Abstract
The phenotypic and genetic analysis results for 84 isolates of Lactococcus garvieae (including 62 strains from trout with lactococcosis from four different countries, 7 strains from cows and water buffalos with subclinical mastitis, 3 from water, and 10 from human clinical samples) are presented. There was great phenotypic heterogeneity (13 different biotypes) based on the acidification of saccharose, tagatose, mannitol, and cyclodextrin and the presence of the enzymes pyroglutamic acid arylamidase and N-acetyl-β-glucosaminidase. L. garvieae also exhibited high genetic diversity by pulsed-field gel electrophoresis (PFGE), with 19 different pulsotypes among the isolates of L. garvieae studied. Only epidemiologically related strains, like the Spanish and Italian fish isolates and the cow and water buffalo isolates, displayed a close genetic relationship by PFGE, while the strains isolated from sporadic clinical cases, like the human isolates, were genetically unrelated. Overall, a general correlation between phenotypic and genetic data was observed. Epidemiological analysis of biotype and PFGE results indicated that the trout lactococcosis outbreaks in Spain and Portugal and those in France and Italy were produced by genetically unrelated clones. In Spain, two different clones were detected; the outbreaks diagnosed from 1995 onward were produced by a clone (biotype 2, pulsotype A1) which, although genetically related, was different from the one that was responsible for the outbreaks studied between 1991 and 1994 (biotype 1, pulsotype B). The Portuguese isolate had a biochemical profile identical to that of the Spanish strain isolated from 1995 onward and is also genetically closely related to this strain (pulsotype A2). There was a close relationship between the two pulsotypes (E and F) found in the Italian isolates. The French isolate (biotype 3, pulsotype D) was not genetically related to any other L. garvieae fish isolate. These results suggest the existence of diverse infection sources for the different lactococcosis outbreaks.
Lactococcus garvieae, L. lactis subsp. lactis, and L. piscium are the species of the genus Lactococcus with clinical significance in humans and animals (1, 15). L. garvieae is responsible for mastitis in cows and buffalos (9, 32), and it has been isolated from clinical specimens of human blood, urine, and skin (14–16). For this reason, L. garvieae is considered to be an emerging pathogen of increased clinical significance in both veterinary and human medicine.
L. garvieae is also a well-recognized bacterial fish pathogen. The first description in Europe of L. garvieae as a fish pathogen was in 1993 (27). Bacteriologic and molecular studies confirmed L. garvieae as the etiological agent of a hemorrhagic septicemia in farmed trout that was characterized by bilateral exophthalmos; darkening of the skin; congestion of the intestine, liver, kidney, spleen, and brain; and a characteristic hemorrhagic enteritis (10). Previously, in 1991, a new enterococcal species, Enterococcus seriolicida, was described as a new pathogen responsible for an infection of eels and yellowtail with symptoms identical to those produced by L. garvieae in trout (19). Further biochemical, protein profile, 16S rRNA sequencing, and DNA hybridization studies confirmed that L. garvieae and E. seriolicida are the same species (10, 12, 32). The septicemic infection produced by L. garvieae was termed lactococcosis (27) to differentiate it from infections produced by other taxa of gram-positive, catalase-negative cocci, such as Streptococcus iniae, S. parauberis, or Vagococcus salmoninarum, that are usually referred to by the generic term streptococcosis (3, 11, 23, 30, 35). During the last decade, infections by L. garvieae, or the synonymous bacterium E. seriolicida, have been diagnosed in many countries (12, 13, 19, 20). Lactococcosis is actually a worldwide bacterial disease affecting different fish species such as eels, yellowtail, or prawns, although the highest sanitary and economic impact is that on the trout farming industry (8, 12, 20, 26). Despite the increased clinical significance of L. garvieae, studies on the characterization and epidemiological relationship of isolates of this microorganism from different species and/or clinical samples are very limited (13, 32).
This report describes the phenotypic and genetic characterization of L. garvieae isolates from trout with lactococcosis in Spain between 1992 and 1998 and their comparison to the strains of L. garvieae isolated from cases of lactococcosis in other European countries, as well as with L. garvieae isolates from human clinical samples and from cows and water buffalos with subclinical mastitis.
MATERIALS AND METHODS
L. garvieae isolates.
Eighty-four isolates of L. garvieae were studied (Table 1). Sixty-two isolates were recovered from diseased trout with lactococcosis. The 54 Spanish isolates were collected between 1992 and 1998 from fish farms in different geographic areas. The Portuguese, the French, and the six Italian isolates were also recovered from trout with lactococcosis. Three isolates of L. garvieae were recovered from the water pond of a fish farm with chronic lactococcosis. Three L. garvieae isolates from cows, 4 from water buffalo with subclinical mastitis, and 10 from humans and two type strains (L. garvieae ATCC 43921T and E. seriolicida ATCC 491561T) were also included in the study. E. seriolicida ATCC 491561T was purchased from the American Type Culture Collection. All isolates, stored frozen (−80°C), were grown on Columbia blood agar (bioMérieux España, S.A.) at 30°C for 24 h.
TABLE 1.
Data on the L. garvieae strains analyzed in this study
| Isolate(s) | Source | Countrya | Yr of isolation | Phenotypic characteristicsb
|
Biotype | Pulsotype | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sac | Tag | Man | Cedex | Pyra | β-Nag | ||||||
| CP1 | Trout | Spain/SF1 | 1991 | + | + | + | − | + | − | 1 | B |
| CP2 | Trout | Spain/SF1 | 1992 | + | + | + | − | + | − | 1 | B |
| 94/32 | Trout | Spain/SF1 | 1994 | + | + | + | − | + | − | 1 | B |
| 1581, 95/861 | Trout | Spain/SF2 | 1995 | + | + | + | + | + | − | 2 | A1 |
| 1336 | Trout | Spain/SF3 | 1996 | + | + | + | + | + | − | 2 | A1 |
| 1182 | Trout | Spain/SF4 | 1996 | + | + | + | + | + | − | 2 | A1 |
| 1184, 1185 | Trout | Spain/SF5 | 1996 | + | + | + | + | + | − | 2 | A1 |
| 1987, 1999, 2000, 2013, 2014, 2001, 2002, 2007, 2005, 2006, 4316 | Trout | Spain/SF1 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 1982 | Trout | Spain/SF6 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 1989, 1990, 1919, 1758, 1964, 1759, 1940, 1944, 1957, 1967 | Trout | Spain/SF7 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 1935, 1936, 1909, 1893, 1934, 1955, 1954, 1924, 1925 | Trout | Spain/SF8 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 1958, 1981, 1956, 1975, 2016, 2017 | Trout | Spain/SF9 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 1960, 1961, 1988 | Trout | Spain/SF10 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 1966 | Trout | Spain/SF11 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 1959, 1986, 1985, 4294 | Trout | Spain/SF12 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 864 | Trout | Portugal | 1995 | + | + | + | + | + | − | 2 | A2 |
| 2398 | Trout | France | 1998 | − | + | + | − | + | − | 3 | D |
| 1691 | Trout | Italy/IF1 | 1997 | − | − | + | − | − | − | 4 | F2 |
| 1683 | Trout | Italy/IF2 | 1997 | − | − | + | − | − | − | 4 | F1 |
| 1684 | Trout | Italy/IF3 | 1997 | − | − | + | − | − | + | 5 | E |
| 1685, 1689 | Trout | Italy/IF4 | 1997 | − | − | + | + | − | + | 6 | F1 |
| 1687 | Trout | Italy/IF5 | 1997 | − | − | + | + | − | + | 6 | E |
| 2008, 4284 | Water | Spain/SF1 | 1998 | + | + | + | + | + | − | 2 | A1 |
| 4289 | Water | Spain/SF1 | 1998 | − | + | + | − | + | − | 7 | K |
| 34, 41, 42 | Cow | Spain | 1999 | − | + | + | − | + | − | 7 | J |
| 1205, 1193, 1183 | Buffalo | Brazil | 1998 | − | − | − | − | + | − | 8 | P1 |
| 1195 | Buffalo | Brazil | 1998 | − | − | − | − | + | + | 9 | P2 |
| IM-86 | Human | Brazil | 1998 | − | − | + | + | − | − | 10 | Q |
| 306-79 | Human | United States | 1997 | − | + | + | + | + | − | 11 | S |
| 673-80, 588-80 | Human | United States | 1980 | − | − | + | + | − | − | 10 | R |
| 2182-81 | Human | United States | 1981 | + | + | + | − | + | − | 1 | H |
| 1108-86 | Human | United States | 1986 | + | + | + | − | + | − | 1 | N |
| 2486-87 | Human | United States | 1987 | + | + | + | + | + | − | 2 | O |
| 364-88 | Human | United States | 1988 | + | + | + | − | − | − | 12 | G |
| 240-88 | Human | United States | 1988 | + | + | + | − | − | − | 12 | L |
| 66-90 | Human | United States | 1990 | − | + | + | − | − | − | 13 | M |
| L. garvieae ATCC 43931T | Cow | United Kingdom | 1984 | − | − | + | − | − | − | 4 | I |
| E. seriolicida ATCC 49156T | Yellowtail | Japan | 1991 | − | − | + | + | − | − | 10 | C |
SF and IF are different fish farms in Spain and Italy, respectively. Fish farms SF1, SF3, SF4, and SF6 are located in the central region of Spain; SF2, SF5, and SF12 are located in the north of Spain; SF7 is in the northwest of Spain; SF8, SF9, and SF10 are located in the west of Spain; SF11 is located in the south of Spain. The IF fish farms are located in the north of Italy.
Sac, Tag, Man, and Cedex, acidification of saccharose, tagatose, mannitol, and cyclodextrin, respectively. Pyra and β-Nag, presence of the respective enzymes.
Biochemical and enzymatic characterization and PCR assay.
Biochemical and enzymatic tests were performed with the Rapid ID 32 Strep and API 50CH systems (bioMérieux España, S.A.) by following the manufacturer's instructions, except for the temperature of incubation, which was always 30°C. The identification of the L. garvieae isolates was confirmed by PCR assay as described by Zlotkin et al. (36).
Analysis of chromosomal DNA restriction patterns by PFGE.
Lactococcus cells were grown aerobically in brain heart infusion broth (Difco) at 37°C to an A610 of approximately 0.6. The cells were harvested by centrifugation (3,500 × g, 10 min, 4°C) and washed twice with buffer (10 mM Tris-HCl, 10 mM EDTA, 10 mM EGTA, 1 M NaCl [pH 8]). Agarose plugs were made from a 1:1 mixture of 2% low-melting-point agarose and the cell suspension. The plugs were lysed in buffer (6 mM Tris-HCl, 100 mM EDTA, 1 M NaCl, 0.5% [wt/vol] Brij 58, 0.2% [wt/vol] sodium deoxycholate, 0.5% [vol/vol] lauroyl sarcosine, lysozyme at 5 mg/ml) for 24 h at 37°C. The cells were treated for 50 h at 56°C with the same volume of a solution (0.25 M EDTA, 20 mM NaCl, 1% lauroyl sarcosine [pH 9]) containing proteinase K at 2.5 mg/ml and washed three times with Tris-EDTA buffer for 1 h at 4°C. ApaI (Promega Co.) was used for restriction endonuclease digestion in accordance with the manufacturer's instructions. The fragments were resolved by pulsed-field gel electrophoresis (PFGE) with electrophoresis- grade agarose (1%; Boehringer Mannheim) by using a CHEF-DR III System (Bio-Rad). The following parameters were used: running time, 21 h; temperature, 14°C; voltage gradient, 200 V; initial pulse time, 0.1 s; final pulse time, 25 s; included angle, 120°. The gels were stained with ethidium bromide (0.5 μg/ml) for 15 min, destained in distilled water, and photographed under UV light. A lambda ladder PFGE marker (Boehringer Mannheim) was used for molecular size determination.
PFGE pattern analysis.
The similarities between restriction endonuclease digestion profiles (REDPs) were expressed as Jaccard similarity indexes determined by the numerical taxonomy program TAXAN (Information Resources Group, Maryland Biotechnology Institute, University of Maryland, College Park). A similarity matrix was computed and transformed into an agglomerative cluster using the unweighted pair group method with arithmetic averages (UPGMA) (31).
RESULTS
Phenotypic studies and PCR identification.
All of the L. garvieae isolates studied gave the expected 1,100-bp PCR amplification product, which is specific for this microorganism (36), confirming the preliminary biochemical identification. There was great biodiversity among the L. garvieae isolates, with 13 different biotypes, based on the acidification of saccharose, tagatose, mannitol, and cyclodextrin and the presence of the enzymes pyroglutamic acid arylamidase (Pyra) and N-acetyl-β-glucosaminidase (β-Nag) (Table 1). All of the strains of L. garvieae, except the water buffalo isolates, produced acid from mannitol. The enzyme Pyra was present in all of the trout isolates except the Italian strains. The cow and water buffalo isolates, as well as 6 out of the 10 human isolates analyzed, were Pyra negative. All of the Spanish fish isolates and the Portuguese isolate produced acid from sucrose and tagatose. The Spanish isolates recovered in 1991, 1992, and 1994 did not produce acid from cyclodextrin, and the enzyme β-Nag was not present (biotype 1). On the other hand, the Portuguese isolate and Spanish strains isolated after 1995 produced acid from cyclodextrin (biotype 2). The French isolate produced acid only from tagatose, and it did not produce acid from cyclodextrin, nor did it have the enzyme β-Nag (biotype 3). The Italian strains did not produce acid from either sucrose or tagatose. Two strains neither produced acid from cyclodextrin nor had the enzyme β-Nag (biotype 4), one strain did not produce acid from cyclodextrin but did have the enzyme β-Nag (biotype 5), and the other three strains produced acid from cyclodextrin and had the enzyme β-Nag (biotype 6).
The strains isolated from cows produced acid from tagatose but not from sucrose and cyclodextrin, and the enzyme β-Nag was not present (biotype 7). The water buffalo strains did not acidify sucrose, tagatose, or cyclodextrin. Three strains did not have the enzyme β-Nag (biotype 8), while the remaining strain produced this enzyme (biotype 9). Human isolates exhibited the greatest diversity, with eight different biochemical-enzymatic profiles (biotypes 1, 2, and 10 to 13). Three of the L. garvieae strains from water exhibited biotype 2, and the other strain had biotype 7.
PFGE analysis.
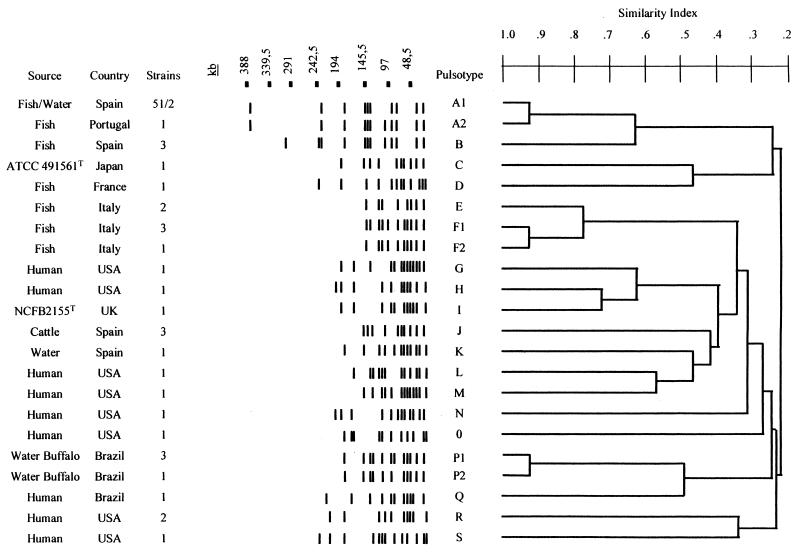
The ApaI enzyme generated 19 different DNA fragment profiles, designated pulsotypes A to S, with 7 to 13 bands over a size ranging of about 25 to 343 kb. Figure 1 shows the dendrogram obtained with the 19 different patterns after UPGMA clustering. The Spanish clinical isolates exhibited two different ApaI patterns: pulsotype B for the strains isolated in 1991, 1992, and 1994 and pulsotype A (subtype A1) for all of the L. garvieae strains isolated from 1995 onward, irrespective of geographic origin. The Portuguese strain was closely related to the Spanish isolates of pulsotype A, differing from the pattern displayed by these strains by only one restriction fragment and referred to as subtype A2. The Italian strains displayed two different patterns, designated pulsotypes E and F, that clustered in a genetically related cluster (0.8 index similarity). Pulsotype F had two subtypes (F1 and F2). The French isolate belonged to a different group (pulsotype D).
FIG. 1.
Dendrogram of L. garvieae isolates based on UPGMA cluster analysis of the 20 different ApaI pulsotypes obtained in this study.
The strains isolated from water buffalos with subclinical mastitis displayed two very similar REDPs with a similarity index of 0.95. The two REDPs differed by only one band and were considered to be two subtypes (P1 and P2) of the same group (pulsotype P). The three strains isolated from cows had the same pulsotype (J). Two of the strains isolated from water exhibited the same pulsotype as the Spanish clinical isolates (A1); the other one was not genetically related (pulsotype K). The human isolates exhibited great diversity, with nine different REDPs (pulsotypes G, H, L, M, N, O, Q, R, and S) with a low level of similarity, which agrees with the phenotypic diversity of these isolates.
DISCUSSION
Lactococcosis is one of the infectious diseases with significant economic and sanitary repercussions for trout farms in Mediterranean countries during the summer months (12, 13, 19, 20, 26, 30). Only two studies concerned with the epidemiological characterization of L. garvieae have been published until now. In the first one, isolates responsible for subclinical mastitis in water buffalos, together with human clinical isolates, were characterized by PFGE (6) and in the second one, a total of 15 strains isolated from diseased fish from different countries were analyzed by ribotyping (13). This epidemiological study was based on PFGE results because of the high discriminative power of PFGE (2, 18, 21, 22), and the phenotypic characterization was done to correlate the phenotypic and genetic data. With a total of 84 isolates of L. garvieae analyzed, this is the largest phenotypic and genetic comparative study trying to correlate these data and it is also the first one including isolates representative of the main diseases caused by L. garvieae and the species in which they have been diagnosed.
Most of the papers published to date have used the API 50CH and API 20 Strep systems for the phenotypic characterization of L. garvieae (7, 8, 13, 26, 34). Our results for those tests included in both of these commercial systems are identical to those reported by the authors of those papers, as well as to those of studies in which conventional biochemical characterization was used (5). The presence of the enzymes Pyra (83% of positive strains), β-Nag (6% of positive strains), alanine-phenylalanine-proline arylamidase (100% of positive strains), glycyl-tryptophan arylamidase (0% of positive strains), and urease (0% of positive strains) and the acidification of cyclodextrin (75% of positive strains), pullulan (0% of positive strains), methyl-β-d-glucopyranoside (100% of positive strains), and β-mannosidase (0% of positive strains) are included only in the Rapid ID 32 Strep system. Although it is not possible to compare the results of these tests with those obtained by other authors, our results agree with those of the bioMérieux database for L. garvieae. With the Rapid ID 32 Strep identification system, all of the Spanish, French, and Portuguese isolates, as well as the type strains of L. garvieae and E. seriolicida, gave negative results for the acidification of ribose. However, all of the strains were systematically positive for this test with the API 50CH system, which agrees with previous descriptions of this species (8–10, 12, 32). These results should be taken into account when this system is used for the routine identification of clinical isolates of L. garvieae.
Our results indicated great phenotypic heterogeneity and genetic diversity among the L. garvieae isolates studied, with a generally good correlation between the phenotypic and genetic properties of L. garvieae. This correlation was especially evident for the trout, cow, and water buffalo isolates. However, two human strains (2182-81 and 1108-86) had the same biotype as the 1991 to 1994 Spanish fish isolates but very different pulsotypes. Similarly, human isolate 2486-87 and the Spanish fish isolates recovered in 1995 also had the same biotype but different pulsotypes (Table 1; Fig. 1). The existence of different isolates that have the same biotype but differ by genetic methods has also been observed in other microorganisms (25, 28) and can be explained by the much higher discriminatory power of PFGE than biotyping (2).
Only epidemiologically related strains, like the Spanish and Italian fish isolates and the cow and water buffalo isolates, displayed a close genetic relationship by PFGE (Fig. 1). The close relationship of the Italian isolates found by us with PFGE is similar to that of other Italian L. garvieae strains found by ribotyping (13). The single French isolate was not genetically related to any other L. garvieae fish isolate. In Spain, two different clones were implicated in the lactococcosis outbreaks between 1991 and 1998. The outbreaks diagnosed since 1995 were produced by a clone (biotype 2, pulsotype A1) which, according to Tenover et al. (33), although genetically related, was different from the one responsible for the outbreaks studied between 1991 and 1994 (biotype 1, pulsotype B). The single Portuguese isolate analyzed had a biochemical profile identical to that of the Spanish strain isolated from 1995 onward and is also closely genetically related to this strain, indicating a clonal relationship between them. Three biotypes had been established for the L. garvieae strains isolated from fish based on their ability to acidify sucrose and/or tagatose (13). The 62 Spanish trout isolates studied by us all produced acid from both sugars, while the 6 Italian isolates did not acidify either sugar. Identical results have been described for other Spanish and Italian L. garvieae fish isolates (13, 26). Thus, the phenotypic differences observed between the clinical fish isolates from different geographical areas, in correlation with the PFGE results, could be useful as phenotypic epidemiological markers for lactococcosis outbreaks. On the other hand, the strains isolated from sporadic clinical cases, like the human isolates, were genetically unrelated, with a similarity index usually lower than 0.5 (33), results that agree with previous reports for other widely distributed pathogens (17). The low genetic relatedness observed between different isolates may suggest that some of the most diverse strains are not L. garvieae. This low genetic relatedness might reflect bacterial populations genetically isolated but showing a high recombination rate. So, after long-term genetic isolation, produced by a difference in geographical or habitat distribution, the bacterial populations show a great divergence. Similar low genetic relatedness has been observed in Neisseria isolates by using PFGE analysis (4). Nevertheless, the identities of those most diverse isolates had been previously confirmed. Thus, the identities of the CP1 and CP2 Spanish trout isolates of pulsotype B were confirmed by sequencing of their 16S rRNA genes (10), and those of the buffalo isolates (pulsotypes P1 and P2) were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of whole-cell protein and DNA-DNA hybridization analysis (32). In addition, all of the strains included in the study gave the expected 1,100-bp PCR amplification product with the specific L. garvieae PCR assay (36), all which precludes any doubt about the identity of all of the strains included in the study as L. garvieae.
Unlike other highly host-adapted bacterial fish pathogens, L. garvieae is pathogenic for several fish species, ruminants, and humans and could therefore be considered potentially zoonotic. However, the low genetic relatedness between the animal and human isolates included in this study (Fig. 1) does not allow confirmation of this hypothesis. Asymptomatic infected fish can act as carriers (24), contributing to the dissemination and transmission of L. garvieae through the trade of livestock. This fact could explain the rapid spread of the L. garvieae pulsotype A strain, first isolated in the north of Spain in 1995, throughout different Spanish regions in only 4 years. L. garvieae is eliminated through the feces of diseased or carrier fish (24). This is probably the origin of the 98/4284 and 2008 L. garvieae strains, which had the same biotype and pulsotype as the clinical isolates (2; A1) and were isolated from the water of a fish farm in which lactococcosis was endemic during the latter years. Therefore, the contaminated water seems to play an important role in the horizontal transmission of lactococcosis, probably through the fecal-oral route (29). L. garvieae is also widespread in the environment (19), and this could be the origin of strain 98/4289. However, from the facts that this strain had the same biotype (biotype 7) as the cow isolates and was genetically more closely related to these strains and some of the human isolates than to the Spanish fish clinical strains (Table 1; Fig. 1) arise the possibilities that some environmental L. garvieae strains evolved from mammals and that they are hypothetically implicated in the epidemiology of fish lactococcosis. However, our results cannot substantiate this hypothesis. In order to do that, it will be necessary to carry out larger molecular epidemiological studies including environmental, human, and animal L. garvieae isolates, as well as experiments to assess the pathogenicity of animal or human isolates from different fish species, mainly for trout.
In summary, the analysis of phenotypic and genetic results indicated that trout lactococcosis outbreaks in Spain and Portugal and those in France and Italy were produced by genetically unrelated clones, suggesting the existence of diverse infection sources for the different lactococcosis outbreaks.
ACKNOWLEDGMENTS
This work was supported by Dibaq-Diproteq S.A.
We thank L. M. Teixeira of the Instituto de Microbiología, Universidade Federal do Rio de Janeiro, for providing the water buffalo and human isolates and L. garvieae ATCC 43921T; C. Michel of the Unité de Virologie et Immunologie Moléculaires, INRA, Jouy-en-Josas, France, for providing the French isolate; A. Rodríguez of Dibaq-Diproteg for the Italian isolates; and M. T. Cutuli, A. Doménech, and J. A. García of the Departamento de Patología Animal I, Facultad de Veterinaria de Madrid, for isolating some of the Spanish trout isolates.
REFERENCES
- 1.Aguirre M, Collins M D. Lactic acid bacteria and human clinical infection. J Appl Bacteriol. 1993;75:95–107. doi: 10.1111/j.1365-2672.1993.tb02753.x. [DOI] [PubMed] [Google Scholar]
- 2.Arbeit R D. Laboratory procedures for the epidemiological analysis of microorganisms. In: Murray P R, Baron E J O, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 190–208. [Google Scholar]
- 3.Austin B, Austin D A. Bacterial fish pathogens: diseases in farmed and wild fish. 2nd ed. Chichester, United Kingdom: Ellis Horwood Limited; 1993. pp. 208–216. [Google Scholar]
- 4.Berrón S, de la Fuente L, Martín E, Vázquez J A. Increasing incidence of meningococcal disease in Spain associated with a new variant of serogroup C. Eur J Clin Microbiol Infect Dis. 1998;17:85–89. doi: 10.1007/BF01682161. [DOI] [PubMed] [Google Scholar]
- 5.Carson J, Gudkovs N, Austin B. Characteristics of an Enterococcus-like bacterium from Australia and South Africa, pathogenic for rainbow trout, Oncorhynchus mykiss (Walbaum) J Fish Dis. 1993;16:381–388. [Google Scholar]
- 6.Carvalho M G, Vianni M C, Elliot J A, Reeves M, Facklam R R, Teixeira L M. Molecular analysis of Lactococcus garvieae and Enterococcus gallinarum isolated from water buffalos with subclinical mastitis. Adv Exp Med Biol. 1997;418:401–404. doi: 10.1007/978-1-4899-1825-3_96. [DOI] [PubMed] [Google Scholar]
- 7.Ceschia G, Giorgetti G, Giavenni R, Sarti M. A new problem for Italian trout farms: streptococcosis in rainbow trout (Oncorhynchus mykiss) Bull Eur Assn Fish Pathol. 1992;12:71–72. [Google Scholar]
- 8.Cheng W, Chen J. Isolation and characterization of an Enterococcus-like bacterium causing muscle necrosis and mortality in Macrobrachium rosenbergii in Taiwan. Dis Aquat Org. 1998;34:93–101. doi: 10.3354/dao034093. [DOI] [PubMed] [Google Scholar]
- 9.Collins M D, Farrow J A E, Phillips B A, Kandler O. Streptococcus garvieae and Streptococcus plantarum sp. nov. J Gen Microbiol. 1983;129:3427–3431. doi: 10.1099/00221287-129-11-3427. [DOI] [PubMed] [Google Scholar]
- 10.Doménech A, Prieta J, Fernández-Garayzábal J F, Collins M D, Jones D, Domínguez L. Phenotypic and phylogenetic evidence for a close relationship between Lactococcus garvieae and Enterococcus seriolicida. Microbiologia. 1993;9:63–68. [PubMed] [Google Scholar]
- 11.Doménech A, Fernández-Garayzábal J F, Pascual C, García J A, Cutuli M T, Moreno M A, Collins M D, Domínguez L. Streptococcosis in cultured turbot (Schophtalmus maximus) associated with Streptococcus parauberis. J Fish Dis. 1996;19:33–38. [Google Scholar]
- 12.Eldar A, Ghittino C, Asanta L, Bozzetta E, Gloria M, Prearo M, Bercovier H. Enterococcus seriolicida is a junior synonym of Lactococcus garvieae, a causative agent of septicemia and meningoencephalitis in fish. Curr Microbiol. 1996;32:85–88. doi: 10.1007/s002849900015. [DOI] [PubMed] [Google Scholar]
- 13.Eldar A, Gloria M, Ghittino C, Zlotkin A, Bercovier H. Biodiversity of Lactococcus garvieae strains isolated from fish in Europe, Asia, and Australia. Appl Environ Microbiol. 1999;65:1005–1008. doi: 10.1128/aem.65.3.1005-1008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliot J A, Collins M D, Pigott N E, Facklam R R. Differentiation of Lactococcus lactis and Lactococcus garvieae from human by comparison of whole-cell protein patterns. J Clin Microbiol. 1991;29:2731–2734. doi: 10.1128/jcm.29.12.2731-2734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facklam R R, Elliot J A. Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin Microbiol Rev. 1995;8:479–495. doi: 10.1128/cmr.8.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fefer J J, Ratzan K R, Sharp S E, Saiz E. Lactococcus garvieae endocarditis: report of a case and review of the literature. Diagn Microbiol Infect Dis. 1998;32:127–130. doi: 10.1016/s0732-8893(98)00065-0. [DOI] [PubMed] [Google Scholar]
- 17.Hytiä E, Hielm S, Björkroth J, Korreala H. Biodiversity of Clostridium botulinum type E strains isolated from fish and fishery products. Appl Environ Microbiol. 1999;65:2057–2064. doi: 10.1128/aem.65.5.2057-2064.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly W J, Davey G P, Ward L J. Characterization of lactococci isolated from minimally processed fresh fruit and vegetables. Int J Food Microbiol. 1998;45:85–92. doi: 10.1016/s0168-1605(98)00135-4. [DOI] [PubMed] [Google Scholar]
- 19.Kusuda R, Kawai K, Salati F, Banner C R, Fryer J L. Enterococcus seriolicida sp. nov., a fish pathogen. Int J Syst Bacteriol. 1991;41:406–409. doi: 10.1099/00207713-41-3-406. [DOI] [PubMed] [Google Scholar]
- 20.Kusuda R, Salati F. Enterococcus seriolicida and Streptococcus iniae. In: Woo P T K, Bruno D W, editors. Fish diseases and disorders, vol. 3. Viral, bacterial, and fungal infections. Wallingford, United Kingdom: CAB International Publishing; 1999. pp. 303–317. [Google Scholar]
- 21.Le Bourgeois P, Mata M, Rizenthaler P. Genome comparison of Lactococcus strains by pulsed-field gel electrophoresis. FEMS Microbiol. 1989;50:65–69. doi: 10.1016/0378-1097(89)90460-6. [DOI] [PubMed] [Google Scholar]
- 22.Louie M, Jayaratne P, Luchsinger I, Devenish J, Yao J, Schlech W, Simor A. Comparison of ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for molecular typing of Listeria monocytogenes. J Clin Microbiol. 1996;34:15–19. doi: 10.1128/jcm.34.1.15-19.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel C, Nougayrede P, Eldar A, Sochon E, De Kinkelin P. Vagococcus salmoninarum, a bacterium of pathological significance in rainbow trout Oncorhynchus mykiss farming. Dis Aquat Org. 1997;30:199–208. [Google Scholar]
- 24.Múzquiz J L, Royo F M, Ortega C, de Blas I, Ruiz I, Alonso J L. Pathogenicity of streptococcosis in rainbow trout (Oncorhynchus mykiss): dependence on age and diseased fish. Bull Eur Assn Fish Pathol. 1999;19:114–119. [Google Scholar]
- 25.Nazarowec-White M, Farber J M. Phenotypic and genotypic typing of food and clinical isolates of Enterobacter sakazakii. J Med Microbiol. 1999;48:559–567. doi: 10.1099/00222615-48-6-559. [DOI] [PubMed] [Google Scholar]
- 26.Palacios M A, Zamora M J, Vázquez J, Zamora E, Duran A. Streptococcosis in rainbow trout (Oncorhynchus mykiss) in Spain. Boll Soc It Patol Ittica. 1993;13:11–16. [Google Scholar]
- 27.Prieta J, Doménech A, Fernández-Garayzábal J F, Collins M D, Rodríguez U M, Jones D, Rodríguez A, Domínguez L. Lactococcosis de la trucha arco iris. Med Vet. 1993;10:367–373. [Google Scholar]
- 28.Privitera A, Licciardello L, Giannino V, Agodi A, Rappazzo G, Nicoletti G, Stefani S. Molecular epidemiology and phylogenetic analysis of Haemophilus parainfluenzae from chronic obstructive pulmonary disease exacerbations. Eur J Epidemiol. 1998;14:405–412. doi: 10.1023/a:1007405406617. [DOI] [PubMed] [Google Scholar]
- 29.Romalde J L, Magariños B, Núñez S, Barja J L, Toranzo E. Host range susceptibility of Enterococcus sp. strains isolated from diseased turbot: possible routes of infection. Appl Environ Microbiol. 1996;62:607–611. doi: 10.1128/aem.62.2.607-611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schäperclaus W. Diseases caused by pathogens. In: Schäperclaus W, Kulow H, Schreckenbach K, editors. Fish diseases. Vol. 1. Rotterdam, The Netherlands: A. A. Balkema; 1992. pp. 578–584. [Google Scholar]
- 31.Sneath P H, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman & Co.; 1973. [Google Scholar]
- 32.Teixeira L M, Merquior V L C, Vianni M C E, Carvalho M G S, Fracalanzza S E L, Steigerwalt A G, Brenner D J, Facklam R R. Phenotypic and genotypic characterization of atypical Lactococcus garvieae strains isolated from water buffalos with subclinical mastitis and confirmation of L. garvieae as a senior subjective synonym of Enterococcus seriolicida. Int J Syst Bacteriol. 1996;46:664–668. doi: 10.1099/00207713-46-3-664. [DOI] [PubMed] [Google Scholar]
- 33.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toranzo A E, Devesa S, Heinen P, Riaza A, Nuñez S, Barja J L. Streptococcosis in cultured turbot caused by an enterococcus-like bacterium. Bull Eur Assn Fish Pathol. 1994;14:19–23. [Google Scholar]
- 35.Wallbanks S, Martínez-Murcia A J, Fryer J L, Phillips B A, Collins M D. 16S rRNA sequence determination for members of the genus Carnobacterium and related lactic acid bacteria and description of Vagococcus salmoninarum. Int J Syst Bacteriol. 1990;40:224–230. doi: 10.1099/00207713-40-3-224. [DOI] [PubMed] [Google Scholar]
- 36.Zlotkin A, Eldar A, Ghittino C, Bercovier H. Identification of Lactococcus garvieae by PCR. J Clin Microbiol. 1998;36:983–985. doi: 10.1128/jcm.36.4.983-985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]