Abstract
The Kruger National Park (KNP), South Africa, has a recorded history of periodic anthrax epidemics causing widespread disease among wild animals. Bacillus anthracis is the causative agent of anthrax, a disease primarily affecting ungulate herbivores. Worldwide there is little diversity among B. anthracis isolates, but examination of variable-number tandem repeat (VNTR) loci has identified six major clones, with the most dissimilar types split into the A and B branches. Both the A and B types are found in southern Africa, giving this region the greatest genetic diversity of B. anthracis worldwide. Consequently, southern Africa has been hypothesized to be the geographic origin of B. anthracis. In this study, we identify the genotypic types of 98 KNP B. anthracis isolates using multiple-locus VNTR analysis. Two major types are evident, the A branch and the B branch. The spatial and temporal distribution of the different genotypes indicates that anthrax epidemic foci are independent, though correlated through environmental cues. Kruger B isolates were found on significantly higher-calcium and higher-pH soils than were Kruger type A. This relationship between genotype and soil chemistry may be due to adaptive differences among divergent anthrax strains. While this association may be simply fortuitous, adaptation of A types to diverse environmental conditions is consistent with their greater geographic dispersal and genetic dissimilarity.
Bacillus anthracis is a gram-positive, rod-shaped, spore-forming bacterium. It is the causative agent of anthrax, a disease primarily affecting ungulate herbivores, occasionally carnivores, and less frequently humans (14). Anthrax is a disease well documented in human history with suggestive reports even in the Bible and Sanskrit manuscripts (5). The field of microbiology was revolutionized by the anthrax studies of Koch, Pasteur, and others (13). Though anthrax was never eradicated, the development of an effective animal vaccine has reduced its importance for humans and animals in developed countries over the last century. Recently, however, anthrax research has been increasingly important due to this pathogen's central position in biological warfare and biological terrorism (6).
It is generally thought that B. anthracis is an obligate pathogen and that little propagation occurs in soil (2, 4, 10). With the exception of scavengers, anthrax is almost never transmitted directly from victim to victim but is rather ingested by herbivores while grazing or browsing. In such a model, the environmental spore reservoir becomes very important to the ecology and evolution of this pathogen. Survival in the soil is crucial for initiating subsequent anthrax epidemics, and these relatively long periods of quiescence may greatly reduce evolutionary change. Thus, any adaptive mutation altering spore survival will be under great selection. Differential environmental selection will lead to the most rapid adaptive changes in this pathogen.
Worldwide there is little diversity among B. anthracis isolates, but examination of variable-number tandem repeat (VNTR) loci has identified six major clones (9). The low diversity among these clonal lineages is consistent with a slowly evolving organism or one that recently derived from a common ancestor. The most dissimilar types are split into two major groups (A and B). Type A strains are found around the world and are responsible for most epidemics and outbreaks. In contrast, type B strains are almost exclusively restricted to southern Africa. It is possible that one or more adaptations allowed the greater geographic range (and diversity) of type A, or the loss of adaptations has restricted type B. Both A and B are found in southern Africa, giving this region the greatest genetic diversity of B. anthracis worldwide. Consequently, southern Africa has been hypothesized to be the geographic origin of B. anthracis (7).
Southern Africa has experienced many anthrax epidemics in the past (1). The wide expanses of savannah and large populations of wild ungulate herbivores are ideally suited for anthrax endemicity. Vestiges of the large wildlife populations and savannah ecosystems still exist today in the parks of southern Africa. One such area is the Kruger National Park (KNP), South Africa.
The KNP has a recorded history of periodic anthrax epidemics (3). There are areas within the KNP known to be anthrax foci, where epidemics have been observed to begin. Two regions, the northernmost tip of the park (since 1959) and the central region (since 1990), are most notable, although the entire KNP region may be affected during an epidemic. Previous investigations (8, 9, 11) show that both A and B isolates occur in the KNP. Because of the close proximity of these different types, the KNP provides a unique opportunity to characterize B. anthracis in an ecological setting and examine possible environmental associations specific to the genotypic groups. In this study, we identified the genotypes of 98 KNP B. anthracis isolates using multiple-locus VNTR analysis (MLVA) (9). Two major types were evident, branch A and branch B. The spatial and temporal distribution of the different genotypes indicates that anthrax epidemic foci are independent, though correlated through environmental cues. Soil chemistry (calcium and pH) differs among outbreak foci, as does the genotypic composition. The relationship between genotype and soil chemistry may be due to adaptive differences among divergent anthrax strains.
MATERIALS AND METHODS
Study site, B. anthracis isolates, and soil parameters.
The KNP is an elongated conservation area consisting of almost 2,000,000 ha of subtropical savannah woodland from 20°19′ to 25°32′ S and 31°0′ to 32°0′ E, situated in the northeasternmost corner of the Republic of South Africa. The eastern border of the park adjoins Mozambique; the northern border adjoins Zimbabwe. The entire area is enclosed with gameproof fencing and adjoins commercial farms, traditional communal grazing areas, and private nature reserves. The KNP contains approximately 2,300 kg of biomass of large mammals per km2, made up of 20 different species. The B. anthracis isolates selected for this study were isolated within the park. The 98 isolates in the study range in date of isolation from 1970 to 1997. The sources of isolates were animal carcasses, soil, bone, water, dung, and diagnostic blood smears taken in the field. The soil pH and calcium values from the site of isolation were determined by reference to soil survey data collected previously (15).
MLVA.
The MLVA strain typing procedure is described in detail elsewhere (9). Briefly, a single colony of B. anthracis was “heat lysed” in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA) at 95°C for 20 min. Cellular debris was removed by centrifugation. The supernatant was placed in a clean tube for storage. The MLVA PCR amplification of the eight selected loci required four PCRs: two multiplexed and two single amplifications. Each amplicon had a single fluorescently labeled primer. Three different dyes were used for the amplicons, while one was reserved for molecular weight standards. Products from both reactions were pooled and then separated by polyacrylamide gel electrophoresis under denaturing conditions on an ABI 377 automated DNA sequencer. The resulting fragment profiles were analyzed using ABI Genescan and Genotyper software to determine allele classification. Diversity group clusters were analyzed using sequential agglomerative hierarchichal nested cluster analysis (SAHN) and unweighted pair group method analysis using average linkages (UPGMA) clustering routines (12) within the NTSYSpc software package (F. J. Rohlf, NTSYSpc, ed. 2.02, Exeter Software, Stony Brook, N.Y., 1998). A simple percent distance dissimilarity matrix of the allele values for the isolates was constructed prior to cluster analysis. Individual marker diversity (diversity index value [D]) is calculated as being equal to 1 − Σ(allele frequency)2 (16) and is based upon the entire isolate collection in the study.
Statistical data analysis.
Space-time cluster calculations were performed using the SatScan software Bernoulli model and are described elsewhere in detail (M. Kulldorff et al., SatScan, ed. v2.1, National Cancer Institute, Bethesda, Md., 1998). In brief, a space-time scan statistic is defined by a cylindrical window with a circular geographic base and with height corresponding to time. The base is defined by an infinite number of distinct geographic circles, with different sets of neighboring census areas within them and each being a possible candidate for a cluster, while the height reflects the time period of potential clusters. The cylindrical window is then moved in space and time, so that for each possible geographic location and size, it visits each possible time period. In effect, we obtain an infinite number of overlapping cylinders different in size and shape, jointly covering the entire study region, where each cylinder reflects a possible cluster. For each location and size of the scanning window, the alternative hypothesis is that there is an elevated rate within the window compared to outside. A likelihood function for the Bernoulli model (used here) is calculated as (n/m)n(1-n/m)(m-n)[(N-m)/(M-m)](N-n){1-[(N-m)/(M-m)]}(M-m)-(N-n)I(x), where I(x) is the indicator function, set at 1 when the space-time scan window has more cases than expected under the null hypothesis and set at 0 otherwise. The likelihood function is maximized over all windows, identifying the window that constitutes the likeliest cluster. This is the cluster that is least likely to have occurred by chance. The likelihood ratio for this window is noted and constitutes the maximum likelihood ratio test statistic. Its distribution under the null hypothesis and its corresponding P value are obtained by repeating the same analytic exercise on a large number of random replications of the data set generated under the null hypothesis in a Monte Carlo simulation.
Probability contour kernel density maps were produced using ArcView GIS software and extensions. These maps portray the probability that a sample taken will be a member of the underlying point population for which a kernel density function was applied from a given geographic area. The kernel density is a moving, three-dimensional function calculated for each map cell by distributing outward the value found in the point population field for each point found within the search radius and then dividing by the area of the circle in area units. The output density values are the occurrences of the measured quantity per specified area unit. In this analysis, the map units were square kilometers and the search radius was 10 km.
The Wilcoxon rank-sum test is a nonparametric analysis of variance and was performed for this study using SAS software. Soil calcium values and pH were set as the dependent variables and classified by genotype group affiliation.
RESULTS
Genetic diversity of B. anthracis in the KNP.
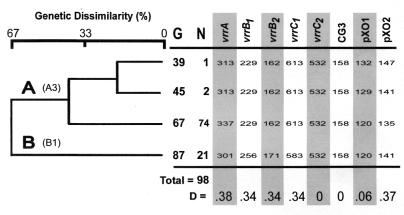
We examined the genetic diversity of 98 B. anthracis isolates from the KNP by using MLVA (9) to better understand the dynamics of anthrax in this region. The isolates included in the study were collected over more than 20 years and spanned multiple epidemics. Alleles detected are listed by size for each of the eight VNTR markers in Fig. 1, along with the numerical designations of their global diversity groups, based on our worldwide strain analysis (9).
FIG. 1.
Genetic relationships among KNP B. anthracis isolates. Eight VNTR marker loci (9) were used to estimate genetic relationships among the 98 B. anthracis isolates in the study. UPGMA cluster analysis generated a dendrogram to graphically represent dissimilarity among the unique types observed. On average, the A and B isolates have different alleles at two-thirds of the eight loci. The allele size at each VNTR locus is shown along with the number of isolates (N) in each genotypic group (G). Genotype number and VNTR marker alleles are consistent with a previous report (9). The major diversity group (9) represented in each of the branches is shown in parentheses beside the branch designation. The D calculated for each of the eight markers is listed below the dendrogram.
We found two dominant and four distinct MLVA strain types (genotypes) in this collection of 98 isolates. Kruger A (number 67) is represented by the most isolates (74 of 98). The Kruger B genotype (number 87) is the second most common (21 of 98), while the other genotypes (number 45, 2 of 98; number 39, 1 of 98) are rare. Kruger A is a member of major cluster A3, which was previously found to be the most widely distributed worldwide, though the Kruger A genotype itself has thus far been isolated and identified only in the KNP. The rare genotype 39, also a member of cluster A3, has been isolated in Namibia in addition to South Africa (9). Kruger B is a member of the B1 diversity group, which is found almost exclusively in southern African countries. The rare genotype 45 is from the A3 cluster and has also been found in Argentina, Turkey, and the United States. It is identical with the U.S. vaccine strain (V770-NPI-R), according to examination of seven of the eight VNTR loci (9).
The dominant KNP strain types, Kruger A and B, are genetically quite dissimilar. Five of the eight MLVA markers had allele differences between these genotypes. This considerable genetic distance is equivalent to the greatest distance reported in the entire worldwide collection (9).
In this set of isolates, it is interesting to note that the most informative marker was not from the pXO1 plasmid VNTR (Fig. 1), in contrast to what was found in the worldwide study (9). In fact, the pXO1 VNTR marker had a low diversity index value (D = 0.06) in this collection. The pXO2 marker was the second most informative marker (D = 0.37) and closer to previous observations. The genomic marker vrrA (D = 0.38) showed the greatest diversity index value. The genomic markers vrrB1, vrrB2, and vrrC1 provided equal discriminatory power (D = 0.34), while the markers vrrC2 and CG3 revealed no diversity and therefore contributed no discriminatory information to the analysis. The mean diversity index value of the eight VNTR markers used in this study was 0.23, much lower than for the worldwide study, where the average was 0.52. And yet, two of the most dissimilar strains known are found in overlapping spatial distributions in the KNP.
Geographic and temporal distribution of isolates.
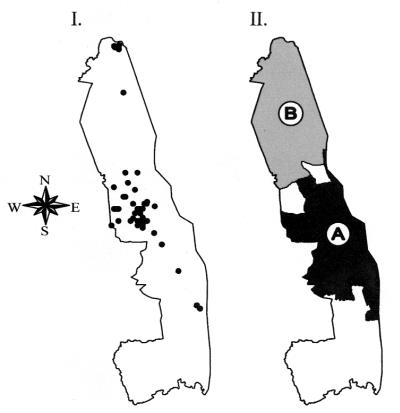
Although the northernmost and central regions within the KNP are known to be foci where epidemics have begun, anthrax cases were found throughout the geographic extent of the park while we collected isolates. Figure 2I indicates the geographic locations of sample isolates, but it alone inadequately represents the true isolate sample density due to close or overlapping locations. As shown in Fig. 3, it is apparent that Kruger A and B were present during the same year, showing temporal overlap. Kruger A and B both contributed to mortality during the same epidemics in the KNP. This was especially true in 1970 to 1981, when about half of the isolates were from each major strain type. This is in contrast to the expectation that a single outbreak will emanate from a single source and that pathogen isolates will have great similarity or even be indistinguishable.
FIG. 2.
Isolate locations and Bernoulli regression clusters. (I) This map shows the individual isolate locations within the KNP. At the scale presented, many isolate locations overlap and may appear less dense than the actual total. (II) This map shows the geographic areas included in the Bernoulli regression for time-space cluster detection. The secondary time-space cluster for Kruger A isolates is in the middle of the park (the primary cluster included the entire park), and the primary time-space cluster for Kruger B isolates includes all of the north of the park.
FIG. 3.
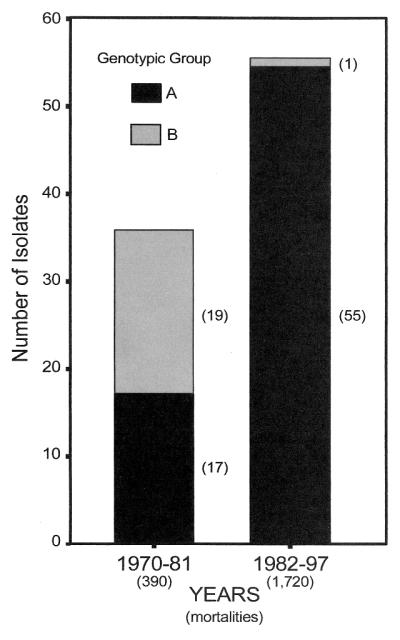
Isolate-year relationships. This histogram shows the number of isolates (in parentheses) and branch affiliation by year of field isolation.
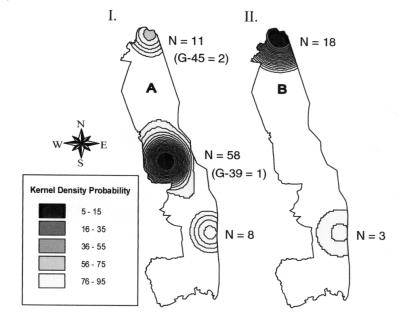
In a previous investigation, we showed that all isolates, when considered together, exhibit space-time clustering (as would be expected in epidemic situations) (11). In this investigation, we used MLVA first to define individual samples as either Kruger A or B and then to demonstrate that differences existed between the spatial and temporal distributions of the two groups. When we used a Bernoulli regression model for space-time clustering, Kruger A and B showed statistically significant independence from each other (Fig. 2II, log-likelihood ratio [LLR] = 21.39, P < 0.001). The differently shaded areas are the management blocks included in clusters detected for Kruger A and B (in the northern region of the KNP for B and in the south for A). The contour maps of kernel density probability (Fig. 4) provide a clearer indication of the geographic distribution of Kruger A and B, which are overlapping yet distinctly different.
FIG. 4.
Kernel density probability of Kruger isolates. (I) This map shows the kernel density probabilities estimated for the Kruger A strains included in this study. Representation of minor strain subtypes (genotypes [G] 39 and 45) is indicated. (II) This map shows the kernel density probabilities for the Kruger B strains. The actual number of samples for each area (N) is also shown for both maps.
Correlation of calcium content and pH with strain distributions.
Soil calcium and pH are believed to be associated ecologically with regions of anthrax endemicity (4). We performed a nonparametric analysis of variance in order to explore the strain-specific associations to these two environmental factors. The results of a Wilcoxon rank-sum analysis show that Kruger A and B differ statistically significantly in their association with both soil calcium and pH (for pH, Z = 4.870 and P < 0.0001; for calcium, Z = 3.999 and P < 0.0001). Kruger B isolates were found on significantly higher-calcium and higher-pH soils than were Kruger type A isolates (Fig. 5). The different geographic distributions and distinct associations with soil calcium and pH suggest unique environmental requirements and different ecological constraints acting on each group.
FIG. 5.
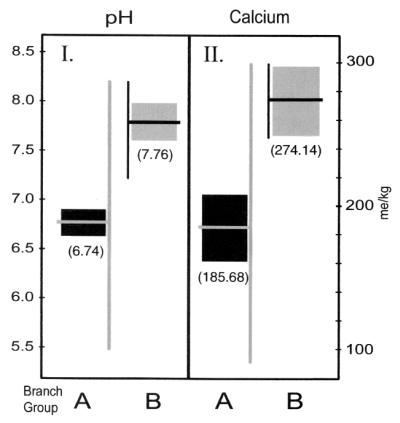
Genotype relationships with soil pH and calcium values. (I) Mean soil pH in parentheses (95% confidence interval and range of values for Kruger A and B genotype groups at the field isolate sites). (II) Mean soil calcium content (95% confidence interval and range of values for Kruger A and B genotype groups). In a nonparametric analysis of variance, the two strain type groups were statistically significantly different with respect to both soil pH (Z = 4.870 and P < 0.0001) and soil calcium (Z = 3.999 and P < 0.0001).
DISCUSSION
We describe the genetic diversity of 98 B. anthracis isolates from the KNP, which we determined by using MLVA, a new, rapid, and robust molecular typing system (9). We have extended previous ecological analyses (11) by classifying the isolates into particular strain types (Kruger A and B) and showed that the two dominant types are clustered in time and space, though not identically. Lastly, we demonstrated that Kruger A and B have significantly different environmental associations in the KNP.
The B. anthracis types in the KNP have the greatest genetic distance among dominant isolates observed in a relatively small geographic area. The genetic distance between Kruger A and B (Fig. 1) is equal to the greatest observed in an extensive worldwide collection (9). Given the inherent homogeneity among B. anthracis strains, we must conclude that the great KNP genetic distances are the result neither of intraepidemic evolution nor of genetic divergence within the time period of this study. Rather, the evolutionary time separating Kruger A and B represents either ancient divergent evolution in the KNP or more recent dispersal of divergent B. anthracis into this area.
The Kruger A strains may represent a diverse B. anthracis type that evolved in southern Africa. Keim et al. hypothesized that southern Africa was the geographic origin of B. anthracis, precisely because of the large genetic diversity observed in the region (7). The rare Kruger A genotypes 39 and 45 may represent older displaced progenitors of the more common genotype 67. Alternatively, it is possible that Kruger A genotype 67 represents an ancient, divergent endemic strain, while the rare genotypes 39 and 45 are recent, independent reintroductions to the area. We cannot, however, rule out the possibility that all Kruger A isolates found in the KNP are the result of one or more introductions via historic or contemporary commerce. While genotype 45 is identical to isolates from Turkey, Argentina, and the United States, it is impossible to discern from these data whether this genotype evolved in southern Africa or was transported to the region.
All of the Kruger B isolates were identical in the study, showing no diversity. Indeed, in a worldwide collection, all B1 genotypes analyzed to date are identical or closely related to those found in the KNP (9). In contrast to type A B. anthracis, the type B isolates are relatively rare and almost exclusive to southern Africa (9). If one of these isolates migrated to the KNP, it would seem more likely that the Kruger A's are more recent introductions and that Kruger B is truly endemic to the proximate KNP region.
The temporal and spatial distributions of the Kruger A and B isolates are overlapping, yet distinct. While analyses performed prior to genotypic group classification yielded positive results for time-space clustering over all isolates (11), classifying the isolates into their genotypic groups reveals the independent time-space clustering of Kruger A and B. This correlated distinction becomes apparent when associations with the environmental factors, soil pH and calcium, are examined.
Kruger A and B have had opportunity over multiple epidemics to be distributed geographically throughout the KNP. However, the genotypic groups show neither a random nor totally overlapping distribution since 1970. Instead, they show distinctly different cluster patterns. The different cluster patterns displayed by Kruger A and B isolates can be attributed to distinct associations with factors in their environment.
Kruger B B. anthracis was very important in the epidemics of 1970 to 1981 and in the northern area of endemicity of the KNP. This epidemic began in the northern KNP and then proceeded to the central and southern areas of the park (3). In the north, isolates that were collected from 1970 to 1981 are both Kruger A and B, while the central isolates are predominantly Kruger A. From these data, we conclude that the earlier anthrax spread was an ecological or environmental progression more than a classic epidemic in which the actual pathogen dispersed. The independence of these foci was masked by correlated environmental cues.
In contrast, the major anthrax outbreak in 1990 was almost entirely composed of Kruger A B. anthracis. This outbreak began in the central regions and progressed to the north. In this epidemic, either environmental induction of independent foci or pathogen dispersal is consistent with this single-strain outbreak. The lack of Kruger B in the northern region in 1990 is puzzling due to its dominance in 1970. However, in early 1990, torrential rains scoured the northern regions and may have severely impacted the spores of the endemic strains. As the anthrax epidemic began in the central region, Kruger A may have physically migrated into an open niche in the northern region. When strain typing is coupled with the geographic and temporal data, it is obvious that anthrax is a dynamic disease that may emanate from several independent foci in close proximity and that these foci may vary across the years. The presence of both types in the northern outbreak of 1970 suggests that independent but environmentally coordinated (induced) foci may exist within regions thought to be single foci.
The important link of calcium and pH to the ecology of B. anthracis has been previously noted in the scientific literature (4). However, the exact nature of this relationship remains the subject of theory and speculation. Regardless of the specific role that these two environmental factors play in the ecology of anthrax, it is seemingly well established that endemicity of B. anthracis is associated with elevated calcium and neutral-to-alkaline soils (4). In this study we have shown a differential association of strain type with these two important environmental factors.
Kruger A strains are associated with broader ranges of soil calcium and pH than Kruger B's (Fig. 5). In the KNP, Kruger A isolates have a wider geographic range and greater genetic diversity than Kruger B isolates (Fig. 1 and 4). Locally and globally, B. anthracis type A isolates reflect a broader geographic range (worldwide) and greater genetic diversity than type B isolates, which are restricted to southern Africa. While this association may be simply fortuitous, adaptation of type A to diverse environmental conditions is consistent with its greater geographic dispersal and genetic dissimilarity. Conversely, type B may have lost the adaptive ability to survive diverse environmental conditions, limiting its geographic dispersal and opportunity to evolve the genetic dissimilarity of type A.
ACKNOWLEDGMENTS
This work was supported by funding from the U.S. Department of Energy (NN20-CBNP), the National Institutes of Health (RO1GM60795), and the Cowden Endowment in Microbiology.
REFERENCES
- 1.De Vos V. Anthrax. In: Coetzer J A W, Thomson G R, Tustin R C, editors. Infectious diseases of livestock with special reference to Southern Africa. Vol. 2. Oxford, United Kingdom: Oxford University Press; 1994. pp. 1262–1289. [Google Scholar]
- 2.De Vos V. The ecology of anthrax in the Kruger National Park. Salisbury Med Bull. 1990;68(Spec. Suppl.):19–23. [Google Scholar]
- 3.De Vos V, Bryden H B. Anthrax in the Kruger National Park: temporal and spatial patterns of disease occurrence. Salisbury Med Bull. 1996;87(Spec. Suppl.):26–30. [Google Scholar]
- 4.Dragon D C, Rennie R P. The ecology of anthrax spores: tough but not invincible. Can Vet J. 1995;36:295–301. [PMC free article] [PubMed] [Google Scholar]
- 5.Farrar W E. Anthrax: from Mesopotamia to molecular biology. Pharos. 1995;58:35–38. [PubMed] [Google Scholar]
- 6.Kaufmann A F, Meltzer M I, Schmid G P. The economic impact of a bioterrorist attack: are prevention and postattack intervention programs justifiable? Emerg Infect Dis. 1997;3:83–94. doi: 10.3201/eid0302.970201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keim P, Kalif A, Schupp J, Hill K, Travis S E, Richmond K, Adair D M, Hugh-Jones M, Kuske C R, Jackson P. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–824. doi: 10.1128/jb.179.3.818-824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keim P, Klevytska A M, Price L B, Schupp J M, Zinser G, Smith K L, Hugh-Jones M E, Okinaka R, Hill K K, Jackson P J. Molecular diversity in Bacillus anthracis. J Appl Microbiol. 1999;87:215–217. doi: 10.1046/j.1365-2672.1999.00873.x. [DOI] [PubMed] [Google Scholar]
- 9.Keim P, Price L B, Klevytska A M, Smith K L, Schupp J M, Okinaka R, Jackson P J, Hugh-Jones M E. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindeque P M, Turnbull P C. Ecology and epidemiology of anthrax in the Etosha National Park, Namibia. Onderstepoort J Vet Res. 1994;61:71–83. [PubMed] [Google Scholar]
- 11.Smith K L, De Vos V, Bryden H B, Hugh-Jones M E, Klevytska A, Price L B, Keim P, Scholl D T. Meso-scale ecology of anthrax in southern Africa: a pilot study of diversity and clustering. J Appl Microbiol. 1999;87:204–207. doi: 10.1046/j.1365-2672.1999.00871.x. [DOI] [PubMed] [Google Scholar]
- 12.Sneath P H A, Sokal R R. Numerical taxonomy. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 13.Turnbull P C. Anthrax vaccines: past, present and future. Vaccine. 1991;9:533–539. doi: 10.1016/0264-410x(91)90237-z. [DOI] [PubMed] [Google Scholar]
- 14.Turnbull P C B, Kramer J M. Bacillus. In: Murray P R, Baron M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 349–356. [Google Scholar]
- 15.Ventor F J. Dissertation. Pretoria, South Africa: University of South Africa; 1990. [Google Scholar]
- 16.Weir B S. Genetic data analysis. Sunderland, Mass: Sinauer Associates, Inc.; 1990. [Google Scholar]