Abstract
The routine identification of Aspergillus fumigatus in clinical samples involves, apart from direct examination, the isolation of the organism on a plate followed by its microscopic characterization. This approach lacks sensitivity, specificity, and speed. A new procedure has been developed combining microcolony formation on a nylon membrane filter at 45°C with the detection of a specific 4-methylumbelliferyl-α-l-arabinopyranoside cleaving enzyme activity in digitonin permeabilized cells. The test takes approximately 14 h and has an efficiency of 98.2% and false-positive and -negative rates of 0 and 3.1%, respectively. When applied to 188 clinical samples taken from patients with proven or nonproven presence of Aspergillus species, a good agreement with the conventional plate-microscopy method was obtained.
Invasive aspergillosis, mainly caused by Aspergillus fumigatus, is a life-threatening condition that requires prompt antifungal therapy (6, 8). Rapid and accurate diagnosis of aspergillosis is still hampered by shortcomings in the currently available methodology. Several PCR and enzyme-linked immunosorbent assay methods have recently been reported (10, 13, 16). However, isolation of the organism on an agar plate followed by microscopic identification of the growth remains the backbone of the laboratory diagnosis in daily routine (11, 12). This type of procedure may take up to 5 days, and other mold species—e.g., Fusarium sp., Scedosporum sp., and Pseudallescheria sp.—may exhibit some of the morphological characteristics of Aspergillus sp. (11). False-negative cultures of blood and bronchoalveolar lavage fluid are common (8, 12, 13).
To improve the sensitivity, specificity, and rapidity of the routine culture approach, a new procedure has been developed combining membrane filtration, microcolony formation on a selective medium at 45°C, and the detection of a specific enzyme activity in digitonin-permeabilized cells. The result is a simple and cost-effective procedure that is capable of detecting A. fumigatus in a filtered sample in approximately 14 h. The present paper describes the development and the validation of this method with 188 clinical samples from hospitalized patients. (The method is subject to a pending patent) [H. J. Nelis and T. G. M. Bauters, 10 March 2000, European Patent Application EP 00870041.1].)
(This work has been presented in part at the 99th General Meeting of the American Society for Microbiology, Chicago, Ill., 29 May to 3 June 1999.)
MATERIALS AND METHODS
Fungal strains for method development.
A total of 134 Aspergillus and 30 non-Aspergillus sp. laboratory strains were used for the method development. These included A. fumigatus (n = 48), Aspergillus flavus (n = 29), Aspergillus niger (n = 22), Aspergillus clavatus (n = 4), Aspergillus nidulans (n = 13), Aspergillus terreus (n = 12), Aspergillus versicolor (n = 6), Fusarium oxysporum (n = 4), Fusarium solani (n = 4), Rhizopus oryzae (n = 4), Rhizopus microsporus (n = 3), Absidia corymbifera (n = 2), Rhizomucor pusillus (n = 4), Penicillium marneffei (n = 5), and Pseudallescheria boydii (n = 4). The strains were obtained from the Mycothèque de l'Université Catholique de Louvain-La-Neuve (Louvain-La-Neuve, Belgium) (A. fumigatus 978 and 15821; A. flavus 1032 and 19007; and A. niger 13608, 19001, and 30113) and from the Institute of Hygiene and Epidemiology, Division of Mycology (Brussels, Belgium) (A. fumigatus 1995, 1997 to 2001, 2003, 2494, 2943, 2945, 2952, 3007, 3125, 3131, 3242, 3768, and 4182 to 4189; A. flavus 306, 627, 2262, 2465, 2647, 2700, 3018, 3719, 3790, 4390, 5094, 5285, 5669, 5675, 5721, 5730, 5901 to 5904, 5906 to 5908, 6741, 9407, and 9408; A. niger 2312, 2864, 2951, 3019, 3415, 3766, 3797, 4023, 4461, 4781, 5296, 5788, 6147, 6350, 6727, 9673, 9709, and 14389; A. clavatus 5138, 6078, 7944, and 9776; A. nidulans 1502, 1503, 1548, 2059, 3563, 3793, 4190, 5137, 5231, 5589, 6365, 9353, and 9679; A. terreus 307, 349, 1945, 2499, 3280, 4395, 5677, 5918, 6640, 6946, and 7940; A. versicolor 2157, 2158, 2646, 2916, 2983, and 3202; F. oxysporum 9571, 3545, and 14513; F. solani 591, 4831, 5601, and 6743; R. oryzae 5215, 5233, 6016, and 6017; R. microsporus 5208, 5234, and 7978; A. corymbifera 3106 and 5092; R. pusillus 1387, 2020, 4897, and 10343; P. marneffei 3272, 4176, and 6051 to 6053; and P. boydii 1055, 3724, 3725, and 3747). The remaining strains were clinical isolates originating from the Laboratory of Bacteriology and Virology of the University Hospital of Ghent, Ghent, Belgium. Isolates were subcultured on Sabouraud glucose agar (SGA) (Difco Laboratories, Detroit, Mich.) and were incubated for at least 72 h at 37°C.
Clinical specimens.
Ear swabs (n = 26), sputa (n = 69), bronchoalveolar liquids (n = 29), sera (n = 4), a tube culture (n = 1), nose swabs (n = 57), and blood samples (n = 2) were provided by the Laboratory of Bacteriology and Virology, and the departments of Otorhinolaryngology and Hematology of the University Hospital of Ghent.
Filters, growth media, chemicals, and reagents.
Nylon membrane filters (MFs) (47-mm diameter; 0.45-μm pore size) (Gelman Sciences, Ann Arbor, Mich.) were used for filtration of the samples. Other filters, including nitrocellulose, polyamide, polysulfone, mixed cellulose ester, and polytetrafluoroethylene filters, were obtained from Millipore Corp. (Bedford, Mass.). Absorbent fiberglass pads were from Gelman Sciences.
SGA was used unmodified and supplemented with an antibacterial mixture (SGA-T), extra growth factors (SGA*), or a combination of both (SGA*-T). The antibacterial mixture consisted of ticarcillin (2,344 μg/ml)-clavulanic acid (156 μg/ml) (Timentin; SmithKline Beecham Pharma, Genval, Belgium). SGA* was an enriched SGA containing pyridoxine (10 μg/ml), thiamine (1.0 μg/ml), nicotinic acid (0.1 μg/ml), riboflavin (10 μg/ml), pantothenate (1.0 μg/ml), para-aminobenzoic acid (1.0 μg/ml), and inositol (10 μg/ml) (all from Sigma Chemical Co., St. Louis, Mo.). SGA*-T contained both the antibacterial mixture and the growth factors. Other growth media included potato glucose agar (PGA) (Difco), SGA supplemented with chloramphenicol (SGA+) (bioMérieux Vitek, Hazelwood, Mo.), SGA supplemented with gentamicin and vancomycin (Sigma), cornmeal agar with 0.5% Tween 80 (CMT) (Difco), and Czapek-Dox agar (CDA) (1).
Screening for enzymatic activities.
The following 4-methylumbelliferyl (4-MU) derivatives were tested as enzyme substrates: acetate, N-acetyl-β-d-galactosaminide, α-l-arabinofuranoside, α-l-arabinopyranoside, α-d,d′-diacetylchitobioside, β-d-galactoside, β-d-glucoside, β-d-glucuronic acid, heptanoate, α-l-iduronide, β-d-lactoside, laurate, oleate, palmitate, phosphate, pyrophosphate, and α-l-rhamnopyranoside. They were purchased from Sigma and Melford Laboratories (Ipswich, United Kingdom).
A 0.1- to 1-mg quantity of each substrate was dissolved in 1 ml of dimethyl sulfoxide (Fluka, Buchs, Switserland), except 4-MU-palmitate and 4-MU-oleate, which were dissolved in dimethylformamide (Merck, Darmstadt, Germany). Stock solutions were diluted in 0.1 M phosphate buffer (pH 4.5) and sterilized by filtration over Nalgene disposable units (0.45-μm pore size; 250 ml; Nalge Co., Rochester, N.Y.). The solutions were dispensed in test tubes and frozen at −20°C until use.
Procedure.
Sputum and other mucous samples were first treated with 1% N-acetyl-l-cysteine (Sigma) for 30 min at 37°C to reduce their viscosity. In case of an insufficient filtration volume, they were diluted in 10 ml of physiological saline. The samples were divided in two equal parts and separately filtered over a 47-mm-diameter nylon MF, with a pore size of 0.45 μm. After filtration, each of the membranes was placed on SGA-T* medium and incubated at 45 and 37°C, respectively, for at least 12 h to yield microcolonies. As soon as colorless microcolonies became visible to the naked eye in daylight, the filters were removed and placed on an absorbent fiberglass pad, impregnated with 340 μl of a buffered solution containing 0.1% (wt/vol) 4-MU-α-l-arabinopyranoside, 0.1% (wt/vol) digitonin, acting as a membrane permeabilizer; and 1 mM MgCl2, acting possibly as a cofactor for the enzyme. After incubation for 30 min at 30°C, the filters were sprayed with 1.2 M sodium hydroxide and inspected under a UV lamp (366 nm). Blue fluorescent microcolonies indicated a positive reaction.
Confirmation and reference method.
To confirm the identity of the fluorescent microcolonies, the membrane was reincubated at 45°C on the original SGA*-T medium for at least 3 days and the mycelial growth was examined macroscopically and microscopically after lactophenol cotton-blue staining. Two reference procedures were used, one in another laboratory immediately after sampling (reference 1), the other one concurrently with the two-step method (reference 2). Reference procedure 1 included direct microscopy and culturing on SGA and blood agar. In reference procedure 2, part of the sample was filtered over a second nylon MF, and this was followed by incubation on SGA+ and microscopy.
Method evaluation.
The following equations were used to calculate the validity parameters of the method (where TP is true positives, TN is true negatives, FP is false positives, FN is false negatives, and n is total number of samples): sensitivity = TP × 100/(TP + FN), specificity = TN × 100/(TN + FP), positive predictive value = TP × 100/(TP + FP), negative predictive value = TN × 100/(TN + FN), and efficiency = (TP + TN) × 100/n.
RESULTS
Enzyme activities in A. fumigatus and other fungal species were assessed from their ability to cleave a series of 4-MU glycosides and esters. In order to be useful for diagnostic purposes, the sensitivity of a given enzymatic reaction had to exceed 95%; i.e., fewer than 5% of strains of the target species should be negative. Likewise, the specificity had to be ≥95%; i.e., the enzyme should occur in ≤5% of other fungal species.
Only the cleavage of 4-MU-α-l-arabinopyranoside satisfied both criteria (Table 1). The corresponding enzyme, tentatively called “arabinopyranosidase,” occurred in 98% of A. fumigatus strains (n = 48) but was absent in other clinically important molds (n = 116) (Table 1). Among the yeasts, 5% of Candida albicans strains tested (n = 129) showed a positive reaction with 4-MU-α-l-arabinopyranoside, whereas Candida glabrata (n = 39), Candida krusei (n = 25), Candida tropicalis (n = 26), and Candida parapsilosis (n = 14) were negative.
TABLE 1.
Cleavage of 4-MU-glycosides and esters by medically important fungi
| Species | % Positive strains
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Growth at 45°C | Arabinopyranoside | Arabinofuranoside | Acetate | Galactoside | Glucoside | Heptanoate | Laurate | NAc-galactosaminide | Phosphate | |
| A. fumigatusa (n = 48) | 100 | 98 | 100 | 100 | 100 | 100 | 100 | 94 | 100 | 100 |
| A. flavus (n = 29) | 0 | 0 | 98 | 100 | 100 | 100 | 100 | 93 | 100 | 100 |
| A. niger (n = 22) | 0 | 0 | 95 | 100 | 95 | 100 | 100 | 97 | 100 | 100 |
| A. clavatus (n = 4) | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| A. nidulans (n = 13) | 0 | 0 | 100 | 100 | 100 | 0 | 100 | 100 | 100 | 100 |
| A. terreus (n = 12) | 0 | 0 | 8 | 100 | 100 | 0 | 100 | 100 | 100 | 100 |
| A. versicolor (n = 6) | 0 | 0 | 0 | 100 | 100 | 0 | 100 | 100 | 0 | 100 |
| F. oxysporum (n = 4) | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 50 | 100 |
| F. solani (n = 4) | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 50 | 100 |
| R. microsporus (n = 3) | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| R. oryzae (n = 4) | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| R. pusillus (n = 4) | 100 | 0 | 0 | 100 | 100 | 0 | 100 | 100 | 100 | 100 |
| A. corymbifera (n = 2) | 100 | 0 | 0 | 100 | 100 | 0 | 100 | 100 | 100 | 100 |
| P. marneffei (n = 5) | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| P. boydii (n = 4) | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 80 | 100 |
Additional reactions were performed for A. fumigatus only with the following 4-MU substrates (% positive strains): diacetylchitobioside (44), glucuronide (0), iduronide (4), lactoside (4), oleate (0), palmitate (0), pyrophosphate (14), and rhamnopyranoside (0).
In physiological saline after ultrasonic lysis of cells or on SGA, fluorescence was weak. However, it was significantly more pronounced on an MF. This was demonstrated at the cellular level by epifluorescence microscopy and after microcolony formation by inspection under a UV lamp (366 nm). Among the MFs compared, nylon proved clearly superior to nitrocellulose, mixed cellulose esters, polyamide, polysulfone, and polytetrafluoroethylene in terms of visually scored intensity of fluorescence.
The rate of microcolony formation on the nylon MF depended on the composition of the growth medium and the temperature but was also influenced by the cellular stage of growth initially present in the sample (mycelium or spores) and the administration of antimycotics to the patient before sampling. The growth-promoting activities of the various media were compared in broth on the basis of turbidity measurements as a function of time. The enriched SGA* yielded faster growth than SGA, CMT, PGA, and CDA (results not shown). However, when using clinical samples, it was necessary to add an antibiotic to the medium to suppress bacterial growth. A combination of ticarcillin-clavulanic acid (Timentin) has previously proven effective to this end (2). To ensure that antibacterials had no negative effect on the proliferation of A. fumigatus, mycelial growth was monitored in broth for 44 h by turbidimetry. To this end, 102 conidia of three A. fumigatus strains were inoculated in 10 ml of SGA* and SGA*-T broth and incubated (results not shown). No differences in growth rate were observed in the two media. Growth at 45°C was superior to that at 37°C and entails additional selectivity by suppressing the multiplication of bacteria and yeasts. However, some other molds were also able to proliferate at this temperature (Table 1). The positive contribution of the nylon MF to the growth rate was demonstrated as follows. First, 100 μl of a suspension of 102 conidia of A. fumigatus per ml (n = 5) was filtered over a nylon MF and placed on SGA-T* medium, supplemented with 1% 4-MU-α-l-arabinopyranoside. The same amount of conidia was inoculated by streaking (no MF) on a second plate with SGA-T* medium, also supplemented with 1% 4-MU-α-l-arabinopyranoside. Both plates were incubated at 45°C, and the different stages of growth were observed microscopically (7). Hyphae started to grow and differentiate after an average time of 10 h (MF) and 14 h (no MF), and vesicles were observed after 14 and 17 h, while the end point of the growth phase, i.e., the presence of phialides and conidia, was observed after 17 and 23 h, respectively. Microcolony formation was strongly affected by the developmental stage of the fungus in the sample. When starting from a sample containing mycelium, as in sputum or bronchoalveolar liquid, microcolonies appeared after approximately 11 to 14 h. However, in the case of spores the detection time was extended to at least 24 h. Delayed growth was also observed in samples from patients treated with amphotericin B or itraconazole.
With the enzyme substrate added to the medium, the first fluorescent microcolonies on the MF could be visualized after approximately 17 h. However, in a two-step procedure where the MF, after microcolony formation, is removed from the SGA*-T and placed on an absorbent pad containing the substrate and 0.1% digitonin as a membrane permeabilizer, a further reduction of 3 h in detection time was obtained.
The method has been applied to 188 clinical specimens, both positive and negative for A. fumigatus. The results with reference to a conventional plate isolation method with microscopic confirmation are listed in Table 2. Sensitivity and specificity were 96.9 and 100%, respectively, while the positive and negative predictive values were 100 and 95.8%, respectively. The overall efficiency of the method was 98.2%. No false positives were observed. The false-negative rates were 3.1 and 0% relative to reference procedures 1 and 2, respectively. Other Aspergillus spp. (A. flavus, n = 8; A. niger, n = 9; A. nidulans, n = 2), were isolated at 37°C but not at 45°C and gave no reaction with 4-MU-arabinopyranoside.
TABLE 2.
Agreement between the two-step and the reference methods for A. fumigatus
| Method | No. of responses
|
|||
|---|---|---|---|---|
| Positive for A. fumigatus | Negative (no molds) | Negative (other moldsa) | Total | |
| Two step | 94 | 72 | 22 | 188 |
| Reference 1 | 97 | 69 | 22 | 188 |
| Reference 2 | 94 | 72 | 22 | 188 |
Aspergillus spp., Penicillium sp.
DISCUSSION
Current research on the diagnosis of aspergillosis is mostly concerned with the development of non-culture-based molecular and immunological methods (8, 11–13). Contrary to this trend, the approach of the present study has been to improve the plate method with regard to specificity, sensitivity, and speed. This has been accomplished by membrane filtration of the sample, microcolony formation on a dedicated selective growth medium, and the demonstration of a specific enzyme activity in permeabilized cells.
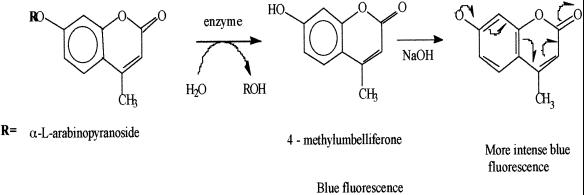
The cleavage of fluorogenic 4-MU glycosides is commonly used to demonstrate enzyme activities in microorganisms (2, 3, 15, 15a). The two consecutive reactions leading to fluorescence are depicted in Fig. 1. However, for A. fumigatus a specific enzyme activity had not been reported. Of all 4-MU substrates tested, only 4-MU-α-l-arabinopyranoside was cleaved with ≥95% species specificity and sensitivity. Figure 2 shows pictures of positive and negative readings, respectively. A. fumigatus was also found to cleave 4-MU-α-l-arabinofuranoside with 100% species sensitivity, but this arabinofuranosidase also occurs in A. niger, A. terreus, and A. nidulans (4, 9, 14).
FIG. 1.
Reactions involved in the enzymatic cleavage of 4-MU-α-l-arabinopyranoside and the subsequent amplification of fluorescence.
FIG. 2.
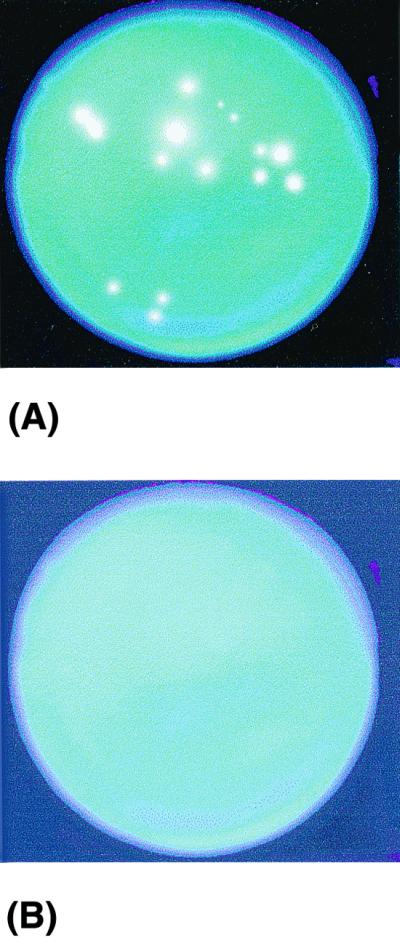
Fluorescent microcolonies generated from A. fumigatus-positive (A) and -negative (B) samples.
Method specificity is further ensured by the use of a selective medium and incubation at 45°C. SGA*-T is an enriched version of SGA-T, which has previously shown its usefulness for the enzymatic detection of Candida spp. (2). It allows optimal proliferation of fungi, while effectively suppressing bacterial growth. Furthermore, most yeasts and bacteria will not proliferate at 45°C.
The sensitivity and the speed of the method depend on the sample size, the rate of formation of microcolonies, and their fluorescent intensity. The use of a nylon MF is crucial in this respect. First, membrane filtration permits the processing of a high sample volume. In a conventional procedure, an agar plate is directly inoculated by streaking the sample, so that only a limited amount of it is transferred. Assuming a detection limit of 1 CFU per membrane filter, the detectability in the present method is limited only by the filterable sample volume and the ability of the cells to multiply sufficiently in the course of the short incubation. Although part of the population will obviously fail to do so, this relative loss of sensitivity is offset by the large sample size, so that a net gain is still achieved. Second, membrane filtration enhances the detectability of the fluorescent microcolonies. On the nylon MF contacted with SGA*-T, fluorescent microcolonies became visible 4 to 5 h sooner than on the same medium after direct inoculation. This difference is the result of two factors, i.e., a higher growth rate of the fungus on a nylon surface and the amplification of fluorescence. The latter phenomenon has previously been demonstrated in connection with the detection of fluorescent microcolonies of Candida spp. and Escherichia coli using 4-MU glycosides (2, 15a).
Growth and, hence, detectability were further influenced by the composition of the medium and the initial physiology of the fungus in the sample. SGA*-T promoted the growth of A. fumigatus more efficiently than the other broth media tested. Mycelium in the sample grows out more readily to microcolonies than spores.
A last factor contributing to the enhanced speed of our method is its two-step concept of dissociating a preliminary growth phase from the actual demonstration of the enzyme. This approach permitted us to permeabilize the cells with digitonin without compromising their viability during propagation. The purpose of permeabilization is to accelerate the uptake and, as a result, the cellular turnover of the substrate. The documented slow entrance of 4-MU glycosides in nonpermeabilized microorganisms supposedly becomes speed limiting in procedures with very short incubation times (15, 15a). In the presence of digitonin, an effective membrane permeabilizer for fungal (5) microcolonies could be visualized roughly 3 h sooner than in its absence. The difference in speed between a >24-h and a 14-h plate method for A. fumigatus is thus the added result of three factors, i.e., the use of a nylon MF, of a favorable growth medium, and of a two-step procedure for the demonstration of a specific enzyme in digitonin-permeabilized cells using a fluorogenic substrate.
The application of the new method to 188 clinical samples has indicated excellent agreement with a conventional method and a low false-negative rate. Depending on whether the result was compared with that of reference procedure 1 or 2, carried out at different points of time, the false-negative rate was either 3.1 or 0%. With two diagnostic criteria, i.e., thermotolerance and a specific enzyme activity, the method exhibits good specificity for A. fumigatus but should yet be considered as presumptive. However, despite the action of digitonin, cells continue to divide upon further incubation of the membrane filter on SGA*-T, so that subsequent macroscopic and microscopic confirmation is feasible. As A. fumigatus is the main cause of aspergillosis, a differentiation from other Aspergillus spp. on the basis of its thermotolerance appears reasonable. A disadvantage of the test is its relative higher cost (estimated at $4 per test) compared to conventional methods (costing less than $1). However, we believe that the cost might be outweighed by the significant reduction in detection time, especially when it comes to diagnosis of life-threatening diseases such as aspergillosis.
ACKNOWLEDGMENTS
We are grateful to R. Aerts for excellent technical assistance. We thank G. Verschraegen (Laboratory of Bacteriology and Virology) for critical review of the manuscript. For providing us with clinical samples, we thank P. Van Cauwenberghe (Department of Otorhinolaryngology), L. Noens (Department of Hematology), G. Verschraegen, and G. Claeys (Laboratory of Bacteriology and Virology) from the University Hospital of Ghent.
REFERENCES
- 1.Atlas R M. Handbook of microbiological media. Boca Raton, Fla: CRC Press; 1993. [Google Scholar]
- 2.Bauters T G, Peleman R, Moerman M, Vermeersch H, Noens L, Nelis H J. Membrane filtration test for rapid presumptive differentiation of four Candida species. J Clin Microbiol. 1999;37:1498–1502. doi: 10.1128/jcm.37.5.1498-1502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobey D G, Ederer G M. Rapid detection of yeast enzymes by using 4-methylumbelliferyl substrates. J Clin Microbiol. 1981;13:393–394. doi: 10.1128/jcm.13.2.393-394.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinar F, Pena J L, Pinaga F, Valles S. Alpha-L-arabinofuranosidase production by Aspergillus nidulans. FEMS Microbiol Lett. 1994;115:107–112. doi: 10.1111/j.1574-6968.1994.tb06622.x. [DOI] [PubMed] [Google Scholar]
- 5.Felix H. Permeabilized cells. Anal Biochem. 1982;120:211–234. doi: 10.1016/0003-2697(82)90340-2. [DOI] [PubMed] [Google Scholar]
- 6.Fridkin S K, Jarvis W. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin D H. The physical environment and growth. In: Griffin D H, editor. Fungal physiology. 2nd ed. New York, N.Y: Wiley-Liss Publisher; 1993. pp. 195–214. [Google Scholar]
- 8.Latgé J-P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luonteri E, Siikaaho M, Tenkanen M, Viikari L. Purification and characterization of three alpha-arabinosidases from Aspergillus terreus. J Bio/Technology. 1995;38:279–291. [Google Scholar]
- 10.Paugam A, Sarfati J, Romieu R, Viguier M, Dupouy-Camet J, Latgé J P. Detection of Aspergillus galactomannan: comparison of an enzyme-linked immunoassay and a europium-linked time-resolved fluoroimmunoassay. J Clin Microbiol. 1998;36:3079–3080. doi: 10.1128/jcm.36.10.3079-3080.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson M D, Warnock D W. Laboratory diagnosis of fungal infection. In: Richardson M D, editor. Fungal infections. 2nd ed. Oxford, United Kingdom: Blackwell Science; 1997. pp. 16–19. [Google Scholar]
- 12.Richardson M D. Aspergillus and Penicillium species. In: Ajello L, Hay R J, editors. Topley and Wilson's microbiology and microbial infections. Medical mycology. 9th ed. London, United Kingdom: Arnold; 1998. pp. 281–312. [Google Scholar]
- 13.Van Burik J-A, Myerson D, Schreckhise R, Bowden R. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van de Veen P, Flipphi M J, Voragen A G, Visser J. Induction of extracellular arabinases on monomeric substrates in Aspergillus niger. Arch Microbiol. 1993;159:66–71. doi: 10.1007/BF00244266. [DOI] [PubMed] [Google Scholar]
- 15.Van Poucke S O, Nelis H J. Development of a sensitive chemiluminometric assay for the detection of β-galactosidase in permeabilized coliform bacteria and comparison with fluorometry and colorimetry. Appl Environ Microbiol. 1995;61:4505–4509. doi: 10.1128/aem.61.12.4505-4509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Van Poucke, S. O., and H. J. Nelis. Rapid detection of fluorescent and chemiluminescent total coliforms and Escherichia coli on membrane filters. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 16.Verweij P E, Latgé J-P, Rijs A J, Melchiers W J, De Pauw B E, Hoogkamp-Korstanje J A, Meis J F. Comparison of antigen detection and PCR assay using bronchoalveolar lavage fluid for diagnosing invasive pulmonary aspergillosis in patients receiving treatment for hematological malignancies. J Clin Microbiol. 1995;33:3150–3153. doi: 10.1128/jcm.33.12.3150-3153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]