Abstract
Introduction
Cell line derived from fish has been established as a promising tool for studying many key issues of aquaculture covering fish growth, disease, reproduction, genetics, and biotechnology. In addition, fish cell lines are very useful in vitro models for toxicological, pathological, and immunological studies. The easier maintenance of fish cell lines in flexible temperature regimes and hypoxic conditions make them preferable in vitro tools over mammalian cell lines. Great excitement has been observed in establishing and characterizing new fish cell lines representing diverse fish species and tissue types. The well-characterized and authenticated cell lines are of utmost essential as these represent cellular functions very similar to in vivo state of an organism otherwise it would affect the reproducibility of scientific research.
Conclusion
The fish cell lines have exhibited encouraging results in several key aspects of in vitro research in aquaculture including virology, nutrition and metabolism, production of vaccines, and transgenic fish production. The review paper reports the cell lines developed from fish, their characterization, and biobanking along with their potential applications and challenges in in vitro research.
Keywords: Applications, Bio-banking, Fish cell line, Cell-based aquaculture, In vitro research
Introduction
The Development of chemically defined cell culture medium like Leibovitz -15 (L-15) and the development of antibiotics with gradual improvisation of cell culture techniques eventually made the generation of cultured cells for deriving continuous cell lines. In addition to being an important biomedical tool like any other cell line, cell cultures prepared from fish, shellfish and seaweeds can provide a significant contribution to the growth of aquaculture. The scientific knowledge gained through the cell culture system can be utilized for manipulating the whole organism to enhance its usefulness for aquaculture. Their cell line could be useful for providing basic insights into growth, reproduction, and health, creating opportunities for manipulation and thus the cell lines could be used as sources of biochemical products in place of the whole organism [1]. Cell-based aquaculture systems using cell cultures could be a game-changing practice to produce seafood and other aqua food across multiple species for meeting the demand of the burgeoning world population [2]. A cell-based aqua food production system utilizing cells in place of whole fish could also lead to greater preservation of the aquatic environments. This practice has to meet the regulatory framework/guidelines developed by the FDA [3] for the safety of food produced using such animal cell culture technology.
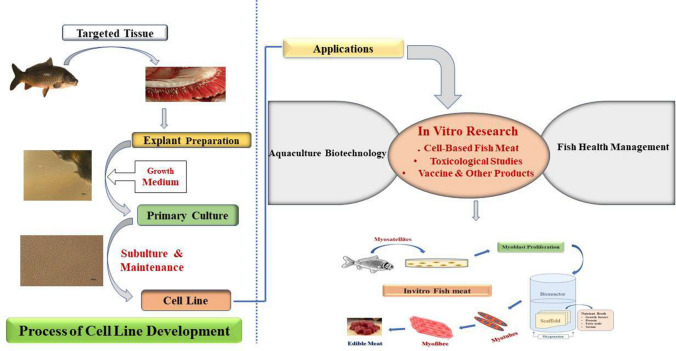
Fish cell culture offers several advantages over mammalian cell culture in terms of adaptation to a broad range of temperature, higher tolerance to hypoxia, easier maintenance of cell culture for longer periods. Cell lines from fish have been increasingly established from different aquaculture species and they are being used in in vitro research related to aquaculture and other interdisciplinary areas. However, there are emerging issues regarding standardization of cell line nomenclature, characterization of cell lines following SOP/recommended guidelines, and conservation of cell lines in separate biobanks across the world-which we review below is of utmost essential to maintain scientific reproducibility in cell-based biological research using fish cell lines. The key areas of aquaculture like fish health, disease diagnosis, safety, and nutritional aspects challenging aquaculture production can be studied using fish cell lines without scarifying whole live fish (Fig. 1). The scientific knowledge generated using fish cell lines would be immensely useful for quality fish production in a sustainable manner. Cell lines would facilitate in vitro research for developing climate-resilient and sustainable aquaculture systems to minimize the key challenges and provide nutritional security to the burgeoning world population.
Fig. 1.
Implications of fish cell line in aquaculture
Global status of fish cell lines
An increasing trend has been observed for the development of fish cell lines from a wider range of tissues covering both tropical and temperate water since the first establishment of the RTG-2 cell line in 1962 [4]. Bairoch enlisted 883 fish cell lines out of 104,421 cell lines from > 590 species in Cellulosaurus; a knowledge resource on cell lines [5]. In general, cell lines have been developed globally using different types of fish tissue samples including gill, caudal fin, eye, liver, and kidney. Fish cell lines have also been established using tissue samples like intestine [66], brain [95], vertebra [105], and snout [121]. Spontaneous differentiation is one of the most challenging for the development of embryonic stem cell culture from fish and this is the main cause behind a very limited number of stem cell lines. Few embryonic fish stem cell lines were developed from sea bream Sparus aurata [6], sea perch Lateolabrax [7], sea bass; Lates calcarifer [8], Catla catla [9], Labeo rohita [10]. A feeder-free cell culture system used for the development of Embryonic Stem (ES) cell lines from medaka and zebrafish has boosted fish stem cell research by replacing the use of feeder layers to inhibit spontaneous differentiation in fish stem cell culture [11].
Cell line characterization
The numbers of cell lines developed from fish have been increasing rapidly which raises the concern for accurate authentication and characterization of fish cell lines to provide reproducible scientific data. The comprehensive guidelines for using cell lines highlight various aspects of cell culture, issues of misidentification, contamination with microbes along with recommendations to overcome these problems [12]. Although these guidelines are meant for scientists in the UK, the basic principles remain the same for international implications. Research and development using cell lines need detailed knowledge on the purity and originality of the cell line [13]. The characterized cell lines are indispensable as they facilitate the researchers to perform in vitro research and standard guidelines are available for their characterization. However, many fish cell lines don’t meet uniform international standards. The Food and Drug Administration has described the steps to be considered while characterizing a cell line used to produce biological products [14]. Such standard protocol for the characterization and authentication of fish cell lines should be practiced throughout the world. Standard protocols for authentication of cell line have been reported [15] wherein standard methods like cytochrome c oxidase subunit1 (CO1) barcode, Short Tandem Repeat (STR) profiling, karyotyping, Single Nucleotide Polymorphisms (SNP) profiling, use of species-specific primers, whole-genome sequencing (WGS), etc. are described as ideal approaches for authentication and maintenance of quality cell lines. Several STR databases of cell lines are maintained by ATCC, DSMZ across the world. CLASTR: The Cellosaurus STR similarity search tool is now in the public domain for comparing STR profiles of the cell lines [16]. Cross-contamination also causes a disastrous feature of the cell line as the cell line losses its originality and hence cross-contamination needs to be avoided by following standard operating procedure (SOP). Development of a framework for cell line annotation linked to STR and SNP profiles in the form of a catalog of synonymous cell lines to avoid or detect cross-contamination [17].
Misidentification of cell lines leads to irreproducible data and hence proper authentication of cell lines using molecular markers is essential. It was obligatory to provide DNA-based certification of the cell line developed [18]. Mitochondrial DNA genes like 16S rRNA and CO1 are used for the authentication of cell lines. Cox I gene has been used as a molecular identification system for animal species which is popularly referred to as ‘‘DNA barcoding’’ [19]. The cox I gene was used as DNA barcodes for the authentication of 67 cell lines [20]. Similarly, many fish cell lines have been DNA barcoded using cox I [21–23]. Cell line repositories like DSMZ, ATCC use DNA barcoding as a standard method for cell line identification. Protein expression signature has also been used for the identification of cell lines derived from fish [24, 25].
Cell lines developed from fish are mostly applied in basic, biomedical and toxicological research in addition to their potential applications in aquaculture. Several key issues in aquaculture can be addressed by cell culture technology and they are reviewed below.
Fish health management
The fish disease has been considered as one of the most critical challenges for sustainable aquaculture production due to the economic loss and widespread use of antibiotics and other compounds causing great risk to the aquatic environment. Fish cell culture has great potential to provide tools and strategies for disease control in aquaculture. In vitro models that use cell culture methods and experimental systems facilitate a deeper understanding of the complex interactions underlying disease outbreaks and its advancement in which the interactions between aggressors and the host can be dissected [26]. Fish cell lines have potential applications in understanding disease mechanisms, developing assays for disease diagnosis, developing drugs and vaccines for the control of the fish disease. The export trade of seafood depends upon the quality and health status of the seafood. The fish cell line model has been considered useful for detecting viral pathogens and strategies need to be implemented accordingly for the health protection of major aquaculture species. Zoonotic disease associated with fish is another concern where consumption of unhealthy fish might be a risk to a human being. Associated in vitro assays would be useful in detecting such harmful pathogens and allergens so that the quality of seafood can be augmented. In vitro methods using cell cultures for addressing health issues in molluscs and crustaceans are equally important. Department of Biotechnology, Govt. of India has funded a national programme on the isolation and characterization of finfish and shellfish viruses using cell lines in India. In vitro approach using permanent cell lines needs to be validated for fish and shellfish disease surveillance and health certification. Transboundary movements of live aquatic animals have greatly increased concern for spreading disease in the aquaculture system.
The viral disease used to cause devastating loss to the aquaculture industry. The entire world has witnessed the deadliest effect of the spread of the virus Covid-19. The isolation of the novel Covid-19 using the animal model has begun and the successful isolation would be useful in understanding the biology and evolution of the Covid-19 in developing drugs, vaccines, and rapid diagnosis kits. Isolation of viruses using fish cell lines is one of the most sensitive techniques for the discernment of the important pathogens causing viral disease in many fish and other species. Hence, the development of control measures to halt the spread of the viral disease depends on the unitisation of fish cell lines for such purposes. A comprehensive list of fish cell lines used in virus susceptibility studies is given in Table 1. Research on the avoidance of infectious fish disease in aquaculture necessitates a cell culture- based approach for understanding the underlying disease mechanism. Fish cell culture-based isolation and propagation of virus has provided momentum to virological studies and facilitated research on viral diseases in important aquaculture species. Propagation of viruses in a cell culture system is one of the bases of a virus surveillance system using cell culture. Ariel et al. developed standard methods to reduce false negatives in cell culture-based surveillance systems in testing fish cell line susceptibility for the viruses [27].
Table 1.
List of some fish cell lines susceptible to the virus
| Fish cell line & tissue/species of origin | Susceptible to virus | Reference | |
|---|---|---|---|
| Fish cell line | Tissue/species of origin | ||
| Bluegill fry (BF-2) | Lymphocystis virus | Wolf et al., 1966 | |
| Haemorrhagic Septicaemia Virus (VHSV) | Skall et al., 2005, Ito et al., 2016 | ||
| Ranavirus isolates and of Epizootic haematopoietic necrosis virus (EHNV) | Ariel et al., 2009b | ||
| Rainbow trout gonad (RTG-2) | Haemorrhagic Septicaemia Virus (VHSV) | Wolf and Quimby, 1962, Ito et al., 2016 | |
| Ranavirus isolates and of Epizootic haematopoietic necrosis virus (EHNV) | Ariel et al., 2009b | ||
| Fathead minnow (FHM) | Haemorrhagic Septicaemia Virus (VHSV) | Gravell and Malsberger, 1965, Ito et al., 2016 | |
| Ranavirus isolates and of Epizootic haematopoietic necrosis virus (EHNV) | Ariel et al., 2009b | ||
|
Epithelioma papillosum cyprini (EPC) |
Haemorrhagic Septicaemia Virus (VHSV) | Fijan et al., 1983 | |
| Ranavirus isolates and of Epizootic haematopoietic necrosis virus (EHNV) | Ariel et al., 2009b | ||
|
Chinook salmon embryo CHSE-214 |
Infectious pancreatic necrosis virus (IPNV) and salmonid alphavirus |
Fryer et al., 1965 | |
| Salmon pancreas disease virus | Lopez et al. 2001 | ||
| MEF | Mandarin fish | Megalocytiviruses | Dong et al., 2013 |
| koi her-pesvirus (KHV), salmonid rhabdoviruses like spring viraemia of carp virus, (SVCV), Chum salmon reovirus and some infectious pancreatic necro-sis (IPN) viruses | Neukirch and Haenen, 2004 | ||
| Clonal-cell lines developed from Siberian sturgeon Acipensar baerii head kidney showed susceptibility to VHSV | Ryu et al., 2018 | ||
| bmGH | Heart from brown-marbled grouper, Epinephelus fuscoguttatus | Turbot reddish body iridovirus (TRBIV) and Lymphocystis disease virus (LCDV) | Wei et al. 2010 |
| TK | Kidney of Scophthalmus maximus | Turbot reddish body iridovirus (TRBIV) | Wang et al. 2010b |
| SFK | kidney-derived cell line from southern flounder, Paralichthys lethostigma | Lymphocystis disease virus (LCDV), | Wang et al. 2011 |
| Flounder fin (FFN) cells from fin tissue and flounder spleen (FSP) cells from spleen tissue | Paralichthys olivaceus | Pancreatic necrosis virus, marine birnavirus, chum salmon virus, infectious haematopoietic necrosis virus, spring viraemia of carp virus and hirame rhabdovirus | Kang et al. 2003 |
| SHK from spotted halibut head kidney | Head kidney of spotted halibut Verasper variegates | Lymphosystis disease virus (LCDV) | Wang et al. 2010a |
| CRF-1 | from red sea bream fin, Pagrus major | Red sea bream iridovirus (RSIV) | Imajoh et al. 2007 |
| RSBF-2 | from red sea bream fin, Pagrus major | RSIV | Ku et al. 2010 |
| SPB – a fishneural progenitor cell line | Brain of snubnosepompano,Trachinotus blochii | Giant sea-perch iridovirus (GSIV-K1), red sea breamiridovirus (RSIV-Ku), grouper nervous necrosisvirus (GNNV-K1), chum salmon reovirus (CSV) and eel herpesvirus (HVA) | Wen et al. 2013 |
| MFF-1 | from mandarin fish fry, Synchiropus splendidus | Infectious spleen and kidney necrosis virus (ISKNV) | Dong et al. 2008 |
| BM | muscle from barramundi, Lates calcarifer | Grouper iridovirus (GIV) | Lai et al. 2008 |
| BSB | swim bladder from barramundi, Lates calcarifer | Grouper iridovirus (GIV | Lai et al. 2008 |
| CB from cobia brain | Brain of Cobia (Rachycentron canadum) | Grouper iridovirus (GIV) | Cheng et al. 2010 |
| CF | Fin of Cobia | Grouper iridovirus (GIV) | Cheng et al. 2010 |
| GBC1 | Clonal cell line from brain of orange-spotted grouper, Epinephelus coioides | Grouper nerve necrosis virus (GNNV) | Wen et al. 2008 |
| GBC4 | Clonal cell line from brain of orange-spotted grouper, Epinephelus coioides | Giant seaperch iridovirus (GSIV) | Wen et al. 2008 |
| FEC |
Embryo of Japanese flounder Paralichthys olivaceus |
Lymphocryptovirus (LCV) and Turbot reddish body virus (TRBV) | Chen et al. 2004 |
| EAGL | Liver of red-spotted grouper, Epinephelus akaara | Singapore grouper iridovirus (SGIV) | Ou-Yang et al. 2010 |
| GS-1 | Spleen of the orange-spotted grouper, Epinephelus coioides | Fish ranavirus: Singapore grouper iridovirus (SGIV) and megalocytivirus: spleen and kidney necrosis virus (ISKNV) | Huang et al., 2018 |
| GS | Spleen of orange spotted grouper, Epinephelus coioides | Singapore grouper iridovirus (SGIV) and Epinephelus tauvina nervous necrosis virus (ETNNV) | Qin et al., 2006 |
| GH (Heart), GE (from eye), GSB (swimbladder), GL (liver) | Tropical Grouper, Epinephelus awoara | Grouper iridovirus (GIV) | Lai et al. 2003 |
| GB from Brain, GF from fin and GL from liver cell lines from | Tropical Grouper, Epinephelus awoara | Yellow grouper nervous necrosis virus (a nodavirus) (YGNNV) | Lai et al. 2003 |
| SISK | kidney of Lates calcarifer | Nodavirus | Babu et al. 2013 |
| SISS | spleen of L. calcarifer | Nodavirus | |
| SIGE | eye of Epinephelus coioides | Nodavirus | |
| CSK | kidney of Channa striatus | Marine fish nodavirus | Abdul Mazeed et al., 2013 |
| SIGE | Eye of grouper (Epinephelus coioides) | Nodavirus, MABV NC-1 and Y6 | Parameswaran et al., 2007 |
| SSN-1 | Striped snakehead (Ophicephalus striatus) | Nodavirus | Frerichs et al. 1996 |
| Simian COS-1 cells | Nodavirus | Delsert et al., 1997 | |
| GF-1 | Nodavirus | Chi et al., 1999 | |
| SF | Asian sea bass (Lates calcarifer) | Nodavirus | Chang et al., 2001 |
| TF | Scophthalmus maximus | Nodavirus | Aranguren et al., 2002 |
| SAF-1 | Fins of gilt-head seabream | Lymphocystis disease virus (LDV) and to several salmonid fish viruses, such as infectious haematopoietic necrosis virus (IHNV), viral haemorrhagic septicemia virus (VHSV) and several strains of infectious pancreatic necrosis virus (IPNV) | Perez-Prieto et al., 1999 |
| TF | Fin of Scophthalmusmaximus, | Turbot reddish body iridovirus (TRBIV) | Fan et al., 2010 |
| SKF-9 | Potted knifejaw (Oplegnathus punctatus) | RSIV (Red sea bream iridovirus (RSIV) | Kawato et al. 2017 |
| Fat head minnow (FHM) | RSIV (Red sea bream iridovirus (RSIV) | Clem et al., 1961 | |
| Striped snakehead (SSN-1) | RSIV (Red sea bream iridovirus (RSIV) | ||
| GF cells grunt fin (GF) | Fin of blue striped grunt (Haemulon sciurus) | RSIV (Red sea bream iridovirus (RSIV) | |
| CFF | Pristolepis fasciatus | Tilapia Lake Virus (TiLV) | Behera et al., 2018 |
| SPB | Brain of snubnose pompano, Trachinotus blochiito | Giant seaperch iridovirus (GSIV-K1), red sea bream iridovirus (RSIV-Ku) and chum salmon reovirus (CSV) (HVA) | Wen et al., 2013 |
| OCF | Caudal fin of Amphiprion ocellaris | Nervous necrosis virus | Yashwanth et al., 2020 |
Highly specific cell lines are used for investigating unique virus which otherwise doesn’t propagate in any normal cell line. The susceptibility of the fish cell lines to the virus varies with species as well as tissue from where the cell line is developed. This raises the importance of the development of species-specific and tissue-specific cell lines from various important aquaculture species. Some fish cell lines like bluegill fry (BF-2), chinook salmon embryo (CHSE-214), epithelioma papulosum cyprinid (EPC), fathead minnow (FHM), rainbow trout gonad (RTG-2), and SAF-1 have shown susceptibility to some of the most commonly available viruses like Infectious pancreatic necrosis (IPN), VHSV, IHNV, IPNV, SVC, koi herpesvirus (KHV) and Channel catfish virus (CCV) that have severely affected several aquaculture species [28, 29]. MEF-8C1 cloned cells obtained from the MEF cell line from mandarin fish suitably propagated megalocytiviruses that cause major problems in finfish aquaculture in China [30]. A transgenic fish cell line RTG-P1 was applied to estimate viremia of Salmonid alphavirus (SAV) which causes a serious viral disease i.e. Salmon Pancreas Disease (SPD) in Atlantic salmon farming [31]. SISK and SISS cell lines developed from the kidney and spleen of Lates calcarifer respectively and SIGE cell line developed from the eye of Epinephelus coioides showed their ability to propagate a nodavirus strain [32]. Yashwanth et al. reported the susceptibility of the OCF cell line to NNV [23]. SSN-1 cell line supported the replication of snakehead fish vesiculovirus (SHVV) which causes great economic loss in fish culture in East Asian countries [33]. Understanding the transmission of viral infection between the two most important aquaculture species mandarin fish and snakehead fish, it would be useful to develop control measures to prevent the spreading of such viral diseases. Fish cell cultures or cell lines could be used for investigating viral pathogenesis and host–pathogen interactions.
In the past, such in vitro methods were used for some bacterial pathogens like mycobacterial host–pathogen interactions using the carp monocytic cell line CLC (carp leukocyte culture) [34]. Recently, Cardiac Primary Cultures (SCPCs) from Atlantic salmon pre-hatch embryos were used to investigate viral host–pathogen interactions and pathogenesis [35]. A blend of cell culture and molecular biology methods will provide deeper insights into host–pathogen interactions. Although advanced antibody-based techniques are being developed in disease control in aquaculture, fish cell culture continues to be an indispensable technique for isolation and characterization of the pathogenic virus and intracellular bacteria and studying their pathogenicity [26]. These fish cell lines are going to play a crucial role in virus isolation and understanding viral pathogenicity and thereby controlling these viral diseases to enhance sustainable aquaculture production.
Pathological & immunological studies
Several fish health-related issues can be studied in vitro using fish cell lines. The most prominent is the application of fish cell lines in disease diagnostics and immunological studies. Some intracellular bacterial fish pathogens like Rickettsiae spp and Renibacterium salmoninarum have been detected in fish using cell cultures [36, 37]. The in vitro investigation using fish cell line (CHSE-214) has improved the knowledge of the infection process by Yersinia ruckeri in salmonid fish as well as the interaction between the pathogen and host cells [38].
Cell cultures are promising in vitro tools in studying the host defense mechanism and thereby help in exploiting the immunological information for the health protection of fish and shellfish used in aquaculture. Fish leukocyte cell lines and macrophages developed from many aquaculture species like carp, catfish have been used for immunological studies. Several monocyte-like cell lines have also been developed using peripheral blood leukocytes of channel catfish [39]. Cell lines developed from gut, skin, and gill are promising in vitro tools for studying the defense mechanism in fish. The immunological potentials of DNA vaccines, synthetic peptides and immunostimulants, and other products can be tested using these fish cell lines. A continuous blood cell line developed from peripheral blood mononuclear cells of Cyprinus carpio was useful in understanding the fundamental aspects of fish immunology [40]. Fish macrophage cell lines are found to be very useful in numerous research applications including immunological studies. Two macrophage cell lines i.e., CTM and CCM [41] developed from Catla catla could be useful in investigating the importance of these cell lines in the differentiation and maturation of thymocytes and other fish immunological studies. SHK-1 macrophage-like cell line developed from Atlantic salmon showed the reaction to monoclonal antibodies against Atlantic salmon peripheral blood leukocytes and the cell line was able to phagocytose bacteria [42]. Macrophage-like cell line RTS11 developed from rainbow trout was used as a promising tool for investigating immune cell-specific responses in vitro [43, 44].
Saprolegniales are considered the most important fungi causing disease in freshwater fish. The cytological response of their piscine hosts is not precisely understood. RTS11 cell line developed from rainbow trout was used to check the response of macrophage to water molds Achlya and Saprolegni [45]. Fish cell lines are a very useful aid in understanding pathogenicity arising due to nutritional issues. Such studies were carried out using fish cell lines to investigate the proinflammatory mechanism underlying the relationship between dietary PUFA and cardiac lesions using a cell line developed from chum salmon [46].
gene-editing and genetic engineering
Genetically edited fish cell lines have enormous biotechnological and clinical applications. CRISPR (Clustered Regularly Interspaced palindromic repeats-Cas9 (CRISPR associated) has revolutionized gene editing. Generation of improved fish cell lines using CRISPR-Cas9 technology would facilitate aquaculture biotechnological research including fish disease studies. Genetically edited cell lines using genome editing technology would be useful to enhance the transfection efficiency of fish cell lines and utilize those cell lines for the efficient production of viruses for vaccine development. This technique has been mostly used for gene editing in mammalian cell lines whereas the use of gene editing for fish cell lines is in the infancy stage. The use of CRISPR-Cas9 based gene editing method has been reported earlier in fish but an efficient method for gene editing was developed in a fish cell line CHSE developed from Chinook salmon Oncorhynchus tshawytscha for the first time [47]. The cell line was genetically engineered to overexpress different forms of CHSE cell line. Although various attempts have been made, a convincing fish knock-out in vitro model has not yet been developed.
A stable trout head kidney cell line was transfected with a variety of plasmids expressing cytokines Interleukin-6 macrophage colony-stimulating factor (MSCF). Rainbow trout head kidney cell line and RTG-2 stable cell lines were engineered in developed conditioned media to express interleukin (IL-2), IL-6, and macrophage colony-stimulating factor (MCSF) [48]. Greasy grouper Epinephelus tauvina liver cell line GL-av was genetically modified to assess the effectiveness of the anti-apoptotic protein Bcl-xL [49]. Fish cell lines have also important applications in in vitro ploidy manipulation. Polyploidization was successfully obtained in a crucian carp induced by a chemical compound and developed an autotetraploid cell line [50].
Genetically engineered fish cell lines have enormous potentials to be used in fish health, genetics, and biotechnological research. The establishment of a stable cell line is the need of the hour for functional genomics studies for fish genetics and health. With the progress of gene delivery methods, the number of stable genetically modified fish cell lines has increased. Not much effort has been made for the functional characterization of immortal fish cell lines towards developing genetically engineered methodologies [51]. Genetical modification of goldfish cell line, Chinook salmon Oncorhynchus tshawytscha embryo CHSE cell line, rainbow trout Oncorhynchus mykiss hepatoma RTH cell line have provided interesting information for fish disease and immunological research [52, 53]. A transformed EPC (Epithelioma Papulosum Cyprini) cell line under the control of the tilapia HSP70 promoter expressed a green fluorescent protein (GFP)-luciferase fusion gene in response to cellular stress [54]. The fish cell lines can be used for studying stressors concerning infectious fish disease in addition to their usage in investigating environmental stressors concerning climate change. A novel in vitro system was developed using genetically modified Chinook salmon embryonic (CHSE)-TOF cell line to measure the sensitivity of some important virus-like Infectious Haematopoietic Necrosis Virus (IHNV), Infectious Pancreatic Necrosis Virus (IPNV), Salmon Alphavirus (SAV), and Epizootic Haematopoietic Necrosis Virus (EHNV) [55].
Transgenic studies and reproductive biotechnology
Gene targeting and transfer of the genes for transgenic fish production become easier with the advancement of cell culture techniques. Transgenic zebrafish produced applying primary cultures of genetically modified zebrafish male germ cells has paved the way for the development of transgenic lines in model organisms or other animal systems [56]. Genetically modified myogenic cell culture was developed from a transgenic trout (Oncorhynchus mykiss) having a construct containing the GFP reporter gene driven by a fast myosin light chain 2 (MlC2f) promoter [57]. The transgenic line can be produced by utilizing the primordial germ cell (PGC) cultures. Vasa marker facilitates isolation and characterization of targeted PGCs for germline-specific expression in fish. Tanaka et al. developed a transgenic line of medaka using GFP expressed germ cells [58]. Successful transplantation of germ cells in fish demonstrated the possibility of surrogate broodstock production in the aquaculture system. Intraperitoneal transplantation of PGCs was used to produce seedlings in rainbow trout for the first time [59]. The progress in stem cell culture and their subsequent applications in vitro basic research, as well as aquaculture biotechnology, will transform the fisheries sector for achieving the blue revolution. Spermatogonial stem cells transplantation offers many scopes for a successful captive breeding programme for aquaculture species. A spermatogonial cell line (SG3) developed from the mature testis of medaka was capable of producing sperm [60]. The production of fertile medaka fish using ES cells proved the possibility of generating nuclear transplants using fish embryonic cells [61]. More research needs to be carried out in aquaculture species utilising these ES cells. Gene transfer through embryonic stem (ES) cell is a promising tool for the production of transgenic animals [62]. Yoshizaki et al. successfully developed a stem cell-mediated gene transfer method to produce transgenic rainbow trout [63]. ES cells along with PGCs and nuclear transfer strategy make the transformation efficient for transgenic fish production.
Fish cell lines as in vitro models
Fish cell lines have enormous potentials to be used model systems for studying fish disease, immunology, biotechnology, nutrition, and toxicity testing of chemicals and therapeutic agents used in aquaculture as they are ideal substitutes for the whole organism which involves increasing questionable ethical issues. In vitro model has been used for investigating the viral replication and genetics and the production of experimental vaccines to be used in aquaculture. Organ culture developed from tilapia, eel, and trout pituitary glands was used as in vitro model for the production of the growth hormone prolactin [64]. Fish cell lines were found to complement in vivo development studies and recognize the involvement of signaling pathways in the developmental processes [65].
Cell cultures developed from fish can be effectively used as model systems to investigate nutrient assimilation and metabolism in fish but rarely such a culture system has been used to study that aspect of fish nutrition. This also raises the need for the development and characterization of the intestinal cell culture systems to support such studies. Cell line developed from the fish intestine is useful in understanding the effect of functional feed ingredients like probiotics and dietary exposure to chemicals in the aquatic system. Kawano et al. reported the use of the intestinal rainbow trout epithelial cell line (RTgutGC) to elucidate the metabolism of environmentally relevant contaminants in the intestinal tract of fish [66]. Recently, RTgutGC was used as an in vitro model for understanding the functional immunity system of the fish gut as well as the effects of functional feed ingredients in the gut cells [67].
Langan et al. investigated the function of spheroid size in the metabolism of propranolol using an RTgutGC cell line as a 3D fish intestinal model [68]. The cell line of the intestinal epithelial region rainbow trout acts as a barrier to study cellular mechanism of immune function, physiological and pathological response, nutrient uptake, and toxicants [118]. The above RTgutGC cells were compared with new cell lines from the proximal and distal intestine of rainbow trout such as RTpi-MI & RTdi-MI and these cells formed a polarized barrier, which was not permeable to larger molecules and absorbed glucose and proline [119]. The RTgill-W1 cells were used as in vitro model for accessing acute toxicity of select chemicals associated with Whole Effluent Toxicity (WET) testing in both marine and freshwater conditions [120]. A physiologically realistic model system- fish-gut-on-chip was developed by combining intestinal cell culture from rainbow trout (Oncorhynchus mykiss) with microchip technology and microfluidic engineering to study its barrier function towards the environment i.e. food & water and to monitor the function in real-time [69]. In vitro models are extremely important to study collagen synthesis and secretion in humans and other higher vertebrates. Very few models have been established to investigate collagen synthesis and secretion in fish. Lee and Bols reviewed the potential applications of fish cell lines to study collagen as in vitro model for evaluation of physic-chemical factors controlling synthesis, secretion, and deposition of collagen [70].
Cell-based aquaculture
Aquaculture has been growing very fast and facing several challenges to meet the rising demand ensuring the safety and quality of fishery products. The concept of producing cell-based seafood has been emerging as a new approach to producing alternate animal protein. This alternative approach of animal protein production from fish would address several key challenges faced by the conventional aquaculture systems and declining marine capture fisheries. This alternative fish production system will reduce pressure on natural resources and the environment. Accordingly, the entire world is moving towards climate-resilient production systems and in vitro meat production has emerged as an area of cutting edge and priority research. The successful launch of the in vitro hamburger in 2013 has accelerated the research focus on cell-based meats [71]. The ease of growing fish cells at a lower temperature compared to mammalian cells may give cost benefits to the production of cellular fish meat as compared to cellular animal meat. Tissue engineering blend with modern aquaculture techniques can be explored to utilize marine cell culture as an attractive opportunity for the production of in vitro fish meat. Fish muscle cell culture can be used for in vitro fish meat production by exploiting their salient physiological properties like tolerance to a hypoxic-conditions, high buffering capacity, and lower temperature [2]. Fish muscle cell cultures are more adaptable to in vitro conditions than mammalian ones and hence in vitro meat production will be more feasible with fish muscle cell cultures. More concerted efforts and investigations are required to generate information on fish and shellfish muscle cell culture systems to suit in vitro fish meat production systems. The fastest possible path to produce cellular fish meat should start with zebrafish for research and development purposes [72].
The importance of genetic modification and closed aquaculture system paves the way for the innovative concept of cell-based fish production i.e. cellular aquaculture [73]. American space organization NASA had supported the first research program on in vitro edible muscle protein production from goldfish for space travelers during long-term manned space exploration [74]. A better understanding of the myogenesis involved in the muscle cell and tissue culture would be essential to trap the benefits of muscle cell culture in promoting cellular aquaculture. In vitro models like C2C12 cell lines have been utilized in understanding molecular mechanisms underlying muscle growth and differentiation in mammals [2]. Such studies are in the infancy stage in teleost due to the unavailability of equivalent permanent muscle cell lines except for a few fish muscle cell lines [75–77, 77]. Most of them are not from aquaculture species except muscle cell lines developed from Wallago attu [21], olive flounder Paralichthys olivaceus [78] and some myosatellite cells developed from the primary culture of muscle-derived from carp [79] and rainbow trout [80]. Prospects of cell-based aquacultures will rely on the development of appropriate cell lines, optimization of growth media, and other factors, mass production of cells. Some institutes like Good Food Institute; New York, USA have taken initiatives to develop cell-based seafood.
The cell-based molecular mechanism studies will provide basic research data for cell-based fish production. Some investigations on harvested native muscle tissues from fresh water and marine fish provide interesting insights into the potentials of developing a muscle cell culture system [81, 82]. The genome editing by CRISPR/Cas9 system is a promising tool that attains targeted gene editing with high efficiency, without the requirement of integrating an exogenous gene. Its potential is yet to be exploited much in aquaculture using fish cell lines. CRISPR/Cas9 system has been used to get higher skeletal muscle/ muscular growth in aquaculture species like red sea bream; Pagrus major [83] and channel catfish; Ictalurus punctatus [84]. Clean meat farm is a million-dollar industry but academic research lags to propagate clean meat production [85]. Academic research focusing on the development of muscle cell culture systems, standardization media, and bioreactor facilities for large-scale cell production would be required to accelerate in vitro fish meat production and bring it to market.
Vaccine and other products developed from fish cell culture
Global aquaculture particularly shrimp farming used to suffer a major economic loss every year due to the occurrence of viral diseases. The development of vaccines has great relevance to the aquaculture industry to mitigate viral diseases. Purified viruses are likely to be the first health product for use as vaccines obtained from piscine cell cultures [1]. Several viral vaccines have been produced with improved techniques for their delivery at affordable prices [86]. Several fish cell lines have been tested for virus replication towards vaccine development. There is a need for scaling up the efforts towards the development of effective vaccines.
Cell-culture-based technology can be used as a robust and reliable alternative for the production of vaccines. The development of the vaccine and its potency testing requires a large number of live fish. Fish cell culture can be used as an alternative to whole live fish for the production and testing efficacy of fish vaccines. Cell lines like Vero, Madin Darby canine kidney (MDCK), chicken embryo fibroblasts (CEFs) have been mainly used for viral vaccine production [87]. Cell lines developed from humans, monkeys, hamsters, dogs, and chickens have so far been used for the development of vaccines. The studies for the development of viral vaccines using fish cell lines are very much limited. Oh et al. reported that the formalin-inactivated RSIV vaccine was developed from the viruses propagated in Grunt Fin (GF) cells [88]. Several inactivated or attenuated fish viral vaccines have been developed for iridovirus and NNV protection [89, 90], and some of them have been commercialized [91]. However, few cell lines are available to replicate megalocytivirus, betanodavirus, herpesvirus, and aquareovirus for vaccine production, and hence more efforts are warranted to develop specific cell lines for the proliferation of these viruses. Fish cell cultures have great applications in modern vaccine technology including recombinant, DNA/RNA particle vaccines. Only a few fish cell lines have been used in viral propagation leading to vaccine development and diagnostics, and many are under trial for vaccine production. Rainbow trout pronephros cells as in vitro model could be used to screen fish DNA vaccine [92]. The anti-VP5 polyclonal antibody was able to neutralize Grass carp reovirus (GCRV) through in vitro micro neutralizing assay in a grass carp cell line CIK [93]. This would be important towards the development of a vaccine to prevent the infection of GCRV in grass carp which causes great damage to grass carp production in China.
The deficiency of treatment options and limited availability of vaccine poses a challenge for control of viral disease control in aquaculture. In this regard, JL122, a broad-spectrum antiviral agent oxazolidine compounds, was proven to inhibit transmission of IHNV, VHSV, and SVCV in the EPC cell line [94]. Another small molecule LJ001, lipophilic thiazolidine derivative also showed broad-spectrum antiviral properties for inhibition of IHNV infection in the EPC cell line. These hold promise as an immersion treatment option for the outbreak of aquatic rhabdoviral infection. The complex interaction between Infectious kidney and spleen necrosis (ISKNV) and its host Chinese Perch Brain (CPB) cells generated new information for understanding viral pathogenesis and developing antiviral treatment strategies [95].
Fish cell lines exhibit less transfection efficiency, unlike mammalian cell lines. Low transfected cell lines are not useful for the production of recombinant protein and other products. In the case of mammalian cell lines, a higher range of transfection efficiency of mammalian cell lines with the aid of the right combination of cell type and method was achieved up to 100% [96]. However, the same methods applied to fish cell-cultured at lower temperatures (5–15 °C) provided the low transfection efficiency which is often below 10% [97] whereas the transfection efficiency of the head kidney cell line was improved from 11.6% to 90.8% using Amaxa's cell line nucleofector solution T and program T-20 [98]. Hence, alternative reagents or methods should be explored to enhance transfection efficiency in fish cell lines. In addition to vaccine production, fish cell lines should also be explored for the production of human pharmaceutical proteins. The ability of the fish cells to grow at as lows as 4 0C could be exploited in this regard. Transformed fish cell line Epithelioma papulosum cyrpini cells (EPC) were used to stably express and secret recombinant pleurocidin (Ple), a linear cationic peptide of 25 amino acids uninterruptedly for more than 2 years [99]. Fibroblast cell plays an important role in increasing collagen synthesis, collagen secretion under the stimulatory influence of ascorbic acid. Some cell lines developed from fish have been reported to be an ideal in vitro source for the synthesis of collagen [100]. Cytokines such as interferon could be considered for their potential therapeutic potential to fill the gap of shortage of fish therapeutics [1]. Fish interferon was partially purified in small quantity from the rainbow trout gonadal cell line, RTG-2. Transfected RTG-2 cell line expressed Interleukin Cytokines (Interleukin (IL)-2, IL-6, and Macrophage Colony Stimulating Factor (MCSF) and the transfected cell line was used to produce conditioned media-rich in these cytokines [48].
Toxicological and environmental monitoring studies
Different inorganic and organic aquatic pollutants influence the quality and health status of farmed fish and shellfish. Proper investigation to know the ill effects of the aquatic pollutants on farmed fish and shellfish is the need of the hour to improve the marketing of quality seafood. The cell lines have been used as alternative tools to replace the use of whole live fish due to a significant correlations observed between in vitro and in vivo data. Cell lines have been applied as a rapid and economic in vitro tool for screening toxicity of chemicals and environmental samples [1, 93]. Fish cell lines have important applications in studying the effects of different aquatic pollutants on the metabolism of aquatic biological systems and hence there is a potential application of fish cell lines in environmental monitoring. Fish cell lines have been adopted as an in vitro tool for ecotoxicological evaluation of chemicals by many international regulatory bodies like Registration, Evaluation, Authorisation, and Restriction of Chemicals (REACH) in Europe, Food, and Drug Administration (FDA) in the USA. Fish cell cultures facilitated in vitro investigation to find toxic effects of polycyclic aromatic hydrocarbons and aflatoxins in farmed fish [101]. Primary cultures have been used in the case of toxicological investigation in invertebrates as permanent invertebrate cell lines are not available. Considerable progress has to be made for the development of invertebrate cell lines to facilitate in vitro investigations in farmed shelf fish and mollusc. In addition to the aquatic pollutants, toxic and residual effects of antimicrobial drugs used in aquaculture need to be investigated where fish cell culture can be utilized to replace the whole live fish model. In vitro studies established a correlation between in vitro immunosuppression and the interference of various antimicrobial drugs [102]. In this regard, in vitro investigation will provide more insights to increase the awareness of global antimicrobial resistance (AMR) initiated World Health Organization.
Fish nutrition and metabolism
Fish cell lines have the potentials to be used in fish feed formation using alternative ingredients as they provide an excellent in vitro model to study nutrient absorption and assimilation. To facilitate such studies, more intestinal fish culture systems need to be developed. Due to the lack of targeted research tools, the current understanding of the underlying effects of feed ingredients on fish nutrition is limited. The application of appropriate fish cell lines would facilitate further research on the basic functions of the digestive tract and the effects of functional feed ingredients on various aspects of fish nutrition [67]. The vital role of cell lines in biological experimentations is to reduce animals, with major three R rules such as reduction, replacement, and refinement [103]. That enhances the interest of researchers to utilize the in vitro model to study cellular environmental conditions of living biological components. The primary cultures of adipocytes or hepatocytes and myoblasts were significantly used to study molecular mechanisms related to fish nutrition [104–108]. This approach provides significant progress to a limited extent because the primary cultures failed to allow the functional genomic analysis to study the specific gene functions.
Morin et al., 2020 studied the role of RTH-149, RT hepatoma-derived cell line to address nutrition-related queries based on major pathways such as macroautophagy (autophagy), general control nonderepressible 2 (GCN2), and mechanistic Target of Rapamycin (mTOR) pathway that regulate cell homeostasis through amino acids to study the nutrient-sensing signalling. These pathways had attention concerning rainbow trout nutrition, which strongly relies on the supply of amino acid and assessing (1) their capacity to be repressed or induced by starvation, (2) their specific regulation by amino acid availabilities, and (3) their related kinetics. They demonstrate that the starvation can be sensed by RTH-149 cells, which then induce the activation of GCN2 and drive the expression of ISR-related genes in an amino acid-dependent manner. The high concentration of HF (1000 nM) upregulates chop but represses the induction of other ISR-related genes. This result corroborates previous findings from different species demonstrating that Chop overexpression contributes to a negative feedback loop responsible for attenuating the starvation-induced GCN2 response. They also demonstrated RT specificities for amino acid dependencies, time response, and the activation levels of their downstream targets [109]. They concluded that RT cell lines could be an alternative in vivo to analyze nutrition-related queries in Rainbow trout and other carnivorous fish using dietary proteins that provide most of energetic metabolism.
The regulations of atg4, lc3b, and sqstm1 observed in RTH-149 cells were previously described in a mouse cell line to be induced following starvation in a GCN2/ATF4-dependent manner [110]. The starvation-induced autophagy kinetics measured in RTH-149 cells matches with the cells of starved mouse embryonic fibroblast (MEF) [111]. That indicates the amino acid sensing and mTOR activation in RTH-149 cells follow the mechanisms shared between trout, human, and mice cell lines [112, 113] and that has been conserved throughout evolution.
Several fish cell lines have been used as in vitro models to study elongation and desaturation of different PUFA. These in vitro models were also useful for unveiling the pro- inflammatory mechanisms underlying the relationship between dietary PUFA and cardiac lesions in salmon [46], the effect of fatty acid diet on fish inflammatory responses [122, 123]. To study fish nutrition and metabolism, cell lines provide a great interest near future, especially advanced methods, such as CRISPR/cas9 and that may certainly work with new feed formulations for the development of sustainable aquaculture.
Biobanking
With global biodiversity rapidly declining, the need to preserve and conserve biological specimens becomes crucial. Cryopreservation techniques have long been used in agriculture for conservation. Proper freezing of cells can generate a bank of genetic material that can remain viable for hundreds or even thousands of years in the future, with the potential not just to act as reference specimens, but the capacity to regenerate live individuals of a species. While classic cryopreservation methods result in frozen sperm, which would need a fresh egg or frozen embryos–which poses challenges for proper freezing–new technology allows for the production of viable offspring from spermatogonial stem cells of fish.
The rapidly increasing number of fish cell lines raises the need for their long-term storage and conservation in different locations. Fish cell lines are not only the source material for in vitro research but also critical for the conservation of fish germplasm. Integrated efforts protect animal populations within their natural habitat (in situ conservation) and outside their natural environments (ex-situ conservation). Similarly, ancillary conservation facilities like repositories of serum, DNA, and cell lines have been supporting basic and applied research [114]. Bio-banking is emerging as one of the most efficient approaches to provide security at the highest level against the loss of diversity of species [115]. Caulfield and Murdoch have critically reviewed various social and technical issues of biobank globally including public perception, biorights, privacy, technology, and commercialization [116]. The standard operating procedure should be followed for the long-term conservation of fish cell lines. The stability of the cell lines and recovery rate of the fish cell lines should be assessed using different replicates in the freezing medium at different passages to minimize the loss of cells. Working and master stocks of the fish cell lines also need to be maintained separately in the cell line repository. An automated controlled-rate programmable freezer would be ideal to provide reproducible cryopreservation with an optimized freezing program as per the cell’s requirement [117]. A simpler device like cryocan filled with liquid nitrogen may also be used for the storage of cell lines. Researchers can access fully characterized and quality-controlled cell lines from a repository without spending time to develop as per their requirement and at a minimal cost. The repository acts as an “insurance” to secure the loss of cell lines developed by a single laboratory. There should be many standby repositories at different places to avoid loss of the cell lines in case of any catastrophic event. Hence, the cell line repository facilitates promote the propagation of in vitro research as well as the conservation of fish germplasm.
The leading cell line repositories in the world like American Type Culture Collection (ATCC), European Collections of Cell Cultures (ECACC), German Collection of Microorganisms and Cell Cultures (DSMZ) have been providing characterized and authenticated cell line to researchers across the world. Details of cell lines maintained in the repositories worldwide are given in Table 2. Some of the cell lines in the repository suffer from misidentification and contamination due to multiple transfers between laboratories. A certificate citing STR profile for each line is essential to guarantee authentic and contamination-free cell lines [12]. A very encouraging progress in the development of cell lines from different fish species including aquaculture species has been observed in India during the last decade. DBT, Govt. of India New Delhi, India has been instrumental in funding various projects in the development and characterization of fish cell lines in India which resulted in a rapid increase in the number of fish cell lines. This raises the need of establishing fish cell line repositories at a national level for the conservation of fish cell lines in secured places. The authors (M Goswami and W S Lakra) have developed a state-of-the-art facility for the development and storage of cell lines at ICAR-National Bureau of Fish Genetic Resources (NBFGR), Lucknow, India. Recognizing the expertise and research contributions of the authors and other colleagues in the country in the fish cell culture area, DBT funded a megaproject to establish a National Repository of Fish Cell lines (NRFC) at NBFGR, Lucknow. This National Repository of Fish Cell Lines (NRFC) has been in operation at NBFGR, Lucknow, since 2010 which is serving as a National Referral Centre of fish cell lines for research use in the country and abroad. More than 50 fish cell lines from 24 different fish species are being maintained and cryopreserved in the NRFC (Table 3). The facility provides services for deposition, characterization, cryopreservation, and distribution of fish cell lines to the scientific community in India. Many cell lines have been supplied to domestic researchers for their research experiments. This cell line repository would play a critical in contributing to the global biobank as many international scientific communities have expressed interest in sharing fish cell lines from India for collaborative in vitro research.
Table 2.
Status of cell line repository in different parts of the world with reference to fish cell line
| Name of the repository | Total cell line deposited | Total fish cell line deposited |
|---|---|---|
| American Type Culture Collection (ATCC), USA; http://www.atcc.org | 4000 | 19 |
|
Cell Bank Australia, Australia info@cellbankaustralia.com; www.cellbankaustralia.com |
1500 | 16 |
|
Health Protection Agency Culture Collection (HPACC) Formerly known as: European Collection of Cell Cultures (ECACC) http://www.hpacultures.org.uk |
1100 | 25 |
|
German Collection of Microorganisms and Cell Cultures (DSMZ), Germany contact@dsmz.de; http://www.dsmz.de |
789 | 17 |
| National Repository of Fish Cell Line (NRFC), Lucknow, India | 50 | 50 |
Table 3.
Details of fish cell lines at national repository of fish cell line (NRFC), India
| Name of cell line | Fish species | Organ | Cell type | Passage number | NRFC code | Depositor/Reference |
|---|---|---|---|---|---|---|
| PCF | Puntius chelynoides | Fin | Fibroblast | 54 | NRFC001 | Goswami et al. 2014a |
| SRF | Schizothorax richardsonii | Fin | Fibroblast | 49 | NRFC002 | Goswami et al. 2013 |
| TTCF | Tor tor | Fin | Fibroblast | 72 | NRFC003 | Yadav et al. 2012 |
| CCF | Cyprinus carpio | Fin | Epithelial | 71 | NRFC004 | Lakra et al. 2010c |
| WAF | Wallago attu | Fin | Fibroblast | 69 | NRFC005 | Dubey et al., 2014 |
| RF | Labeo rohita | Fin | Fibroblast | 100 | NRFC006 | Lakra et al. 2010a |
| WAM | Wallago attu | Muscle | Fibroblast | 38 | NRFC047 | Dubey et al., 2015a |
| WAG | Wallago attu | Gill | Epithelial | 67 | NRFC048 | Dubey et al., 2015b |
| CPG | Channa punctatus | Gill | Epithelial | 52 | NRFC049 | M Goswami |
| DRM | Danio rerio | Muscle | Fibroblast | 40 | NRFC050 | Kumar et al., 2016 |
| CTM | Catla catla | Thymus (macrophage) | Fibroblast | 49 | NRFC028 | Chaudhary et al., 2014 |
| CTE | Catla catla | Thymus (epithelial) | Epithelial | 53 | NRFC029 | Chaudhary et al., 2013 |
| CCM | Catla catla | Blood (lymphocytes) | Epithelial | NA* | NRFC034 | Chaudhary et al., 2012 |
| KCF | Koi carp | Fin | Epithelial | 54 | NRFC007 | Swaminathan et al., 2015 |
| HBF | Horabragus brachysoma | Fin | Epithelial | 50 | NRFC008 | Swaminathan et al., 2016 |
| Puntius denisonii | Fin | Epithelial | NA | NRFC009 | Swaminathan et al., 2012 | |
| CFF | Pristolepis fasciata | Fin | Fibroblast | 51 | NRFC039 | Swaminathan et al., 2013 |
| CFFN2 | Amphiprion sebae | Fin | Epithelial | 194 | NRFC010 | K. Riji John, Fisheries College and Research Institute (FCRI), Tamilnadu Veterinary & Animal Sciences University, Tuticorin, Tamil Nadu |
| CFBR | Brain | Epithelial | 161 | NRFC011 | ||
| CFSP | Spleen | Epithelial | 241 | NRFC012 | ||
| CFCP1 | Caudal peduncle | Epithelial | 200 | NRFC013 | ||
| SISK | Lates calcarifer | Kidney | Epithelial | 78 | NRFC014 | Hameed et al., 2006 |
| SISS | Spleen | Epithelial | 92 | NRFC015 | Parameswaran et al. 2006b | |
| SIGE | Epinephelus coioides | Eye muscle | Epithelial | 89 | NRFC016 | Parameswaran et al. 2007 |
| IGK | Kidney | Epithelial | 93 | NRFC017 | A. S. Sahul Hameed | |
| SICE | Catla catla | Eye muscle | Epithelial | 84 | NRFC018 | Ahmed et al., 2008 |
| SICH | Heart | Epithelial | 83 | NRFC019 | Ahmed et al., 2009b | |
| CB | Brain | Epithelial | 72 | NRFC020 | Ahmed et al., 2009a | |
| ICG | Gill tissue | Epithelial | 84 | NRFC021 | Taju et al., 2013 | |
| ICF | Clarius batrachus | Fin | Epithelial | 84 | NRFC022 | Babu et al., 2011 |
| LRG | Labeo rohita | Gill | Epithelial | 94 | NRFC023 | Abdul et al., 2013a |
| IEE | Etroplus suratensis | Eye | Epithelial | 90 | NRFC040 | Babu et al., 2012 |
| IEK | Kidney | Epithelial | 96 | NRFC041 | ||
| IEG | Gill | Epithelial | 80 | NRFC042 | ||
| IEB | Brain | Fibroblast | 74 | NRFC043 | ||
| RE | Labeo rohita | Eye | Epithelial | 76 | NRFC044 | Ahmed et al., 2009a |
| CSK | Channa straiatus | Kidney | Fibroblast | 96 | NRFC045 | Abdul et al., 2013b |
| CSG | Gill | Epithelial | 79 | NRFC046 | Abdul et al., 2014 | |
| DT1CPEx | Dascyllus trimaculatus | Caudal peduncle | Fibroblast | 78 | NRFC024 | K S Sobhana, Central Marine Fisheries Research Institute, Kochi |
| DT1F4Ex | Fin | Fibroblast | 72 | NRFC025 | ||
| DT1CPTr | Caudal peduncle | Fibroblast | 60 | NRFC026 | ||
| RC4H1Tr | Rachycentron canadum | Heart | Fibroblast | 52 | NRFC027 | |
| EM2HTr | Epinephelus malabaricus | Heart | Fibroblast | 73 | NRFC030 | |
| EM2GEx | Gill | Fibroblast | 84 | NRFC031 | Sobhana et al., 2009 | |
| EM3GEx | Gill | Epithelial | 74 | NRFC032 | ||
| EM4SPEx | Spleen | Epithelial | 92 | NRFC033 | K S Sobhana | |
| PC1CpTr | Pomacentrus caeruleus | Caudal peduncle | Fibroblast | 74 | NRFC035 | |
| PC1F1Ex | Fin | Fibroblast | 61 | NRFC036 | ||
| PC1L1Tr | Liver | Fibroblast | 50 | NRFC037 | ||
| HC2SpEx | Epinephelus merra | Spleen | Epithelial | 71 | NRFC038 |
*NA Not Available
Conclusion
The growing interests in conducting in vitro research using fish cell lines have necessitated intensification of efforts to maintain constant quality and authenticate cell lines using standard protocol throughout their in vitro life. Fish cell culture has been increasingly used in modern biological research. However, fish cell culture research confronts many challenges like misidentification and contamination. As aquaculture continues to grow worldwide, the application of the fish cell lines in addressing fish disease, genetics, and biotechnological interventions will also increase many folds. Although the total number of fish cell lines has been increasing development and characterization of cell lines from crustaceans and other important marine and aquaculture species are still elusive. Preliminary efforts have been made for the development of stem cell cultures from fish but this area needs more focus to explore their use in modern aquaculture and biotechnology. The potentials of fish cell lines in developing vaccines for aquaculture and other derivable products from cells have yet not been explored fully. Hence, scaling up the fish cell culture systems is essential to grab the opportunities of using fish cell cultures in cell-based aquaculture.
There is a need for global networking and collaborations towards applications of fish cell lines for carrying out advanced in vitro research in fisheries and aqua, blue economy, human health, and environmental management. With technological interventions, fish cell lines could be explored to produce several new products. The information provided by the authors in this paper will add new knowledge to the global database of the fish cell lines besides their potential application in the advancement of aquaculture biotechnology and fisheries science research.
Acknowledgements
Director, ICAR-National Bureau of Fish Genetic Resources, Lucknow; Director, ICAR-Central Institute of Fisheries Education, Mumbai are thankfully acknowledged for providing the facilities. The authors gratefully acknowledge the Department of Biotechnology, Govt. of India, New Delhi for financial support.
Author's Contribution
All the authors contributed to the preparation of this manuscript. MG, and YBS were responsible for the literature search and the first draft of this article. VT and WSL were responsible for language polishing and further editing the manuscript. All authors read and approved the final manuscript. Dr. MG Covered a major aspects of fish cell line for in vitro research like, the current status of fish cell line, novel characterization methods for fish cell line, biobanking, role of cell line in fish health management including the pathological and immunological studies, gene editing of fish cell line and the major contributor of the establishment of NRCF, India. YBS Contributed in the studies on vaccine and other products developed from fish cell culture and the studies on cell-based aquaculture. VT Contributed in the studies of fish cell line in transgenic studies and reproductive biology. WSL Contributed in the studies of fish cell line in toxicological research and environmental monitoring.
Funding
No funding was received to assist with the preparation of this manuscript.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Consent to participate
Not Applicable.
Consent to publish
Not Applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bols NC. Biotechnology and aquaculture: the role of cell cultures. Biotechnol Adv. 1991;9(1):31–49. doi: 10.1016/0734-9750(91)90403-I. [DOI] [PubMed] [Google Scholar]
- 2.Rubio N, Datar I, Stachura D, Krueger K. Cell-based fish: a novel approach to seafood production and an opportunity for cellular agriculture. 2019 doi: 10.20944/preprints201811.0326.v2. [DOI] [Google Scholar]
- 3.FDA (2018) Re: Foods Produced Using Animal Cell Culture Technology; Request for Comments; Docket No. FDA-2018-N-2155.
- 4.Wolf K, Quimby MC. Established eurythermic line of fish cells in vitro. Science. 1962;135(3508):1065–1066. doi: 10.1126/science.135.3508.1065. [DOI] [PubMed] [Google Scholar]
- 5.Bairoch A. The Cellosaurus: a cell line knowledge resource. J Biomol Tech. 2019;18:2902–3002. doi: 10.7171/jbt.18-2902-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bejar J, Hong Y, Alvarez MC. An ES-like cell line from the marine fish Sparus aurata: characterization and chimaera production. Transgenic Res. 2002;11(3):279–289. doi: 10.1023/A:1015678416921. [DOI] [PubMed] [Google Scholar]
- 7.Chen SL, Sha ZX, Ye HQ. Establishment of a pluripotent embryonic cell line from sea perch (Lateolabrax japonicus) embryos. Aquaculture. 2003;218(1–4):141–151. doi: 10.1016/S0044-8486(02)00570-7. [DOI] [Google Scholar]
- 8.Parameswaran V, Shukla R, Bhonde R, Hameed AS. Establishment of embryonic cell line from sea bass (Lates calcarifer) for virus isolation. J Virol Methods. 2006;137(2):309–316. doi: 10.1016/j.jviromet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Dash C, Routray P, Tripathy S, Verma DK, Guru BC, Meher PK, Nandi S, Eknath AE. Derivation and characterization of embryonic stem-like cells of Indian major carp Catla catla. J Fish Biol. 2010;77(5):1096–1113. doi: 10.1111/j.1095-8649.2010.02755.x. [DOI] [PubMed] [Google Scholar]
- 10.Goswami M, Lakra WS, Yadav K, Jena JK. Development of an ES-like cell culture system (RESC) from rohu, Labeo rohita (Ham.) Fish Physiol Biochem. 2012;38(6):1775–1783. doi: 10.1007/s10695-012-9674-5. [DOI] [PubMed] [Google Scholar]
- 11.Hong N, Schartl M, Hong Y. Derivation of stable zebrafish ES-like cells in feeder-free culture. Cell Tissue Res. 2014;357(3):623–632. doi: 10.1007/s00441-014-1882-0. [DOI] [PubMed] [Google Scholar]
- 12.Geraghty RJ, Capes-Davis A, Davis JM, Downward J, Freshney RI, Knezevic I, Lovell-Badge R, Masters JR, Meredith J, Stacey GN, Thraves P, Vias M. Guidelines for the use of cell lines in biomedical research. Br J Cancer. 2014;111(6):1021–1046. doi: 10.1038/bjc.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan J, Hukku B. Cell line characterization and authentication. In Methods in cell biology. Academic Press. 1998;57:203–216. doi: 10.1016/S0091-679X(08)61579-4. [DOI] [PubMed] [Google Scholar]
- 14.Food and Drug Administration (1993) Points to consider in the characterization of cell lines used to produce biologicals. https://www.fda.gov/downloads/BiologicsBloodVaccines/SafetyAvailability/UCM162863.pdf.
- 15.Almeida JL, Cole KD, Plant AL. Standards for cell line authentication and beyond. PLoS Biol. 2016;14(6):e1002476. doi: 10.1371/journal.pbio.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robin (2019) The Cellosaurus: a cell line knowledge resource. http://web.expasy.org/cellosaurus/. [DOI] [PMC free article] [PubMed]
- 17.Yu M, Selvaraj SK, Liang-Chu MM, Aghajani S, Busse M, Yuan J, Lee G, Peale F, Klijn C, Bourgon R, Kaminker JS. A resource for cell line authentication, annotation and quality control. Nature. 2015;520(7547):307–311. doi: 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
- 18.Fusenig NE, Capes-Davis A, Bianchini F, Sundell S, Lichter P. The need for a worldwide consensus for cell line authentication: experience implementing a mandatory requirement at the International Journal of Cancer. PLoS Biol. 2017 doi: 10.1371/journal.pbio.2001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hebert PD, Ratnasingham S, De Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B. 2003 doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper JK, Sykes G, King S, Cottrill K, Ivanova NV, Hanner R, Ikonomi P. Species identification in cell culture: a two-pronged molecular approach. In Vitro Cell Develop Biol-Animal. 2007;43(10):344–351. doi: 10.1007/s11626-007-9060-2. [DOI] [PubMed] [Google Scholar]
- 21.Dubey A, Goswami M, Yadav K, Sharma BS. Development and characterization of a cell line WAF from freshwater shark Wallago attu. Mol Biol Rep. 2014;41(2):915–924. doi: 10.1007/s11033-013-2936-1. [DOI] [PubMed] [Google Scholar]
- 22.Goswami M, Sharma BS, Yadav K, Bahuguna SN, Lakra WS. Establishment and characterization of a piscean fibroblastic cell line from Puntius (Tor) chelynoides suitable for toxicity and gene expression studies as in vitro model. Tissue Cell. 2014;46:206–212. doi: 10.1016/j.tice.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Yashwanth BS, Goswami M, Valappil RK, Thakuria D, Chaudhari A. Characterization of a new cell line from ornamental fish Amphiprion ocellaris (Cuvier, 1830) and its susceptibility to nervous necrosis virus. Sci Rep. 2020;10(1):1–13. doi: 10.1038/s41598-020-76807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagg SK, Lee LE. A proteomics approach to identifying fish cell lines. Proteomics. 2005;5(16):4236–4244. doi: 10.1002/pmic.200401290. [DOI] [PubMed] [Google Scholar]
- 25.Goswami M, Dubey A, Yadav K, Sharma BS, Lakra WS. Identification of fish cell lines using 2-D electrophoresis based protein expression signatures. Curr Proteomics. 2016;13(4):245–252. doi: 10.2174/157016461204160119161034. [DOI] [Google Scholar]
- 26.Villena AJ. Applications and needs of fish and shellfish cell culture for disease control in aquaculture. Rev Fish Biol Fisheries. 2003;13(1):111–140. doi: 10.1023/A:1026304212673. [DOI] [Google Scholar]
- 27.Ariel E, Nicolajsen N, Christophersen MB, Holopainen R, Tapiovaara H, Jensen BB. Propagation and isolation of ranaviruses in cell culture. Aquaculture. 2009;294(3–4):159–164. doi: 10.1016/j.aquaculture.2009.05.019. [DOI] [Google Scholar]
- 28.Lorenzen E, Carstensen B, Olesen NJ. Inter-laboratory comparison of cell lines for susceptibility to three viruses: VHSV, IHNV and IPNV. Dis Aquat Org. 1999;37(2):81–88. doi: 10.3354/dao037081. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Prieto SI, Rodriguez-Saint-Jean S, Garcia-Rosado E, Castro D, Alvarez MC, Borrego JJ. Virus susceptibility of the fish cell line SAF-1 derived from gilt-head seabream. Dis Aquat Org. 1999;35(2):149–153. doi: 10.3354/dao035149. [DOI] [PubMed] [Google Scholar]
- 30.Dong C, Shuang F, Weng S, He J. Cloning of a new fibroblast cell line from an early primary culture from mandarin fish (Siniperca chuatsi) fry for efficient proliferation of megalocytiviruses. Cytotechnology. 2014;66(6):883–890. doi: 10.1007/s10616-013-9642-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collet B, Urquhart K, Noguera P, Larsen KH, Lester K, Smail D, Bruno D. A method to measure an indicator of viraemia in Atlantic salmon using a reporter cell line. J Virol Methods. 2013;191(2):113–117. doi: 10.1016/j.jviromet.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Babu VS, Majeed SA, Nambi KSN, Taju G, Madan N, Raj NS, Hameed AS. Comparison of betanodavirus replication efficiency in ten Indian fish cell lines. Adv Virol. 2013;158(6):1367–1375. doi: 10.1007/s00705-013-1617-7. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Wen Y, Hu X, Wang W, Liang X, Li J, Vakharia V, Lin L. Breaking the host range: mandarin fish is susceptible to a vesiculovirus derived from snakehead fish. J Gen Virol. 2015;96(4):775–781. doi: 10.1099/vir.0.000037. [DOI] [PubMed] [Google Scholar]
- 34.El-Etr SH, Yan L, Cirillo JD. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect Immun. 2001;69:7310–7317. doi: 10.1128/IAI.69.12.7310-7317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noguera PA, Grunow B, Klinger M, Lester K, Collet B, Del-Pozo J. Atlantic salmon cardiac primary cultures: An in vitro model to study viral host pathogen interactions and pathogenesis. PLoS ONE. 2017;12(7):e0181058. doi: 10.1371/journal.pone.0181058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fryer JL, Lannan CN. Rickettsial infections of fish. Annu Rev Fish Dis. 1996;6:3–13. doi: 10.1016/S0959-8030(96)90002-2. [DOI] [Google Scholar]
- 37.McIntosh D, Flano E, Grayson TH, Gilpin ML, Austin B, Villena AJ. Production of putative virulence factors by Renibacterium salmoninarum grown in cell culture. Microbiology. 1997;143(10):3349–3356. doi: 10.1099/00221287-143-10-3349. [DOI] [PubMed] [Google Scholar]
- 38.Menanteau-Ledouble S, Nöbauer K, Razzazi-Fazeli E, El-Matbouli M. Effects of Yersinia ruckeri invasion on the proteome of the Chinook salmon cell line CHSE-214. Sci Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-68903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallejo AN, Ellsaesser CF, Miller NW, Clem LW. Spontaneous development of functionally active long-term monocytelike cell lines from channel catfish. In Vitro Cell Develop Biol-Animal. 1991;27(4):279–286. doi: 10.1007/BF02630904. [DOI] [PubMed] [Google Scholar]
- 40.Faisal M, Ahne W. A cell line (CLC) of adherent peripheral blood mononuclear leucocytes of normal common carp Cyprinus carpio. Dev Comp Immunol. 1990;14(2):255–260. doi: 10.1016/0145-305X(90)90097-X. [DOI] [PubMed] [Google Scholar]
- 41.Chaudhary DK, Sood N, Rathore G, Pradhan PK, Punia P, Agarwal NK. Jena JK (2014) Establishment and characterization of macrophage cell line from thymus of Catla catla (Hamilton. Aquac Res. 1822;45(2):299–311. doi: 10.1111/j.1365-2109.2012.03227.x. [DOI] [Google Scholar]
- 42.Koppang EO, Fischer U, Satoh M, Jirillo E. Inflammation in fish as seen from a morphological point of view with special reference to the vascular compartment. Curr Pharm Des. 2007;13(36):3649–3655. doi: 10.2174/138161207783018644. [DOI] [PubMed] [Google Scholar]
- 43.Ganassin RC, Bols NC. Development of a monocyte/macrophage-like cell line, RTS11, from rainbow trout spleen. Fish Shellfish Immunol. 1998;8(6):457–476. doi: 10.1006/fsim.1998.0153. [DOI] [Google Scholar]
- 44.DeWitte-Orr S (2006) A study of innate antiviral mechanisms using fish cell lines. http://hdl.handle.net/10012/1272.
- 45.Kales SC, DeWitte-Orr SJ, Bols NC, Dixon B. Response of the rainbow trout monocyte/macrophage cell line, RTS11 to the water molds Achlya and Saprolegnia. Mol Immunol. 2007;44(9):2303–2314. doi: 10.1016/j.molimm.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Bell JG, Sargent JR. The incorporation and metabolism of polyunsaturated fatty acids in phospholipids of cultured cells from chum salmon (Oncorhynchus keta) Fish Physiol Biochem. 1992;10:99–109. doi: 10.1007/BF00004521. [DOI] [PubMed] [Google Scholar]
- 47.Dehler CE, Boudinot P, Martin SA, Collet B. Development of an efficient genome editing method by CRISPR/Cas9 in a fish cell line. Mar Biotechnol. 2016;18(4):449–452. doi: 10.1007/s10126-016-9708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corripio-Miyar Y, Secombes CJ, Zou J. Long-term stimulation of trout head kidney cells with the cytokines MCSF, IL-2 and IL-6: Gene expression dynamics. Fish Shellfish Immunol. 2012;32(1):35–44. doi: 10.1016/j.fsi.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Chen SP, Yang HL, Lin HY, Chen MC, Wu JL, Hong JR. Enhanced viability of a nervous necrosis virus-infected stable cell line over-expressing a fusion product of the zfBcl-xL and green fluorescent protein genes. J Fish Dis. 2006;29(6):347–354. doi: 10.1111/j.1365-2761.2006.00725.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y, Wang M, Jiang M, Peng L, Wan C, Liu J, Liu W, Zhao R, Zhao X, Hu W, Liu S, Xiao Y. Autotetraploid cell Line induced by SP600125 from crucian carp and its developmental potentiality. Sci Rep. 2016;6:21814. doi: 10.1038/srep21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collet B, Collins C, Lester K. Engineered cell lines for fish health research. Dev Comp Immunol. 2018;80:34–40. doi: 10.1016/j.dci.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 52.Bonham K, Zafarullah M, Gedamu L. The rainbow trout metallothioneins: molecular cloning and characterization of two distinct cDNA sequences. DNA. 1987;6(6):519–528. doi: 10.1089/dna.1987.6.519. [DOI] [PubMed] [Google Scholar]
- 53.Helmrich A, Bailey GS, Barnes DW. Transfection of cultured fish cells with exogenous DNA. Cytotechnology. 1988;1(3):215–221. doi: 10.1007/BF00145024. [DOI] [PubMed] [Google Scholar]
- 54.Molina A, Carpeaux R, Martial JA, Muller M. A transformed fish cell line expressing a green fluorescent protein-luciferase fusion gene responding to cellular stress. Toxicol In Vitro. 2002;16(2):201–207. doi: 10.1016/S0887-2333(01)00106-0. [DOI] [PubMed] [Google Scholar]
- 55.Lester K, Hall M, Urquhart K, Gahlawat S, Collet B. Development of an in vitro system to measure the sensitivity to the antiviral Mx protein of fish viruses. J Virol Methods. 2012;182(1–2):1–8. doi: 10.1016/j.jviromet.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 56.Kurita K, Burgess SM, Sakai N. Transgenic zebrafish produced by retroviral infection of in vitro-cultured sperm. Proc Natl Acad Sci. 2004;101(5):1263–1267. doi: 10.1073/pnas.0304265101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabillard JC, Sabin N, Paboeuf G. In vitro characterization of proliferation and differentiation of trout satellite cells. Cell Tissue Res. 2010;342(3):471–477. doi: 10.1007/s00441-010-1071-8. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka M, Kinoshita M, Kobayashi D, Nagahama Y. Establishment of medaka (Oryzias latipes) transgenic lines with the expression of green fluorescent protein fluorescence exclusively in germ cells: a useful model to monitor germ cells in a live vertebrate. Proc Natl Acad Sci. 2001;98(5):2544–2549. doi: 10.1073/pnas.041315498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi Y, Yoshizaki G, Takeuchi T. Generation of live fry from intraperitoneally transplanted primordial germ cells in rainbow trout. Biol Reprod. 2003;69(4):1142–1149. doi: 10.1095/biolreprod.103.017624. [DOI] [PubMed] [Google Scholar]
- 60.Hong Y, Liu T, Zhao H, Xu H, Wang W, Liu R, Chen T, Deng J, Gui J. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc Natl Acad Sci. 2004;101(21):8011–8016. doi: 10.1073/pnas.0308668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wakamatsu Y, Ju B, Pristyaznhyuk I, Niwa K, Ladygina T, Kinoshita M, Araki K, Ozato K. Fertile and diploid nuclear transplants derived from embryonic cells of a small laboratory fish, medaka (Oryzias latipes) Proc Natl Acad Sci. 2001;98(3):1071–1076. doi: 10.1073/pnas.98.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alvarez MC, Bejar J, Chen S, Hong Y. Fish ES cells and applications to biotechnology. Mar Biotechnol. 2007;9:117–127. doi: 10.1007/s10126-006-6034-4. [DOI] [PubMed] [Google Scholar]
- 63.Yoshizaki G. Gene transfer in salmonidae: applications to aquaculture. Aquaculture Sci. 2001;49(2):137–142. [Google Scholar]
- 64.Baker BI, Ingleton PM. Secretion of prolactin and growth hormone by teleost pituitariesin vitro. J Comp Physiol. 1975;100(4):269–282. doi: 10.1007/BF00691048. [DOI] [Google Scholar]
- 65.Bloch SR, Vo NT, Walsh SK, Chen C, Lee LE, Hodson PV, Bols NC. Development of a cell line from the American eel brain expressing endothelial cell properties. In Vitro Cell Develop Biol-Animal. 2016;52(4):395–409. doi: 10.1007/s11626-015-9986-8. [DOI] [PubMed] [Google Scholar]
- 66.Kawano A, Haiduk C, Schirmer K, Hanner R, Lee LEJ, Dixon B, Bols NC. Development of a rainbow trout intestinal epithelial cell line and its response to lipopolysaccharide. Aquac Nutr. 2011;17(2):e241–e252. doi: 10.1111/j.1365-2095.2010.00757.x. [DOI] [Google Scholar]
- 67.Wang J, Lei P, Gamil AAA, Lagos L, Yue Y, Schirmer K, Mydland LT, Øverland M, Krogdahl Å, Kortner TM. Rainbow Trout (Oncorhynchus Mykiss) intestinal epithelial cells as a model for studying gut immune function and effects of functional feed ingredients. Front Immunol. 2019;10:152. doi: 10.3389/fimmu.2019.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langan LM, Owen SF, Trznadel M, Dodd NJ, Jackson SK, Purcell WM, Jha AN. Spheroid size does not impact metabolism of the β-blocker propranolol in 3D intestinal fish model. Front Pharmacol. 2018;9:947. doi: 10.3389/fphar.2018.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drieschner C, Vo NT, Schug H, Burkard M, Bols NC, Renaud P, Schirmer K. Improving a fish intestinal barrier model by combining two rainbow trout cell lines: epithelial RTgutGC and fibroblastic RTgutF. Cytotechnology. 2019;71(4):835–848. doi: 10.1007/s10616-019-00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee LEJ, Bols NC. Collagen producing fish cell lines and their use in biomedical research. In In Vitro Cell Develop Biol-Animal. 2016;52:17–17. [Google Scholar]
- 71.Zaraska M (2013). Lab‐grown beef taste test: ‘Almost’ like a burger. The Washington post. Retrieved from http://www.washingtonpost.com/national/health-science/lab-grown-beef-taste-test-almost-like-a-burger/2013/08/05/921a5996-fdf4-11e2-96a8-d3b921c0924a_story.html.
- 72.Potter G, Smith AS, Vo NT, Muster J, Weston W, Bertero A, Maves L, Mack DL, Rostain A. A more open approach is needed to develop cell-based fish technology: it starts with Zebrafish. One Earth. 2020;3(1):54–64. doi: 10.1016/j.oneear.2020.06.005. [DOI] [Google Scholar]
- 73.Krueger K, Rubio N, Datar I, Stachura D. Cell-based fish: a novel approach to seafood production and an opportunity for cellular agriculture. Frontiers in Sustainable Food Systems. 2019;3:43. doi: 10.3389/fsufs.2019.00043. [DOI] [Google Scholar]
- 74.Benjaminson M, Gilchriest J, Lorentz M. In vitro edible muscle protein production system (MMPS): stage 1, fish. Acta Astronaut. 2002;51:879–889. doi: 10.1016/S0094-5765(02)00033-4. [DOI] [PubMed] [Google Scholar]
- 75.Zhao Z, Lu Y. Establishment and characterization of two cell lines from bluefin trevally Caranx melampygus. Dis Aquat Org. 2006;68(2):91–100. doi: 10.3354/dao068091. [DOI] [PubMed] [Google Scholar]
- 76.Zhao Z, Montgomery-Brock D, Lee CS, Lu Y. Establishment, characterization and viral susceptibility of 3 new cell lines from snakehead, Channa striatus (Blooch) Methods Cell Sci. 2004;25(3–4):155–166. doi: 10.1007/s11022-004-3804-0. [DOI] [PubMed] [Google Scholar]
- 77.Rougee L, Ostrander GK, Richmond RH, Lu Y. Establishment, characterization, and viral susceptibility of two cell lines derived from goldfish Carassius auratus muscle and swim bladder. Dis Aquat Org. 2007;77(2):127–135. doi: 10.3354/dao01802. [DOI] [PubMed] [Google Scholar]
- 78.Kumar A, Singh N, Goswami M, Srivastava JK, Mishra AK, Lakra WS. Establishment and characterization of a new muscle cell line of Zebrafish (Danio rerio) as an in vitro model for gene expression studies. Anim Biotechnol. 2016;27(3):166–173. doi: 10.1080/10495398.2016.1147455. [DOI] [PubMed] [Google Scholar]
- 79.Peng L, Zheng Y, You F, Wu Z, Zou Y, Zhang P. Establishment and characterization of a testicular Sertoli cell line from olive flounder Paralichthys olivaceus. Chin J Oceanol Limnol. 2016;34(5):1054–1063. doi: 10.1007/s00343-016-5091-4. [DOI] [Google Scholar]
- 80.Koumans JTM, Akster HA, Dulos GJ, Osse JWM. Myosatellite cells of Cyprinus carpio (Teleostei) in vitro: isolation, recognition and differentiation. Cell Tissue Res. 1990;261(1):173–181. doi: 10.1007/BF00329450. [DOI] [Google Scholar]
- 81.Powell RL, Dodson MV, Cloud JG. Cultivation and differentiation of satellite cells from skeletal muscle of the rainbow trout Salmo gairdneri. J Exp Zool. 1989;250(3):333–338. doi: 10.1002/jez.1402500314. [DOI] [Google Scholar]
- 82.Castellini MA, Somero GN. Buffering capacity of vertebrate muscle: correlations with potentials for anaerobic function. J Comp Physiol. 1981;143(2):191–198. doi: 10.1007/BF00797698. [DOI] [Google Scholar]
- 83.Anchelin M, Murcia L, Alcaraz-Pérez F, García-Navarro EM, Cayuela ML. Behaviour of telomere and telomerase during aging and regeneration in zebrafish. PLoS ONE. 2011;6(2):e16955. doi: 10.1371/journal.pone.0016955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kishimoto K, Washio Y, Yoshiura Y, Toyoda A, Ueno T, Fukuyama H, Kato K, Kinoshita M. Production of a breed of red sea bream Pagrus major with an increase of skeletal muscle mass and reduced body length by genome editing with CRISPR/Cas9. Aquaculture. 2018;495:415–427. doi: 10.1016/j.aquaculture.2018.05.055. [DOI] [Google Scholar]
- 85.Khalil K, Elayat M, Khalifa E, Daghash S, Elaswad A, Miller M, Abdelrahman H, Ye Z, Odin R, Drescher D, Vo K. Generation of myostatin gene-edited channel catfish (Ictalurus punctatus) via zygote injection of CRISPR/Cas9 system. Sci Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-07223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dolgin E. Sizzling interest in lab-grown meat belies lack of basic research. Nature. 2019;566:161–162. doi: 10.1038/d41586-019-00373-w. [DOI] [PubMed] [Google Scholar]
- 87.Dhar AK, Manna SK, Allnutt FT. Viral vaccines for farmed finfish Virusdisease. 2014;25(1):1–17. doi: 10.1007/s13337-013-0186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Genzel Y. Designing cell lines for viral vaccine production: Where do we stand? Biotechnol J. 2015;10(5):728–740. doi: 10.1002/biot.201400388. [DOI] [PubMed] [Google Scholar]
- 89.Oh SY, Kim WS, Oh MJ, Nishizawa T. Multiplication rate of red seabream iridovirus (RSIV) in rock bream Oplegnathus fasciatus at different fish rearing temperatures. Fish Pathology. 2016;51(4):194–198. doi: 10.3147/jsfp.51.194. [DOI] [Google Scholar]
- 90.Nakajima K, Ito T, Kurita J, Kawakami H, Itano T, Fukuda Y, Aoi Y, Tooriyama T, Manabe S. Effectiveness of a vaccine against red sea bream iridoviral disease in various cultured marine fish under laboratory conditions. Fish Pathology. 2002;37(2):90–91. doi: 10.3147/jsfp.37.90. [DOI] [Google Scholar]
- 91.Sato A, Okamoto N. Induction of virus-specific cell-mediated cytotoxic responses of isogeneic ginbuna crucian carp, after oral immunization with inactivated virus. Fish Shellfish Immunol. 2010;29(3):414–421. doi: 10.1016/j.fsi.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 92.Biering E, Villoing S, Sommerset I, Christie KE. Update on viral vaccines for fish. Devlopmental Biology (Basel) 2005;121:97–113. [PubMed] [Google Scholar]
- 93.Ortega-Villaizan M, Martinez-Lopez A, Garcia-Valtanen P, Chico V, Perez L, Coll JM, Estepa A. Ex vivo transfection of trout pronephros leukocytes, a model for cell culture screening of fish DNA vaccine candidates. Vaccine. 2012;30(41):5983–5990. doi: 10.1016/j.vaccine.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 94.He Y, Xu H, Yang Q, Xu D, Lu L. The use of an in vitro microneutralization assay to evaluate the potential of recombinant VP5 protein as an antigen for vaccinating against Grass carp reovirus. Virology journal. 2011;8(1):1–6. doi: 10.1186/1743-422X-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Balmer BF, Getchell RG, Powers RL, Lee J, Zhang T, Jung ME, Purcell MK, Snekvik K, Aguilar HC. Broad-spectrum antiviral JL122 blocks infection and inhibits transmission of aquatic rhabdoviruses. Virology. 2018;525:143–149. doi: 10.1016/j.virol.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li C, Fu X, Lin Q, Liu L, Liang H, Huang Z, Li N. Autophagy promoted infectious kidney and spleen necrosis virus replication and decreased infectious virus yields in CPB cell line. Fish Shellfish Immunol. 2017;60:25–32. doi: 10.1016/j.fsi.2016.11.037. [DOI] [PubMed] [Google Scholar]
- 97.Bonetta L. The inside scoop—evaluating gene delivery methods. Nat Meth. 2005;2(11):875–883. doi: 10.1038/nmeth1105-875. [DOI] [Google Scholar]
- 98.Romoren K, Fjeld XT, Poleo AB, Smistad G, Thu BJ, Evensen O. Transfection efficiency and cytotoxicity of cationic liposomes in primary cultures of rainbow trout (Oncorhynchus mykiss) gill cells. Biochim Biophys Acta. 2005;1717(1):50–57. doi: 10.1016/j.bbamem.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 99.Schiotz BL, Rosado EG, Baekkevold ES, Lukacs M, Mjaaland S, Sindre H, Grimholt U, Gjøen T. Enhanced transfection of cell lines from Atlantic salmon through nucoleofection and antibiotic selection. BMC Res Notes. 2011;4(1):136. doi: 10.1186/1756-0500-4-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brocal I, Falco A, Mas V, Rocha A, Perez L, Coll JM, Estepa A. Stable expression of bioactive recombinant pleurocidin in a fish cell line. Appl Microbiol Biotechnol. 2006;72(6):217–1228. doi: 10.1007/s00253-006-0393-7. [DOI] [PubMed] [Google Scholar]
- 101.Spiteri KW (2014) The establishment of a fibroblastic cell line from yellow perch (Perca flavescens) and its potential applications in toxicology. https://scholars.wlu.ca/etd/1630/.
- 102.Behrens A, Schirmer K, Bols NC, Segner H. Polycyclic aromatic hydrocarbons as inducers of cytochrome P4501A enzyme activity in the rainbow trout liver cell line, RTL-W1, and in primary cultures of rainbow trout hepatocytes. Environ Toxicol Chem Int J. 2001;20(3):632–643. doi: 10.1002/etc.5620200324. [DOI] [PubMed] [Google Scholar]
- 103.Lunden T, Miettinen S, Lönnström LG, Lilius EM, Bylund G. Effect of florfenicol on the immune response of rainbow trout (Oncorhynchus mykiss) Vet Immunol Immunopathol. 1999;67(4):317–325. doi: 10.1016/S0165-2427(98)00232-3. [DOI] [PubMed] [Google Scholar]
- 104.Russell WMS, Burch RL. The principles of humane experimental technique. Methuen; 1959. [Google Scholar]
- 105.Segner H, Blair J, Wirtz G, Miller M. Cultured trout liver-cells-utilization of substrates and response to hormones. In Vitro Cellular & Developmental Biology-Animal. 1994;30A:306–311. doi: 10.1007/BF02631451. [DOI] [PubMed] [Google Scholar]
- 106.Froehlich JM, Seiliez I, Gabillard JC, Biga PR. Preparation of primary myogenic precursor cell/myoblast cultures from basal vertebrate lineages. J Vis Exp. 2014 doi: 10.3791/51354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bower NI, Johnston IA. Paralogs of Atlantic salmon myoblast determination factor genes are distinctly regulated in proliferating and differentiating myogenic cells. Am J Physiol-Regul Integr Comp Physiol. 2010;298:R1615–R1626. doi: 10.1152/ajpregu.00114.2010. [DOI] [PubMed] [Google Scholar]
- 108.Garcia de la Serrana D, Codina M, Capilla E, Jimenez-Amilburu V, Navarro I, Du SJ, Johnston IA, Gutierrez J. Characterisation and expression of myogenesis regulatory factors during in vitro myoblast development and in vivo fasting in the gilthead sea bream (Sparus aurata) Comp Biochem Physiol A-Mol Integr Physiol. 2014;167:90–99. doi: 10.1016/j.cbpa.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 109.Cleveland BM. In vitro and in vivo effects of phytoestrogens on protein turnover in rainbow trout (Oncorhynchus mykiss) white muscle. Comp Biochem Physiol C-Toxicol Pharm. 2014;165:9–16. doi: 10.1016/j.cbpc.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 110.Morin G, Pinel K, Dias K, Seiliez I, Beaumatin F. RTH-149 cell line, a useful tool to decipher molecular mechanisms related to fish nutrition. Cells. 2020;9(8):1754. doi: 10.3390/cells9081754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.B’chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2 alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, Kodama A, Ishihara T, Mizushima T, Mizushima N. An autophagic flux probe that releases an internal control. Mol Cell. 2016;64:835–849. doi: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 113.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Beaumatin F, O’Prey J, Barthet VJA, Zunino B, Parvy JP, Bachmann AM, O’Prey M, Kania E, Gonzalez PS, Macintosh R. mTORC1 activation requires DRAM-1 by facilitating lysosomal amino acid efflux. Mol Cell. 2019;76:163–176. doi: 10.1016/j.molcel.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wildt DE. Genome resource banking for wildlife research, management, and conservation. ILAR J. 2000;41(4):228–234. doi: 10.1093/ilar.41.4.228. [DOI] [PubMed] [Google Scholar]
- 116.Goswami M, Mishra A, Ninawe NS, Trudeau VL, Lakra WS. Bio-banking: an emerging approach for conservation of fish germplasm. Poultry Fisheries Wildlife Sci. 2016 doi: 10.4172/2375-446X.1000143. [DOI] [Google Scholar]
- 117.Caulfield T, Murdoch B. Genes, cells, and biobanks: Yes, there’s still a consent problem. PLoS Biol. 2017 doi: 10.1371/journal.pbio.2002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Freshney RI. Culture of animal cells: a manual of basic technique and specialized applications. NJ: Wiley; 2015. [Google Scholar]
- 119.Minghetti M, Drieschner C, Bramaz N, Schug H, Schirmer K. A fish intestinal epithelial barrier model established from the rainbow trout (Oncorhynchus mykiss) cell line. RTgutGC Cell biology and toxicology. 2017;33(6):539–555. doi: 10.1007/s10565-017-9385-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pasquariello R, Verdile N, Pavlovic R, Panseri S, Schirmer K, Brevini TA, Gandolfi F. New stable cell lines derived from the proximal and distal intestine of rainbow trout (Oncorhynchus mykiss) retain several properties observed in vivo. Cells. 2021;10(6):1555. doi: 10.3390/cells10061555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scott J, Belden JB, Minghetti M. Applications of the RTgill-W1 cell line for acute whole-effluent toxicity testing: in vitro–in vivo correlation and optimization of exposure conditions. Environ Toxicol Chem. 2021;40(4):1050–1061. doi: 10.1002/etc.4947. [DOI] [PubMed] [Google Scholar]
- 122.Yu Y, Wei S, Wang Z, Huang X, Huang Y, Cai J, Li C, Qin Q. Establishment of a new cell line from the snout tissue of golden pompano Trachinotus ovatus, and its application in virus susceptibility. J Fish Biol. 2016;88(6):2251–2262. doi: 10.1111/jfb.12986. [DOI] [PubMed] [Google Scholar]
- 123.Ashton I, Clements K, Barrow SE, Secombes CJ, Rowley AF. Effects of dietary fatty acids on eicosanoid-generating capacity, fatty acid composition and chemotactic activity of rainbow trout (Oncorhynchus mykiss) leucocytes. Biochim Biophys Acta. 1994;1214:253–262. doi: 10.1016/0005-2760(94)90071-X. [DOI] [PubMed] [Google Scholar]
- 124.Tocher DR, Bell JG, Sargent JR. Production of eicosanoids derived from 20:4n–6 and 20:5n–3 in primary cultures of turbot (Scophthalmus maximus) brain astrocytes in response to platelet activating factor, substance P and interleukin-1 beta. Comp Biochem Physiol B Biochem Mol Biol. 1996;115:215–222. doi: 10.1016/0305-0491(96)00113-7. [DOI] [PubMed] [Google Scholar]