Abstract
Objective
To explore the main bioactive compounds and investigate the underlying mechanism of Pollen Typhae (PT) against diabetic retinopathy (DR) by network pharmacology and molecular docking analysis.
Methods
Bioactive ingredients and the target proteins of PT were obtained from TCMSP, and the related target genes were acquired from the SwissTargetPrediction database. The target genes of DR were obtained from GeneCards, TTD database, DisGeNET database, and DrugBank. The compound-target interaction network was established based on Cytoscape 3.7.2. The protein-protein interaction (PPI) network was constructed via STRING database and Cytoscape 3.7.2. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were visualized through DAVID database and Bioinformatics. Ingredient-gene-pathway network analysis was conducted to further screen the ingredients, target proteins, and pathways closely related to the biological mechanism on PT for DR, and molecular docking analysis was performed by SYBYL-X 2.1.1 software. Finally, the mechanism and underlying targets of PT in the treatment of DR were predicted.
Results
A total of 8 compounds and 171 intersection targets were obtained based on the online network database. 7 main compounds were screened from compound-target network, and 53 targets including the top six key targets (PTGS2, AKT1, VEGFA, MAPK3, TNF, and EGFR) were further acquired from PPI analysis. The 53 key targets covered 80 signaling pathways, among which PI3K-Akt signaling pathway, focal adhesion, Rap1 signaling pathway, VEGF signaling pathway, and HIF-1 signaling pathway were closely connected with the biological mechanism involved in the alleviation of DR by PT. Ingredient-gene-pathway network shows that AKTI, EGFR, and VEGFA were core genes, kaempferol and isorhamnetin were pivotal ingredients, and VEGF signaling pathway and Rap1 signaling pathway were closely involved in anti-DR. The docking results indicated that five main compounds (arachidonic acid, isorhamnetin, quercetin, kaempferol, and (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one) had good binding activity with EGFR and AKT1 targets.
Conclusion
The active ingredients in PT may regulate the levels of inflammatory factors, suppress the oxidative stress, and inhibit the proliferation, migration, and invasion of retinal pericytes by acting on PTGS2, AKT1, VEGFA, MAPK3, TNF, and EGFR targets through VEGF signaling pathway, PI3K-Akt signaling pathway, Rap1 signaling pathway, and HIF-1 signaling pathway to play a therapeutic role in diabetic retinopathy.
1. Introduction
Diabetic retinopathy (DR), which results from chronic high blood glucose levels and almost occurs in medium and late stage of type 1 and type 2 diabetes, is one of the most serious microvascular complications of diabetes mellitus [1]. Diabetic retinopathy can result in severe visual impairment, vitreous hemorrhage, and even blindness [2]. The World Health Organization estimated that more than 93 million people worldwide are affected by DR. The rapidly rising incidence of DR is a serious threat to public health which has been put into much attention. Moreover, DR is the main cause of adult-acquired blindness [3, 4], which dramatically affects a patient's life and spirit and leads to a massive social economic burden. Currently, a variety of pharmaceutical and surgical treatments have been developed for DR [5]; however, their adverse side effects and limited therapeutic effect have contributed to making DR a challenge for clinical practice. Therefore, it is crucial to develop potential and effective therapeutic strategies to treat DR based on its pathogenesis.
Traditional Chinese medicine (TCM), as a type of alternative drug, is considered a multicomponent and multitarget treatment for diseases corresponding to complex organic targets. Thus, it is necessary to study pharmacological effects and molecular mechanisms of TCMs. Pollen Typhae (PT) is a classical and vital Chinese medicine with well-defined phytochemicals, which has been used for the treatment of hyperlipidemia [6], nonproliferative diabetic retinopathy [7], and vitreous hemorrhage [8]. Previous reports have shown that active ingredients of PT can inhibit the apoptosis [9], improve insulin resistance [10], and increase insulin sensitivity [11]. PT is an effective TCM for the treatment of DR. However, due to the complexity of the active ingredients of PT, its molecular mechanism has not been elucidated clearly. Therefore, the objective of this study is to elucidate the active ingredients of PT correlated to the molecular mechanism of PT on the targets of DR.
Network pharmacology, an emerged discipline of Bioinformatics, is increasingly applied in TCM research in recent years. This approach is in accordance with the holistic theory that TCM emphasizes the treatment of diseases with multiple components and multiple targets [12]. Consequently, systematic pharmacology has been recognized as a suitable method to achieve the interaction between the active ingredients and various targets and explore the underlying mechanisms against complicated disease [13, 14]. Molecular docking technology, a key-and-lock principle, is a theoretical simulation approach to investigate the binding capacity of protein receptors to small medicine molecules using computers to study the receptors' characteristics as well as the interaction between receptors and medicinal molecules. We have referenced the previously published article [15] to investigate the mechanisms of PT on DR.
In this study, the bioactive components of PT and the specific target proteins of PT and DR were combined to construct the pharmacological networks of “components-targets-diseases” and PPI. Moreover, several biologic pathways have been proposed to be involved in DR, for example, PI3K-Akt signaling pathways, VEGF signaling pathways, HIF-1 signaling pathways, and Rap1 and TNF signaling pathways. This suggests that it is of great significance to explore the characteristics of ingredients and targets of PT on DR in biological networks and molecular docking analysis. The workflow is summarized in Figure 1.
Figure 1.
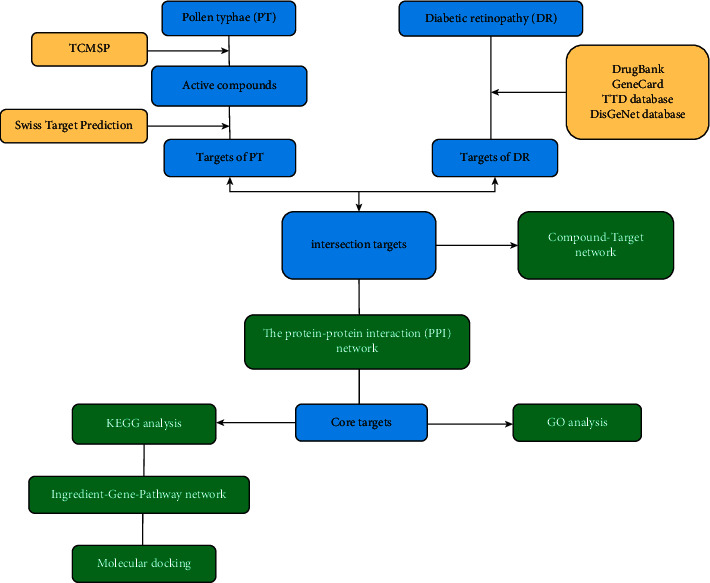
The workflow of this study.
2. Materials and Methods
2.1. Collection of Chemical Ingredients and Related Targets in PT
The chemical ingredients of PT were gathered from TCMSP database (http://old.tcmsp-e.com/tcmsp.php). In order to select the ingredients, which have better pharmacokinetic properties and oral bioavailability in vivo, the ingredients were filtrated by suggested criterion in TCMSP database, and the ingredients whose drug-likeness (DL) ≥0.18 and oral bioavailability (OB) ≥30% were regarded as putative major ingredients and retained [16]. Additionally, the SMILES number of ingredients obtained from PubChem database was imported into the SwissTargetPrediction database through Probability∗ >0 to select the predicted targets.
2.2. Collection of the Targets of Diabetic Retinopathy
The DR-related targets were searched in the DrugBank (https://http://www.drugbank.ca/), GeneCards platform (https://www.genecards.org), Therapeutic Target Database (TTD), and DisGeNET database with “diabetic retinopathy” as the keywords [17, 18]. These collected targets were merged and duplicates were removed.
2.3. Establishment of Compound-Target Network and Analysis
To better demonstrate the anti-DR ingredients and targets of PT, Cytoscape 3.7.2 software was used to visualize the results and analyze the topological properties of the network. In the network, nodes represent ingredients or targets, and edges indicate compound-target gene interactions. In the topological analysis, the topological parameters were significant indexes to judge the importance of nodes, mainly including the degree of nodes and the betweenness centrality. Afterwards, the main active components were screened.
2.4. Protein-Protein Interaction Analysis
In order to explore the underlying mechanism of PT on DR, protein-protein interaction (PPI) networks were constructed using the STRING database and Cytoscape 3.7.2 software [19, 20]. The 171 intersection target genes were imported into the STRING database and the species were selected as “Homo sapiens” to obtain the interaction relationship between the targets, and it was saved in TSV format. Then, the file with TSV format was set into Cytoscape 3.7.2 software and the topology parameters were calculated to build an advanced protein-protein interaction network diagram and filter the core targets.
2.5. Gene Ontology and KEGG Pathway Enrichment Analysis
Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) are important methods to screen the key targets and enriched pathways in DAVID database (https://david.ncifcrf.gov/tools.jsp) setting P < 0.05 as the threshold [21, 22]. In this study, we chose the top 10 functional categories in biological process (BP), cellular component (CC), and molecular function (MF) and the top 20 DR-related KEGG terms, by using Bioinformatics (http://www.bioinformatics.com.cn/), to explore the key targets and the related pathways [23].
2.6. Construction of Ingredient-Gene-Pathway Network and Analysis
The ingredient-gene-pathway network was established by importing top 5 intersecting ingredients, 7 intersecting genes, and the leading 20 KEGG pathways into Cytoscape software. In this network, nodes represent ingredients, targets, and pathways, and edges indicate compound-target and target-pathway interactions. The topological parameters, mainly including the degree of nodes and the betweenness centrality, were utilized to evaluate the centrality features of nodes in ingredient-gene-pathway network.
2.7. Molecular Docking Analysis
Top five molecular compounds based on compound-target network were screened, and their molecular structures were downloaded from TCMSP. The top two key targets were selected by ingredient-gene-pathway network, and their PDB was obtained from Protein Data Bank (PDB) database. Then the PDB formats of proteins and the compounds were docked and visualized in the SYBYL-X 2.1.1 software, and the docking score was calculated to evaluate the binding degree between the bioactive components and the targets of PT. It is generally believed that the CSCORE >4.0 indicates that the docking molecules have good binding activity with the target.
3. Results
3.1. Screening of Active Ingredients and Related Targets
With the values of OB ≥30% and DL ≥0.18 properties, 8 active compounds, (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one, arachidonic acid, isorhamnetin, quercetin, kaempferol, beta-sitosterol, testosterone palmitate, and kaempferol-3-O-α-L-rhamnosyl(1 ⟶ 2)-β-D-glucoside qt were selected from TCMSP database and 283 related targets of the compounds were screened from SwissTargetPrediction database. In addition, a total of 4077 DR-related targets were identified through the screening of the disease target database after removing the duplicate data. Then, the intersection of 171 targets was obtained.
3.2. The Compound-Target Network
In order to identify the interaction relationship between the ingredients and related targets, a compound-target network and topological analysis were made by using Cytoscape 3.7.2 and the degree values of compounds-targets were calculated. The higher the degree is, the closer the relationship between compounds and targets is, and the more important the compounds are in this network. Figure 2 shows that five compounds probably were considered as significant therapeutic compounds in DR, namely, arachidonic acid, isorhamnetin, quercetin, kaempferol, and (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one; and the seven ingredients are listed in Table 1.
Figure 2.
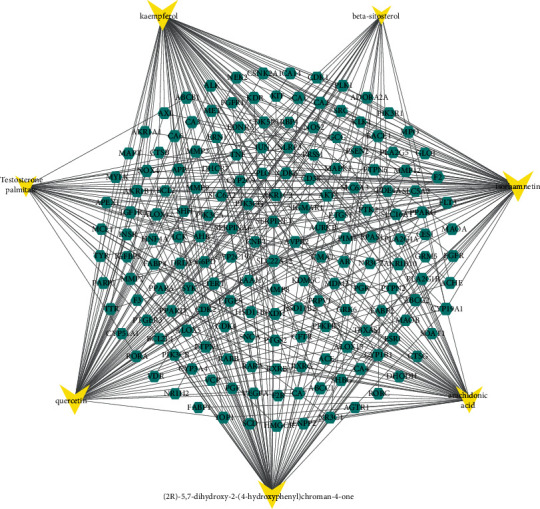
Active component-target network. The triangle node in yellow represents active component, and the hexagon node in green represents the intersection targets. The interaction between the components and the targets is represented by the edge.
Table 1.
Information of 7 compounds in the pharmacological network.
| Molecular ID | Molecular name | Molecular structure | OB (%) | DL |
|---|---|---|---|---|
| MOL001439 | Arachidonic acid |
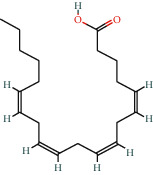
|
45.57 | 0.2 |
|
| ||||
| MOL000358 | Beta-sitosterol |
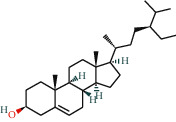
|
36.91 | 0.75 |
|
| ||||
| MOL000354 | Isorhamnetin |
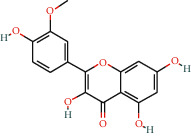
|
49.6 | 0.31 |
|
| ||||
| MOL000422 | Kaempferol |
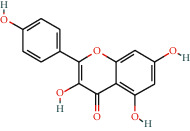
|
41.88 | 0.24 |
|
| ||||
| MOL000098 | Quercetin |
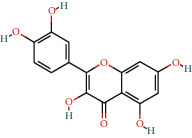
|
46.43 | 0.28 |
|
| ||||
| MOL006111 | Testosterone palmitate |
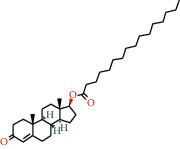
|
34.14 | 0.71 |
|
| ||||
| MOL001040 | (2R)-5,7-Dihydroxy-2-(4-hydroxyphenyl)chroman-4-one |
|
42.36 | 0.21 |
3.3. Construction and Topological Analysis of the PPI Network
The intersection targets PT and DR were inputted into the STRING database to obtain the PPI network, with 171 nodes and 1649 edges. Cytoscape 3.7.2 was used for merging and calculating topological analysis. The higher the degree is, the more important the targets are in this network. As is shown in Figure 3, the 53 key targets including the top six key targets (PTGS2, AKT1, VEGFA, MAPK3, TNF, and EGFR) were acquired, and the details of the 53 key targets are listed in Table 2. The PPI analysis demonstrated that these targets may be the key targets for PT at treating DR. In addition, the related abbreviations are provided in Supplementary Table S1.
Figure 3.
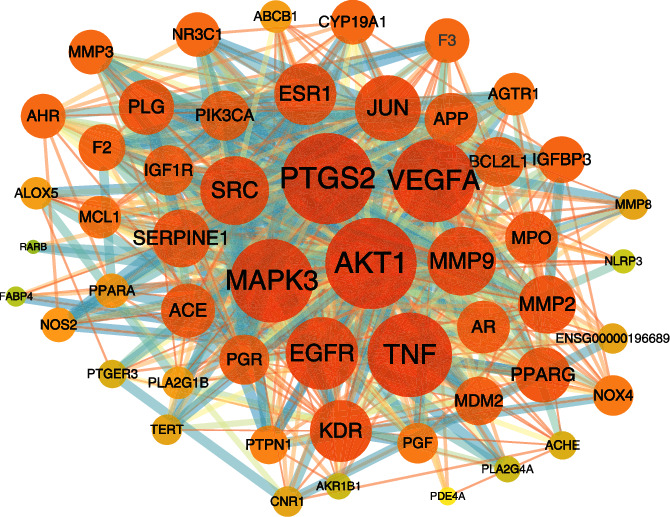
The PPI network. A darker color and larger node indicate a larger degree value, and the interaction between the components and the targets is represented by the edge.
Table 2.
The details of the 53 key targets.
| Core targets | Protein name | UniProt ID |
|---|---|---|
| AKT1 | RAC-alpha serine/threonine-protein kinase | P31749 |
| VEGFA | Vascular endothelial growth factor A | P15692 |
| MAPK3 | Mitogen-activated protein kinase 3 | P27361 |
| TNF | Tumor necrosis factor | P01375 |
| EGFR | Epidermal growth factor receptor | P00533 |
| SRC | Proto-oncogene tyrosine-protein kinase Src | P12931 |
| PTGS2 | Prostaglandin G/H synthase 2 | P35354 |
| MMP9 | Matrix metalloproteinase-9 | P14780 |
| JUN | Transcription factor AP-1 | P05412 |
| ESR1 | Estrogen receptor | P03372 |
| KDR | Vascular endothelial growth factor receptor 2 | P35968 |
| PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | P42336 |
| AR | Androgen receptor | P10275 |
| MDM2 | E3 ubiquitin-protein ligase Mdm2 | Q00987 |
| MMP2 | 72 kDa type IV collagenase | P08253 |
| APP | Amyloid-beta precursor protein | P05067 |
| PPARG | Peroxisome proliferator-activated receptor gamma | P37231 |
| BCL2L1 | Bcl-2-like protein 1 | Q07817 |
| ACE | Angiotensin-converting enzyme | P12821 |
| IGF1R | Insulin-like growth factor 1 receptor | P08069 |
| PGR | Progesterone receptor | P06401 |
| SERPINE1 | Plasminogen activator inhibitor 1 | P05121 |
| F2 | Coagulation factor II | P00734 |
| MPO | Myeloperoxidase | P05164 |
| NR3C1 | Nuclear receptor subfamily 3 group C member 1 | P04150 |
| MCL1 | Induced myeloid leukemia cell differentiation protein Mcl-1 | Q07820 |
| PLG | Plasminogen | P00747 |
| AGTR1 | Type-1 angiotensin II receptor | P30556 |
| CYP19A1 | Cytochrome P450 19A1 | P11511 |
| IGFBP3 | Insulin-like growth factor-binding protein 3 | P17936 |
| AHR | Aryl hydrocarbon receptor | P35869 |
| PGF | Placenta growth factor | P49763 |
| ABCB1 | ATP-dependent translocase ABCB1 | P08183 |
| PTPN1 | Tyrosine-protein phosphatase nonreceptor type 1 | P18031 |
| PPARA | Peroxisome proliferator-activated receptor alpha | Q07869 |
| ACHE | Acetylcholinesterase | P22303 |
| PLA2G4A | Cytosolic phospholipase A2 | P47712 |
| MMP3 | Stromelysin-1 | P08254 |
| PLA2G1B | Phospholipase A2 | P04054 |
| ALOX5 | Polyunsaturated fatty acid 5-lipoxygenase | P09917 |
| TERT | Telomerase reverse transcriptase | O14746 |
| F3 | Coagulation factor III | P13726 |
| CNR1 | Cannabinoid receptor 1 | P21554 |
| NOX4 | NADPH oxidase 4 | Q9NPH5 |
| MMP8 | Matrix metalloproteinase-8 | P22894 |
| NOS2 | Nitric oxide synthase | P35228 |
| AKR1B1 | Aldo-keto reductase family 1 member B1 | P15121 |
| PTGER3 | Prostaglandin E2 receptor EP3 subtype | P43115 |
| ENSG00000196689 | none | none |
| FABP4 | Fatty acid-binding protein 4 | P15090 |
| NLRP3 | NACHT, LRR, and PYD domains-containing protein 3 | Q96P20 |
| PDE4A | cAMP-specific 3′,5′-cyclic phosphodiesterase 4A | P27815 |
| RARB | Retinoic acid receptor beta | P10826 |
3.4. GO Enrichment Analysis of Key Targets
GO enrichment analysis process consists of biological process (BP), cellular component (CC), and molecular function (MF). A total of GO terms, including 227 biological processes, 27 cellular components, and 55 molecular functions with P < 0.05, were obtained through inputting the 53 key genes into the DAVID website. Among them, the top ten most significantly enriched GO terms, respectively, were input into Bioinformatics (http://www.bioinformatics.com.cn/) to obtain the GO analysis diagram. As is shown in Figure 4, these targets were mainly enriched in signal transduction, cell proliferation, and apoptosis in BP, including positive regulation of transcription from RNA polymerase II promoter (GO:0045944), signal transduction (GO:0007165), negative regulation of apoptotic process (GO:0043066), positive regulation of cell proliferation (GO:0008284), and positive regulation of apoptotic process (GO:0043065). In CC analysis, the cellular components of these key targets mainly were nucleus (GO:0005634), plasma membrane (GO:0005886), extracellular space (GO:0005615), cytosol (GO:0005829), and extracellular region (GO:0005576). The MF analysis indicates that these key targets are closely related to protein binding (GO:0005515). These analyses suggested that PT may modulate angiogenesis by regulating the proliferation and apoptosis of retinal vascular cells, as well as the binding of core targets to its receptor, to treat DR. The details of GO enrichment are provided in Supplementary Table S2.
Figure 4.
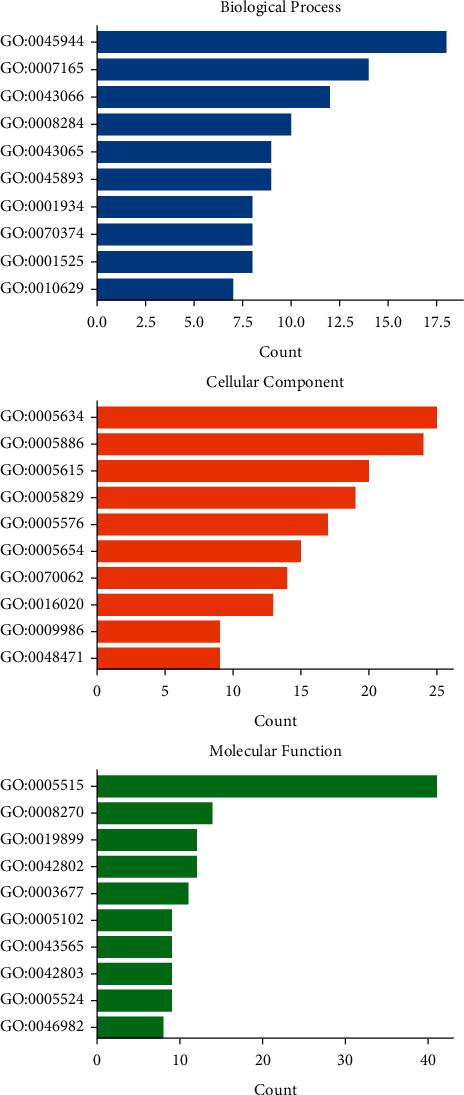
The diagram of the GO enrichment analysis. The Y-axis represents GO terms. The X-axis indicates the number of genes enriched.
3.5. KEGG Enrichment Analysis
There were 80 KEGG signaling pathways obtained by the 53 key genes which were inputted into the DAVID database. Analysis of the KEGG pathway was carried out via Bioinformatics (http://www.bioinformatics.com.cn/), and a bubble diagram was obtained by listing the top 20 signaling pathways according to the enrichment score. As shown in Figure 5, the results of KEGG pathway enrichment included VEGF signaling pathway, PI3K-Akt signaling pathway, Rap1 signaling pathway, and HIF-1 signaling pathway, which were mainly related to oxidative stress, inflammation response, and retinal angiogenesis and proliferation. It is indicated that PT may act through these signaling pathways in treating PT. Furthermore, the top 20 signaling pathways and their specific related genes information are shown in Table 3.
Figure 5.
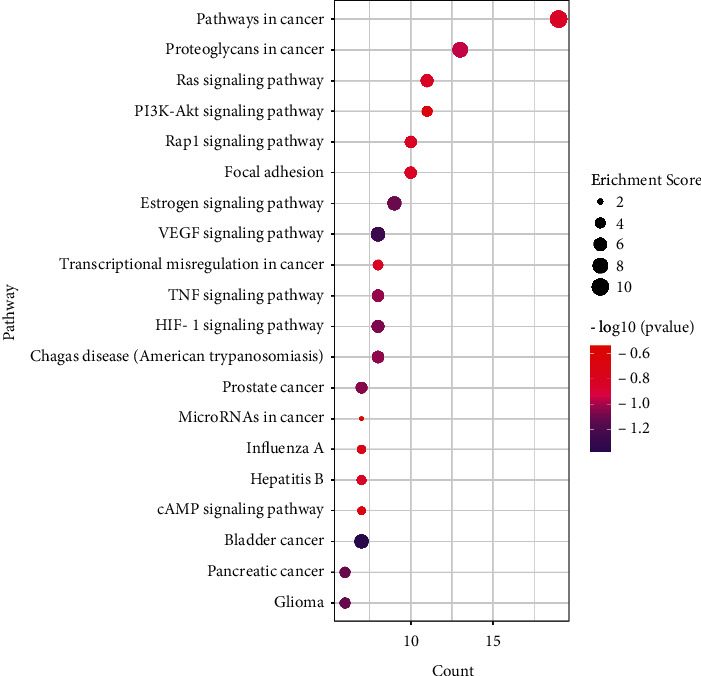
KEGG enrichment analysis. Count and pathway are represented by the x-axis and y-axis, respectively; the size and color of the dots indicate the enrichment score and the level of P value, respectively.
Table 3.
The top 20 signaling pathways with related genes.
| Term | Pathway | Genes | P value |
|---|---|---|---|
| hsa05200 | Pathways in cancer | JUN, NOS2, MMP2, PTGER3, PTGS2, MMP9, EGFR, PGF, IGF1R, VEGFA, AR, PIK3CA, MDM2, AGTR1, AKT1, RARB, PPARG, BCL2L1, MAPK3 | 4.30E − 11 |
| hsa05205 | Proteoglycans in cancer | SRC, MMP2, ESR1, TNF, MMP9, EGFR, IGF1R, VEGFA, PIK3CA, MDM2, KDR, AKT1, MAPK3 | 7.30E − 09 |
| hsa04014 | Ras signaling pathway | PIK3CA, PLA2G1B, KDR, PLA2G4A, AKT1, EGFR, PGF, BCL2L1, IGF1R, MAPK3, VEGFA | 2.62E − 06 |
| hsa04151 | PI3K-Akt signaling pathway | PIK3CA, MDM2, KDR, AKT1, EGFR, PGF, BCL2L1, IGF1R, MCL1, MAPK3, VEGFA | 1.05E − 04 |
| hsa04510 | Focal adhesion | JUN, PIK3CA, SRC, KDR, AKT1, EGFR, PGF, IGF1R, MAPK3, VEGFA | 9.89E − 06 |
| hsa04015 | Rap1 signaling pathway | PIK3CA, CNR1, SRC, KDR, AKT1, EGFR, PGF, IGF1R, MAPK3, VEGFA | 1.16E − 05 |
| hsa04915 | Estrogen signaling pathway | JUN, PIK3CA, SRC, MMP2, AKT1, ESR1, MMP9, EGFR, MAPK3 | 3.25E − 07 |
| hsa04370 | VEGF signaling pathway | PIK3CA, SRC, KDR, PLA2G4A, AKT1, PTGS2, MAPK3, VEGFA | 1.68E − 07 |
| hsa04066 | HIF-1 signaling pathway | PIK3CA, NOS2, SERPINE1, AKT1, EGFR, IGF1R, MAPK3, VEGFA | 3.82E − 06 |
| hsa05142 | Chagas disease (American trypanosomiasis) | JUN, ACE, PIK3CA, NOS2, SERPINE1, AKT1, TNF, MAPK3 | 6.52E − 06 |
| hsa04668 | TNF signaling pathway | JUN, PIK3CA, MMP3, AKT1, PTGS2, TNF, MMP9, MAPK3 | 7.88E − 06 |
| hsa05202 | Transcriptional misregulation in cancer | IGFBP3, MMP3, MDM2, PPARG, MPO, MMP9, BCL2L1, IGF1R | 1.39E − 04 |
| hsa05219 | Bladder cancer | SRC, MMP2, MDM2, MMP9, EGFR, MAPK3, VEGFA | 3.13E − 07 |
| hsa05215 | Prostate cancer | AR, PIK3CA, MDM2, AKT1, EGFR, IGF1R, MAPK3 | 2.94E − 05 |
| hsa05161 | Hepatitis B | JUN, PIK3CA, SRC, AKT1, TNF, MMP9, MAPK3 | 4.68E − 04 |
| hsa05164 | Influenza A | JUN, PIK3CA, NLRP3, AKT1, PLG, TNF, MAPK3 | 0.001220965 |
| hsa04024 | cAMP signaling pathway | JUN, PIK3CA, PTGER3, PDE4A, AKT1, PPARA, MAPK3 | 0.002361912 |
| hsa05206 | MicroRNAs in cancer | ABCB1, MDM2, PTGS2, MMP9, EGFR, MCL1, VEGFA | 0.013841455 |
| hsa05212 | Pancreatic cancer | PIK3CA, AKT1, EGFR, BCL2L1, MAPK3, VEGFA | 8.07E − 05 |
| hsa05214 | Glioma | PIK3CA, MDM2, AKT1, EGFR, IGF1R, MAPK3 | 8.07E − 05 |
3.6. The Ingredient-Gene-Pathway Network
The ingredient-gene-pathway network consists of 29 nodes, including 5 active ingredients, 7 intersecting genes, and 20 pathways. As shown in Figure 6, AKTI, EGFR, and VEGFA are the core genes. According to the topological analysis, kaempferol and isorhamnetin are pivotal ingredients and could be the key effective components of PT for the treatment of DR. In this network, several pathways are involved in anti-DR, mainly including VEGF signaling pathway and Rap1 signaling pathway. It is indicated that PT may act through AKTI, EGFR, and VEGFA via VEGF signaling pathway and Rap1 signaling pathway in treating PT.
Figure 6.
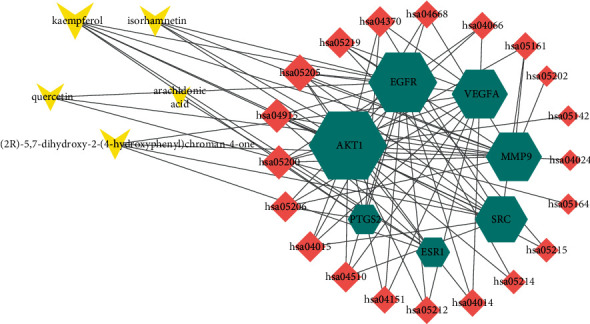
The ingredient-gene-pathway network. The triangle node in yellow represents active component, the hexagon node in green represents the intersecting targets, and the diamond node in red represents the signaling pathways. The interaction between the components and the targets is represented by the edge.
3.7. Molecular Docking Results
Five compounds had a docking score higher than 4.0 with EGFR and AKT1, suggesting good binding activity. All docking scores are enlisted in Table 4. As shown in Figure 7(A), (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one stably bound to AKT1 through ARG23, ASN53, LYS14, ARG86, TYR18, and ILE19; arachidonic acid stably bound to the active site of AKT1 through hydrogen bonding interactions with ASN53, ARG86, and LYS14 on the AKT1 target protein (Figure 7(B)); isorhamnetin bound to the active site of AKT1 stably through hydrogen bond interactions with LYS14, ARG25, ASN53, GLU17, TYR18, and ILE19 on the AKT1 target protein (Figures 7(C)); quercetin and kaempferol stably bound to AKT1 through ASN54, ASN53, ARG86, GLU17, and LYS14 (Figures 7((D) and (E))); (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one stably bound to the active site of EGFR through THR360 and ARG40 on EGFR target protein (Figure 7(a)); arachidonic acid stably bound to the active site of EGFR through hydrogen bonding interactions with THR358 and THR330 on the EGFR target protein (Figure 7(b)); isorhamnetin bound to EGFR stably through hydrogen bond interactions with ASP323, SER324, GLU320, THR358, ASP355, THR360, and ASN328 (Figure 7(c)); quercetin stably bound to EGFR through ASN331, SER324, THR358, HIS359, and PHE351 on the EGFR target protein (Figure 7(d)); kaempferol stably bound to the active site of AKT1 through PHE357, HIS359, ASN331, and ASN328 (Figure 7(e)).
Table 4.
Docking scores of active compounds of PT with core targets.
| Targets | PDB ID | Compound | Molecular ID | CSCORE |
|---|---|---|---|---|
| AKT1 | 1unq | (2R)-5,7-Dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | MOL001040 | 4 |
| AKT1 | 1unq | Arachidonic acid | MOL001439 | 5 |
| AKT1 | 1unq | Isorhamnetin | MOL000354 | 5 |
| AKT1 | 1unq | Quercetin | MOL000098 | 4 |
| AKT1 | 1unq | Kaempferol | MOL000422 | 4 |
| EGFR | 5w7b | (2R)-5,7-Dihydroxy-2-(4-hydroxyphenyl)chroman-4-one | MOL001040 | 4 |
| EGFR | 5w7b | Arachidonic acid | MOL001439 | 4 |
| EGFR | 5w7b | Isorhamnetin | MOL000354 | 4 |
| EGFR | 5w7b | Quercetin | MOL000098 | 3 |
| EGFR | 5w7b | Kaempferol | MOL000422 | 3 |
Figure 7.
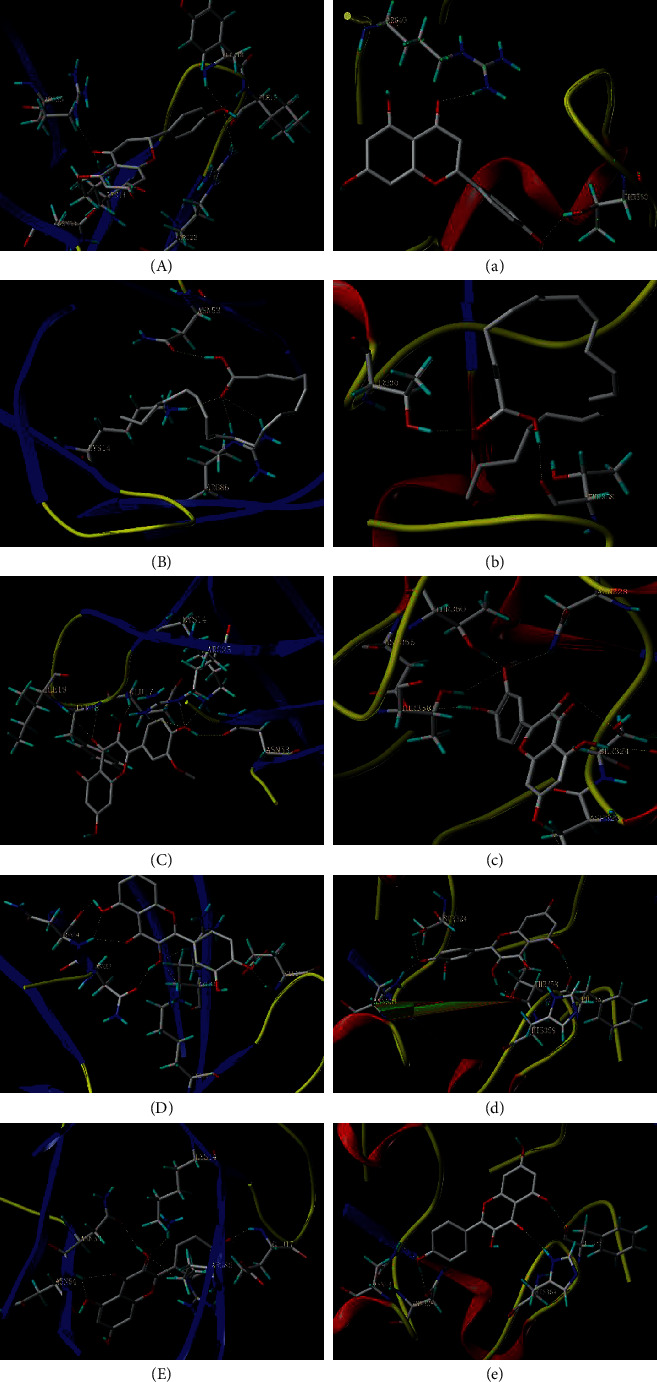
The model of molecular docking. (A) AKTI-(2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one; (B) AKTI-arachidonic acid; (C) AKT1-isorhamnetin; (D) AKT1-quercetin; (E) AKT1-kaempferol; (a) EGFR-(2R)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one; (b) EGFR-arachidonic acid; (c) EGFR-isorhamnetin; (d) EGFR-quercetin; (e) EGFR-kaempferol.
4. Discussion
Diabetic retinopathy progresses through a variety of pathophysiological pathways, including the stimulation of VEGF in the eyes, oxidative stress, inflammation response, and the activation of the hexosamine pathway. The current treatments are monotherapy with limited efficacy, such as intravitreal injection of anti-VEGFs, laser and surgery, and other methods. Pollen Typhae (PT), a multicomponent and multitarget traditional Chinese medicine (TCM), has been clinically used in the treatment of DR and has a significant effect. The previous pharmacological research demonstrated that the components of PT have good antioxidant and anti-inflammatory effects [24]. Therefore, it is of great significance to further explore the active ingredients and molecular mechanism anti-DR. In our study, combining compound-target and ingredient-gene-pathway networks, we predicted that active constituents (arachidonic acid, isorhamnetin, quercetin, and kaempferol) are closely related to PT for DR. Based on the PPI analysis and ingredient-gene-pathway network, core target proteins (PTGS2, AKT1, VEGFA, MAPK3, TNF, and EGFR) were obtained. Considering the results of the KEGG pathway enrichment analysis and ingredient-gene-pathway network, the key signaling pathways (VEGF signaling pathway, PI3K-Akt signaling pathway, Rap1 signaling pathway, and HIF-1 signaling pathway) were selected. Arachidonic acid is one of the essential polyunsaturated fatty acids in human body and plays a key role in regulating inflammatory responses. Previous report indicated that 5,6-dihydroxytrienoic lactone, a stable metabolite of arachidonic acid, could mediate microvascular dilation to contribute to the anti-DR activity [25]. Furthermore, lots of experiments have shown that arachidonic acid and its metabolites can suppress inflammation [26] and reduce free radical generation to participate in the prevention of diabetes and its microvascular complications. Pollen Typhae is particularly rich in flavonoids, especially flavanol triglycosides including derivatives of isorhamnetin, quercetin, and kaempferol. Modern pharmacological studies have confirmed that flavonoids have good effect on the antioxidant and anti‐inflammation activities, regulation of blood glucose and lipids, and regulation of cell cycle and vascular integrity. Several studies have demonstrated that isorhamnetin has good effects on attenuation of diabetes-induced oxidative stress and protection of nerves [27, 28]. Studies have certified that quercetin can protect the nerves in diabetic rat retina and prevent diabetic retinopathy in rats by inducing heme oxygenase-1 expression [29, 30]. Researchers have found that quercetin has significant inhibitory activity against the expressions of MCP-1, MMP9, and VEGF and may have a promising role on the treatment of DR [31], and it also can relieve inflammatory responses by suppression of nuclear factor kappa B in mouse. In this study, ingredient-gene-pathway network shows that kaempferol is one of the core ingredients. Many experiments and findings have confirmed that kaempferol could protect retinal ganglion cells through regulating vasohibin-1 from high-glucose-induced injury [32] and prevent retinal pigment epithelial cell damage from oxidative stress [32]. Another study has indicated that kaempferol can inhibit VEGF and PGF expression and in vitro angiogenesis under high-glucose environment [33]. Therefore, these might demonstrate that the active ingredients of PT play a crucial role in developing microvascular events and it might be promising in the prevention and treatment of DR.
The PPI network analysis illustrates that gene targets (PTGS2, AKT1, VEGFA, MAPK3, and TNF) may be the main targets of PT to DR. The ingredient-gene-pathway network shows that AKT1 and EGFR are the core target genes. AKT1 is a suppressor gene and plays a key role in DR, and pharmacological studies have demonstrated that high glucose promotes retinal endothelial cell migration through activation of AKT1 [34, 35]. The previous studies have indicated that EGFR inhibitors played the important role in attenuating inflammatory infiltration and angiogenesis in mice with diabetic retinopathy [36]. Another in vivo study indicated that it can prevent pathological retinal neovascularization via inhibiting EP (4)R-EGFR-Gab1-AKT signaling pathway [37]. Studies and experiments have certified that regulating the EGFR pathway also could inhibit the proliferation, migration, and invasion of endothelial cells and retinal pericytes of diabetic retinopathy rats [38]. Lots of studies have suggested that PTGS2 could reduce the inflammatory response by downregulating the PTGS2 level [39, 40]. The study suggests that inhibition of p-c-Jun expression could prevent retinal neuronal cell death in NMDA-induced excitotoxicity [41]. Furthermore, inhibiting the activation of the stress response kinase JNK can improve insulin action in retina [42]. Jun was determined as a target of c-Jun N-terminal kinase and plays a critical role in regulating angiogenesis in vitro [43]. Jun was proved to be involved in improving hypoxia in retinal vascular endothelial cells by regulating cooperation of c-Jun/AP-1 and HIF-1alpha [44], which is potentially important for treatment of DR. Less relevant studies have shown that ESR1 has a direct effect on DR; however, some articles previously reported that ESR1 plays a critical role in glucose transporter GLUT4 regulation and maintains glycemic homeostasis [45, 46]. A previous research indicated that MAPK3 is a key target for treatment of rats with diabetic retinopathy [47].
Functional and enrichment analysis shows that these pathways (VEGF signaling pathway, PI3K-Akt signaling pathway, Rap1 signaling pathway, and HIF-1 signaling pathway) are significantly involved in antioxidative stress and anti-inflammation and suppress the angiogenesis of retina. Study suggested that modulating the PI3K/AKT signaling pathway could ameliorate diabetes-induced proliferative retinopathy [48, 49]. In vitro evidence showed that the activation of PI3K/Akt signaling pathway was closely related to oxidative stress inhibition and proinflammatory response of microglial cells associated with diabetic retinopathy [50]. Focal adhesion signaling pathway was important for maintaining the integrity of retinal vascular structure and function [51, 52]. According to the study results, those genes enriched in focal adhesion signaling pathway were considerably upregulated [53–55]. Moreover, proteomic bioinformatic analysis in DR indicated that DR development was closely related to focal adhesion and PI3K-Akt signaling pathway [56]. For the VEGF signaling pathway, one study's results suggested that the inhibition of the VEGF signaling pathway could protect diabetic retina, and this protective effect might be related to anti-inflammatory and antioxidative activities [57]. In vivo studies indicated that downregulation of the VEGF and HIF1 signaling pathway could inhibit the proliferation and angiogenesis of high-glucose-induced human retinal endothelial cells [58–60]. Additionally, lots of studies and experiments results have confirmed that downregulation of the VEGF and HIF1 signaling pathway could also prevent hypoxia-induced retinal angiogenesis [61–63]. One experiment demonstrated that HIF-1 signaling pathway had a protective effect on DR [64], and it is suggested that targeting the HIF-1 signaling pathway could be a therapeutic approach to prevent development of DR.
5. Conclusion
In summary, this study is aimed at illuminating the therapeutic effect on DR through network pharmacology and molecular docking analysis. We predicted that these active ingredients in PT, including arachidonic acid, isorhamnetin, quercetin, and kaempferol, and core targets, such as PTGS2, AKT1, VEGFA, MAPK3, TNF, and EGFR, mainly intervene in core targets through VEGF signaling pathway, PI3K-Akt signaling pathway, Rap1 signaling pathway, HIF-1 signaling pathway, and other signaling pathways, thereby exerting their roles in the treatment DR.
Acknowledgments
This work was funded by the 2015 Traditional Chinese Medicine Scientific Research (no. 201507001-11).
Data Availability
The datasets in this study can be obtained from the corresponding author upon reasonable request. The authors are responsible for providing the final supplementary material files that will be published along with the article.
Disclosure
Rongrong zhou and De Jin are the co-first authors.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Supplementary Materials
Table S1: the abbreviations. Table S2: the GO enrichment with related genes.
References
- 1.Klein R., Klein B. F., Moss S. E., Davis M. D., DeMets D. L., DeMets D. L. The Wisconsin epidemiologic study of diabetic retinopathy. IX. Four-year incidence and progression of diabetic retinopathy when age at diagnosis is less than 30 years. Archives of Ophthalmology . 1989;107(2):237–243. doi: 10.1001/archopht.1989.01070010243030. [DOI] [PubMed] [Google Scholar]
- 2.Vujosevic S., Aldington S. J., Silva P., et al. Screening for diabetic retinopathy: new perspectives and challenges. The Lancet Diabetes & Endocrinology . 2020;8(4):337–347. doi: 10.1016/s2213-8587(19)30411-5. [DOI] [PubMed] [Google Scholar]
- 3.Wong T. Y., Klein R., Islam F. M. A., et al. Diabetic retinopathy in a multi-ethnic cohort in the United States. American Journal of Ophthalmology . 2006;141(3):446–455. doi: 10.1016/j.ajo.2005.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein B. E. Overview of epidemiologic studies of diabetic retinopathy. Ophthalmic Epidemiology . 2007;14(4):179–183. doi: 10.1080/09286580701396720. [DOI] [PubMed] [Google Scholar]
- 5.Wang W., Lo A. Diabetic retinopathy: pathophysiology and treatments. International Journal of Molecular Sciences . 2018;19(6):p. 1816. doi: 10.3390/ijms19061816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J. M. Clinical observation on 60 cases of hyperlipidemia treated with pollen Typhae. Jiujiang Medical Journal . 2002;17(4):p. 218. [Google Scholar]
- 7.Li J. Clinical observation on shengpuhuang decoction in the treatment of non-proliferative diabetic retinopathy. CJGMCM . 2019;34(2):255–257. [Google Scholar]
- 8.Guo J. R. Sheng PuhuangTang treating retinal hemorrhage randomized controlled study. Journal of Practical Traditional Chinese Internal Medicine . 2013;27(7):46–47. [Google Scholar]
- 9.Chen M. T., Huang R. L., Ou L. J., et al. Pollen typhae total flavone inhibits endoplasmic reticulum stress-induced apoptosis in human aortic-vascular smooth muscle cells through down-regulating PERK-eIF2α-ATF4-CHOP pathway. Chinese Journal of Integrative Medicine . 2019;25(8):604–612. doi: 10.1007/s11655-019-3052-4. [DOI] [PubMed] [Google Scholar]
- 10.Feng X. T., Chen Q., Xie Z., et al. Pollen typhae total flavone improves insulin resistance in high-fat diet and low-dose streptozotocin-induced type 2 diabetic rats. Bioscience, Biotechnology, and Biochemistry . 2014;78(10):1738–1742. doi: 10.1080/09168451.2014.930318. [DOI] [PubMed] [Google Scholar]
- 11.He Y. M., Wang W., Chen W.-H., et al. Effects of pollen typhae total flavone on glucose and lipid metabolism in 3T3-L1 adipocytes. Journal of Chinese Integrative Medicine . 2006;6:593–595. doi: 10.3736/jcim20060610. [DOI] [PubMed] [Google Scholar]
- 12.You J.-s., Li C.-y., Chen W., et al. A network pharmacology-based study on Alzheimer disease prevention and treatment of Qiong Yu Gao. BioData Mining . 2020;13(1):p. 2. doi: 10.1186/s13040-020-00212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo T.-t., Lu Y., Yan S.-k., Xiao X., Rong X.-l., Guo J. Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chinese Journal of Integrative Medicine . 2020;26(1):72–80. doi: 10.1007/s11655-019-3064-0. [DOI] [PubMed] [Google Scholar]
- 14.Lyu Y., Chen X., Xia Q., Zhang S., Yao C. Network pharmacology-based study on the mechanism of pinellia ternata in asthma treatment. Evidence-based Complementary and Alternative Medicine . 2020;2020:12. doi: 10.1155/2020/9732626.9732626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y., Yang J., Jing W., et al. Systemic elucidation on the potential bioactive compounds and hypoglycemic mechanism of Polygonum multiflorum based on network pharmacology. Chinese Medicine . 2020;15(1):p. 121. doi: 10.1186/s13020-020-00401-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang P., He H., Xu S., Liu P., Bai X. Potential molecular target prediction and docking verification of hua-feng-dan in stroke based on network pharmacology. Evidence-based Complementary and Alternative Medicine . 2020;2020:12. doi: 10.1155/2020/8872593.8872593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Zhang S., Li F., et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Research . 2020;48(D1):D1031–D41. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo X., Ji J., Feng Z., Hou X., Luo Y., Mei Z. A network pharmacology approach to explore the potential targets underlying the effect of sinomenine on rheumatoid arthritis. International Immunopharmacology . 2020;80 doi: 10.1016/j.intimp.2020.106201.106201 [DOI] [PubMed] [Google Scholar]
- 19.Fan Q., Guo L., Guan J., et al. Network pharmacology-based study on the mechanism of gegen qinlian decoction against colorectal cancer. Evidence-based Complementary and Alternative Medicine . 2020;2020:14. doi: 10.1155/2020/8897879.8897879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W., Fan Y., Tian C., et al. Deciphering the molecular targets and mechanisms of HGWD in the treatment of rheumatoid arthritis via network pharmacology and molecular docking. Evidence-based Complementary and Alternative Medicine . 2020;2020:13. doi: 10.1155/2020/7151634.7151634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song X., Zhang Y., Dai E., Wang L., Du H. Prediction of triptolide targets in rheumatoid arthritis using network pharmacology and molecular docking. International Immunopharmacology . 2020;80 doi: 10.1016/j.intimp.2019.106179.106179 [DOI] [PubMed] [Google Scholar]
- 22.Wang C., Ren Q., Chen X.-T., et al. System pharmacology-based strategy to decode the synergistic mechanism of zhi-zhu wan for functional dyspepsia. Frontiers in Pharmacology . 2018;9:p. 841. doi: 10.3389/fphar.2018.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mou X., Zhou D. Y., Zhou D., Liu K., Chen L. J., Liu W. H. A bioinformatics and network pharmacology approach to the mechanisms of action of Shenxiao decoction for the treatment of diabetic nephropathy. Phytomedicine . 2020;69 doi: 10.1016/j.phymed.2020.153192.153192 [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Liao M., Xiao P. Chemistry and pharmacological activity of a traditional Chinese drug Pollen Typhae. Lishizhen Medicine and Materia Medica Research . 1998;9:49–50. [Google Scholar]
- 25.Levi-Rosenzvig R., Beyer A. M., Hockenberry J., et al. 5,6-δ-DHTL, a stable metabolite of arachidonic acid, is a potential EDHF that mediates microvascular dilation. Free Radical Biology and Medicine . 2017;103:87–94. doi: 10.1016/j.freeradbiomed.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 26.Gundala N. K. V., Das U. N. Arachidonic acid-rich ARASCO oil has anti-inflammatory and antidiabetic actions against streptozotocin + high fat diet induced diabetes mellitus in Wistar rats. Nutrition . 2019;66:203–218. doi: 10.1016/j.nut.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Jamali-Raeufy N., Baluchnejadmojarad T., Roghani M., Keimasi S., Goudarzi M. Isorhamnetin exerts neuroprotective effects in STZ-induced diabetic rats via attenuation of oxidative stress, inflammation and apoptosis. Journal of Chemical Neuroanatomy . 2019;102 doi: 10.1016/j.jchemneu.2019.101709.101709 [DOI] [PubMed] [Google Scholar]
- 28.Chen F., Hu M., Shen Y., et al. Isorhamnetin promotes functional recovery in rats with spinal cord injury by abating oxidative stress and modulating M2 macrophages/microglia polarization. European Journal of Pharmacology . 2021;895 doi: 10.1016/j.ejphar.2021.173878.173878 [DOI] [PubMed] [Google Scholar]
- 29.Chen X.-L., Chai G.-R., Liu S., Yang H.-W. Quercetin protects against diabetic retinopathy in rats by inducing heme oxygenase-1 expression. Neural Regeneration Research . 2021;16(7):1344–1350. doi: 10.4103/1673-5374.301027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ola M. S., Ahmed M. M., Shams S., Al-Rejaie S. S. Neuroprotective effects of quercetin in diabetic rat retina. Saudi Journal of Biological Sciences . 2017;24(6):1186–1194. doi: 10.1016/j.sjbs.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B., He T., Xing Y., Cao T. Effects of quercetin on the expression of MCP-1, MMP-9 and VEGF in rats with diabetic retinopathy. Experimental and Therapeutic Medicine . 2017;14(6):6022–6026. doi: 10.3892/etm.2017.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L., Sun J., Shi S., Qin X., Zhang K., Xu J. Kaempferol protects retinal ganglion ceils from high-glucose-induced injury by regulating vasohibin-1. Neuroscience Letters . 2020;716 doi: 10.1016/j.neulet.2019.134633.134633 [DOI] [PubMed] [Google Scholar]
- 33.Xu X. H., Zhao C., Peng Q., Xie P., Liu Q. H. Kaempferol inhibited VEGF and PGF expression and in vitro angiogenesis of HRECs under diabetic-like environment. Brazilian Journal of Medical and Biological Research . 2017;50(3) doi: 10.1590/1414-431x20165396.e5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Q., Sheibani N. High glucose promotes retinal endothelial cell migration through activation of Src, PI3K/Akt1/eNOS, and ERKs. American Journal of Physiology-Cell Physiology . 2008;295(6):C1647–C1657. doi: 10.1152/ajpcell.00322.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W.-Z., Yang J., Xue L.-P., Xiao L.-B., Li Y. MiR-126 overexpression inhibits high glucose-induced migration and tube formation of rhesus macaque choroid-retinal endothelial cells by obstructing VEGFA and PIK3R2. Journal of Diabetes and Its Complications . 2017;31(4):653–663. doi: 10.1016/j.jdiacomp.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Ju X., Yang X., Yan T., et al. EGFRinhibitor,AG1478, inhibits inflammatory infiltration and angiogenesis in mice with diabetic retinopathy. Clinical and Experimental Pharmacology and Physiology . 2019;46(1):75–85. doi: 10.1111/1440-1681.13029. [DOI] [PubMed] [Google Scholar]
- 37.Xie T., Zhang Z., Cui Y., et al. Prostaglandin E2 promotes pathological retinal neovascularisation via EP4R-EGFR-Gab1-AKT signaling pathway. Experimental Eye Research . 2021;205 doi: 10.1016/j.exer.2021.108507.108507 [DOI] [PubMed] [Google Scholar]
- 38.Zhou L., Zhang S., Zhang L., Li F., Sun H., Feng J. MiR-199a-3p inhibits the proliferation, migration, and invasion of endothelial cells and retinal pericytes of diabetic retinopathy rats through regulating FGF7 via EGFR/PI3K/AKT pathway. Journal of Receptors and Signal Transduction . 2021;41(1):19–31. doi: 10.1080/10799893.2020.1783556. [DOI] [PubMed] [Google Scholar]
- 39.Astakhova A., Chistyakov D., Thomas D., et al. Inhibitors of oxidative phosphorylation modulate astrocyte inflammatory responses through AMPK-dependent Ptgs2 mRNA stabilization. Cells . 2019;8(10):p. 1185. doi: 10.3390/cells8101185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hellmann J., Tang Y., Zhang M. J., et al. Atf3 negatively regulates Ptgs2/Cox2 expression during acute inflammation. Prostaglandins & Other Lipid Mediators . 2015;116-117:49–56. doi: 10.1016/j.prostaglandins.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujita N., Sase K., Tsukahara C., Arizono I., Takagi H., Kitaoka Y. Pemafibrate prevents retinal neuronal cell death in NMDA-induced excitotoxicity via inhibition of p-c-Jun expression. Molecular Biology Reports . 2021;48(1):195–202. doi: 10.1007/s11033-020-06032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller W. P., Ravi S., Martin T. D., Kimball S. R., Dennis M. D. Activation of the stress response kinase JNK (c-Jun N-terminal kinase) attenuates insulin action in retina through a p70S6K1-dependent mechanism. Journal of Biological Chemistry . 2017;292(5):1591–1602. doi: 10.1074/jbc.M116.760868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaikai S., Yuchen S., Lili J., Zhengtao W. Critical role of c-Jun N-terminal kinase in regulating bFGF-induced angiogenesis in vitro. Journal of Biochemistry . 2011;150(2):189–197. doi: 10.1093/jb/mvr060. [DOI] [PubMed] [Google Scholar]
- 44.You J.-J., Yang C.-M., Chen M.-S., Yang C.-H. Regulation of Cyr61/CCN1 expression by hypoxia through cooperation of c-Jun/AP-1 and HIF-1α in retinal vascular endothelial cells. Experimental Eye Research . 2010;91(6):825–836. doi: 10.1016/j.exer.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Barreto-Andrade J. N., de Fátima L. A., Campello R. S., Guedes J. A. C., de Freitas H. S., Machado M. M. F. Estrogen receptor 1 (ESR1) enhances slc2a4/GLUT4 expression by a SP1 cooperative mechanism. International Journal of Medical Sciences . 2018;15(12):1320–1328. doi: 10.7150/ijms.26774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregorio K. C. R., Laurindo C. P., Machado U. F. Estrogen and glycemic homeostasis: the fundamental role of nuclear estrogen receptors ESR1/ESR2 in glucose transporter GLUT4 regulation. Cells . 2021;10(1):p. 99. doi: 10.3390/cells10010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Safaei A., Rezaei Tavirani M., Zamanian Azodi M., et al. Diabetic retinopathy and laser therapy in rats: a protein-protein interaction network analysis. Journal of Lasers in Medical Sciences . 2017;8(1):S20–s21. doi: 10.15171/jlms.2017.s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi J., Lv H., Tang C., Li Y., Huang J., Zhang H. Mangiferin inhibits cell migration and angiogenesis via PI3K/AKT/mTOR signaling in high glucose‑ and hypoxia‑induced RRCECs. Molecular Medicine Reports . 2021;23(6) doi: 10.3892/mmr.2021.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui J., Gong R., Hu S., Cai L., Chen L. Gambogic acid ameliorates diabetes-induced proliferative retinopathy through inhibition of the HIF-1α/VEGF expression via targeting PI3K/AKT pathway. Life Sciences . 2018;192:293–303. doi: 10.1016/j.lfs.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Yang X., Huo F., Liu B., et al. Crocin inhibits oxidative stress and pro-inflammatory response of microglial cells associated with diabetic retinopathy through the activation of PI3K/akt signaling pathway. Journal of Molecular Neuroscience . 2017;61(4):581–589. doi: 10.1007/s12031-017-0899-8. [DOI] [PubMed] [Google Scholar]
- 51.Li Z., Zhang J., Xue Y., et al. Pitavastatin stimulates retinal angiogenesis via HMG-CoA reductase-independent activation of RhoA-mediated pathways and focal adhesion. Graefe’s Archive for Clinical and Experimental Ophthalmology . 2021;259(9):2707–2716. doi: 10.1007/s00417-021-05328-4. [DOI] [PubMed] [Google Scholar]
- 52.Sun Q., Shen Y., Su L., Xu X. Inhibition of pathological retinal neovascularization by a small peptide derived from human tissue-type plasminogen kringle 2. Frontiers in Pharmacology . 2019;10:p. 1639. doi: 10.3389/fphar.2019.01639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H., Li T., Wang H., et al. Diabetes promotes retinal vascular endothelial cell injury by inducing CCN1 expression. Frontiers in Cardiovascular Medicine . 2021;8 doi: 10.3389/fcvm.2021.689318.689318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falero-Perez J., Sorenson C. M., Sheibani N. Retinal astrocytes transcriptome reveals Cyp1b1 regulates the expression of genes involved in cell adhesion and migration. PLoS One . 2020;15(4) doi: 10.1371/journal.pone.0231752.e0231752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schimmel L., Fukuhara D., Richards M., et al. c-Src controls stability of sprouting blood vessels in the developing retina independently of cell-cell adhesion through focal adhesion assembly. Development . 2020;147(7) doi: 10.1242/dev.185405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao H., Xin W., Sun L. M., Li S. S., Zhang T., Ding X. Y. Comprehensive proteomic profiling of aqueous humor proteins in proliferative diabetic retinopathy. Translational Vision Science & Technology . 2021;10(6):p. 3. doi: 10.1167/tvst.10.6.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen M., Lv H., Gan J., Ren J., Liu J. Tang wang ming mu granule attenuates diabetic retinopathy in type 2 diabetes rats. Frontiers in Physiology . 2017;8:p. 1065. doi: 10.3389/fphys.2017.01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang J., Chang X. Celastrol inhibits the proliferation and angiogenesis of high glucose-induced human retinal endothelial cells. BioMedical Engineering Online . 2021;20(1):p. 65. doi: 10.1186/s12938-021-00904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu J.-M., Zhang Z.-Z., Ma X., Fang S.-F., Qin X.-H. Repression of microRNA-21 inhibits retinal vascular endothelial cell growth and angiogenesis via PTEN dependent-PI3K/Akt/VEGF signaling pathway in diabetic retinopathy. Experimental Eye Research . 2020;190 doi: 10.1016/j.exer.2019.107886.107886 [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z.-Z., Qin X.-H., Zhang J. MicroRNA-183 inhibition exerts suppressive effects on diabetic retinopathy by inactivating BTG1-mediated PI3K/Akt/VEGF signaling pathway. American Journal of Physiology-Endocrinology and Metabolism . 2019;316(6):E1050–E1060. doi: 10.1152/ajpendo.00444.2018. [DOI] [PubMed] [Google Scholar]
- 61.Wu J., Ke X., Fu W., et al. Inhibition of hypoxia-induced retinal angiogenesis by specnuezhenide, an effective constituent of ligustrum lucidum ait., through suppression of the HIF-1α/VEGF signaling pathway. Molecules . 2016;21(12):p. 1756. doi: 10.3390/molecules21121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu J., Ke X., Ma N., et al. Formononetin, an active compound of Astragalus membranaceus (Fisch) Bunge, inhibits hypoxia-induced retinal neovascularization via the HIF-1α/VEGF signaling pathway. Drug Design, Development and Therapy . 2016;10:3071–3081. doi: 10.2147/dddt.S114022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J., Ke X., Wang W., et al. Aloe-emodin suppresses hypoxia-induced retinal angiogenesis via inhibition of HIF-1α/VEGF pathway. International Journal of Biological Sciences . 2016;12(11):1363–1371. doi: 10.7150/ijbs.16334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L., Li S., Wang L., et al. Uncovering the protective mechanism of Taohong Siwu decoction against diabetic retinopathy via HIF-1 signaling pathway based on network analysis and experimental validation. BMC Complementary Medicine and Therapies . 2020;20(1):p. 298. doi: 10.1186/s12906-020-03086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: the abbreviations. Table S2: the GO enrichment with related genes.
Data Availability Statement
The datasets in this study can be obtained from the corresponding author upon reasonable request. The authors are responsible for providing the final supplementary material files that will be published along with the article.


