.1007/s10393-020-01503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plowright RK, Becker DJ, McCallum H, Manlove KR. Sampling to elucidate the dynamics of infections in reservoir hosts. Philos Trans R Soc Lond B Biol Sci. 2019;374:20180336. doi: 10.1098/rstb.2018.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sokolow SH, Nova N, Pepin KM, et al. Ecological interventions to prevent and manage zoonotic pathogen spillover. Philos Trans R Soc Lond B Biol Sci. 2019;374:20180342. doi: 10.1098/rstb.2018.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poisot T, Ouellet M-A, Mollentze N, et al. Imputing the mammalian virome with linear filtering and singular value decomposition. arXiv. 2021 http://arxiv.org/abs/2105.14973 published online May 31. (preprint). [Google Scholar]
- 77.Babayan SA, Orton RJ, Streicker DG. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science. 2018;362:577–580. doi: 10.1126/science.aap9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mollentze N, Babayan SA, Streicker DG. Identifying and prioritizing potential human-infecting viruses from their genome sequences. bioRxiv. 2021 doi: 10.1101/2020.11.12.379917. published online June 3. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rannala B, Yang Z. Phylogenetic inference using whole genomes. Annu Rev Genomics Hum Genet. 2008;9:217–231. doi: 10.1146/annurev.genom.9.081307.164407. [DOI] [PubMed] [Google Scholar]
- 80.Pedersen AB, Babayan SA. Wild immunology. Mol Ecol. 2011;20:872–880. doi: 10.1111/j.1365-294X.2010.04938.x. [DOI] [PubMed] [Google Scholar]
- 81.Teeling EC, Vernes SC, Dávalos LM, Ray DA, Gilbert MTP, Myers E. Bat biology, genomes, and the Bat1K project: to generate chromosome-level genomes for all living bat species. Annu Rev Anim Biosci. 2018;6:23–46. doi: 10.1146/annurev-animal-022516-022811. [DOI] [PubMed] [Google Scholar]
- 82.Jebb D, Huang Z, Pippel M, et al. Six reference-quality genomes reveal evolution of bat adaptations. Nature. 2020;583:578–584. doi: 10.1038/s41586-020-2486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang L-F, Gamage AM, Chan WOY, Hiller M, Teeling EC. Decoding bat immunity: the need for a coordinated research approach. Nat Rev Immunol. 2021;21:269–271. doi: 10.1038/s41577-021-00523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skirmuntt EC, Escalera-Zamudio M, Teeling EC, Smith A, Katzourakis A. The potential role of endogenous viral elements in the evolution of bats as reservoirs for zoonotic viruses. Annu Rev Virol. 2020;7:103–119. doi: 10.1146/annurev-virology-092818-015613. [DOI] [PubMed] [Google Scholar]
- 85.Jebb D, Huang Z, Pippel M, et al. Six new reference-quality bat genomes illuminate the molecular basis and evolution of bat adaptations. bioRxiv. 2019 doi: 10.1101/836874. published online Nov 9. (preprint). [DOI] [Google Scholar]
- 86.Kingston T, Aguirre L, Armstrong K, et al. Bats in the anthropocene: conservation of bats in a changing world. Springer; Cham: 2016. Networking networks for global bat conservation; pp. 539–569. [Google Scholar]
- 87.Phelps KL, Hamel L, Alhmoud N, et al. Bat research networks and viral surveillance: gaps and opportunities in western Asia. Viruses. 2019;11:E240. doi: 10.3390/v11030240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cook JA, Arai S, Armién B, et al. Integrating biodiversity infrastructure into pathogen discovery and mitigation of emerging infectious diseases. BioScience. 2020;70:531–534. doi: 10.1093/biosci/biaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frick WF, Pollock JF, Hicks AC, et al. An emerging disease causes regional population collapse of a common North American bat species. Science. 2010;329:679–682. doi: 10.1126/science.1188594. [DOI] [PubMed] [Google Scholar]
- 90.Davy CM, Donaldson ME, Subudhi S, et al. White-nose syndrome is associated with increased replication of a naturally persisting coronaviruses in bats. Sci Rep. 2018;8:15508. doi: 10.1038/s41598-018-33975-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hall JS, Knowles S, Nashold SW, et al. Experimental challenge of a North American bat species, big brown bat (Eptesicus fuscus), with SARS-CoV-2. Transbound Emerg Dis. 2020;68:3443–3452. doi: 10.1111/tbed.13949. [DOI] [PubMed] [Google Scholar]
- 92.Olival KJ, Cryan PM, Amman BR, et al. Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: a case study of bats. PLoS Pathog. 2020;16:e1008758. doi: 10.1371/journal.ppat.1008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cox-Witton K, Baker ML, Edson D, Peel AJ, Welbergen JA, Field H. Risk of SARS-CoV-2 transmission from humans to bats - an Australian assessment. One Health. 2021;13:100247. doi: 10.1016/j.onehlt.2021.100247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Common SM, Shadbolt T, Walsh K, Sainsbury AW. The risk from SARS-CoV-2 to bat species in england and mitigation options for conservation field workers. Transbound Emerg Dis. 2021 doi: 10.1111/tbed.14035. published online Feb 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cook JD, Grant EHC, Coleman JTH, Sleeman JM, Runge MC. Risks posed by SARS-CoV-2 to North American bats during winter fieldwork. Conserv Sci Pract. 2021;3:e410. doi: 10.1111/csp2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kingston T, Frick W, Kading R, et al. IUCN SSC Bat Specialist Group (BSG) recommended strategy for researchers to reduce the risk of transmission of SARS-CoV-2 from humans to bats (version 2·0). IUCN SSC Bat Specialist Group. 2021. https://www.iucnbsg.org/uploads/6/5/0/9/6509077/amp_recommendations_for_researchers_final.pdf
- 97.Zhao H. COVID-19 drives new threat to bats in China. Science. 2020;367:1436. doi: 10.1126/science.abb3088. [DOI] [PubMed] [Google Scholar]
- 98.Fenton MB, Mubareka S, Tsang SM, Simmons NB, Becker DJ. COVID-19 and threats to bats. Facets. 2020;5:349–352. [Google Scholar]
- 99.Aguiar LMS, Brito D, Machado RB. Do current vampire bat (Desmodus rotundus) population control practices pose a threat to Dekeyser's nectar bat's (Lonchophylla dekeyseri) long-term persistence in the Cerrado? Acta Chiropt. 2010;12:275–282. [Google Scholar]
- 100.Streicker DG, Recuenco S, Valderrama W, et al. Ecological and anthropogenic drivers of rabies exposure in vampire bats: implications for transmission and control. Proc Biol Sci. 2012;279:3384–3392. doi: 10.1098/rspb.2012.0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blackwood JC, Streicker DG, Altizer S, Rohani P. Resolving the roles of immunity, pathogenesis, and immigration for rabies persistence in vampire bats. Proc Natl Acad Sci USA. 2013;110:20837–20842. doi: 10.1073/pnas.1308817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The standardised data on betacoronavirus associations, and all associated predictor data, are available from the Viral Emergence Research Initiative consortium's Github (github.com/viralemergence/virionette). All modelling teams contributed an individual repository with their methods, which are available in the organisational directory (github.com/viralemergence). Code for the 2020 analysis, and a working reproduction of each author's contributions, is available from the study repository (github.com/viralemergence/Fresnel), and code to produce the updated analyses from 2021 is available in a separate repository (github.com/viralemergence/Fresnel_Jun). A complete list of the 412 predicted betacoronavirus bat hosts from our final ensemble is provided in the appendix.