Abstract
Objective: To explore whether METTL3 was involved in the pathogenesis of hepatocellular carcinoma (HCC) by modulating the m6A level of USP7. Methods: We performed qRT-PCR and western blot assays to detect the expression level of METTL3 in HCC tissues and paired adjacent normal tissues, as well as HCC cell lines. The level of m6A in HCC tissues and cells was quantitatively analyzed by m6A RNA Methylation Quantitative Kit. We examined the effect of METTL3 on cell proliferation ability by CCK-8 and EdU assays, and examined cell migration and cell invasion ability by Transwell assay. It was predicted via bioinformatics tool that USP7 may undergo methylation in HCC. Subsequently, we performed qRT-PCR assay to detect the expression level of USP7 in HCC tissues and analyzed its correlation with the expression level of METTL3. We verified the regulatory relationship between METTL3/USP7 and transfected USP7 siRNA in cells to detect its effects on cell invasion, migration and proliferation. The regulatory effect of METTL3 on USP7 in HCC was analyzed by corresponding experiments. Results: The qRT-PCR results indicated that METTL3 was highly expressed in HCC tissues and cell lines. The level of m6A was remarkably increased in HCC tissues and cell lines. Besides, the elevated METTL3 expression was related to worse overall survival. The abilities of cell invasion, migration and proliferation were remarkably attenuated by down-regulation of METTL3 expression. Through bioinformatics analysis, it was found that USP7 might be regulated by METTL3 to undergo methylation modification. The qRT-PCR results showed that the USP7 was highly expressed in HCC tissues, and was positively correlated with the level of METTL3. Further experiments showed that down-regulation of USP7 could reduce cell proliferation, migration, and invasion. METTL3 could positively regulate the malignant phenotype of USP7 in HCC. Conclusion: METTL3 might regulate the expression of USP7 through m6A methylation and facilitate the invasion, migration and proliferation of HCC cells.
Keywords: RNA methylation, METTL3, cell proliferation, cell migration, cell invasion
Introduction
Primary liver cancer is one of the most common malignant cancers worldwide [1-3]. According to histology, it can be divided into hepatocellular carcinoma (HCC), combined hepatocellular and cholangiocarcinoma and cholangiocarcinoma, with hepatocellular carcinoma in the majority, accounting for about 90% [4]. The 5-year survival rate of patients with HCC is very poor, and about 600,000 patients die every year [5]. Therefore, it’s of great significance to explore prognostic biomarkers and treatment targets for HCC.
RNA methylation modifications account for more than 60% of RNA modifications, which have many types including m1A (N1-methyladenosine) occurring on the first nitrogen atom of adenine, m6A (N6-methyladenosine) occurring on the sixth nitrogen atom of adenine, and m5C (C5-methylcytidine) occurring on the fifth carbon atom of cytosine [6]. m6A of RNA is the most common type of RNA modifications and is dynamic and reversible [7-11]. M6A mainly enriches in a conserved consensus motif named as RRACH [11,12]. Now, we have a clear understanding of the formation of m6A. The modification function of m6A is mainly determined by demethylases (ALKBH5 and FTO) and methyltransferases (METTL3, METTL14 and WTAP) [13-16]. M6A recognizing proteins are composed of YTH domain protein family and nuclear heterozygous protein HNRNP family, which are responsible for downstream RNA translation and degradation [17]. The degree and the pattern of m6A of mRNAs can affect their variable transport, storage, splicing, translation and stability [11]. Abnormity in regulatory mechanisms of m6A can lead to the occurrence of a variety of human diseases including cancers [18,19].
METTL3, as one of the core components of m6A methyltransferase complex, takes part in mediating writing process of m6A methylation. It was confirmed to be abnormally expressed in many malignancies and have close association with poor prognosis in patients. Interestingly, it can play different or even opposite roles in different types of tumors. Most studies suggest that elevated METTL3 expression levels can promote tumor progression. For instances, Wang et al. showed that METTL3 is remarkably upregulated in gastric cancer (GC) tissues and associated with poor prognosis. Overexpression of METTL3 can promote GC proliferation and liver metastasis in vitro and in vivo. Mechanistically, METTL3 can stimulate m6A modification of HDGF mRNA, and IGF2BP3 can directly recognize and bind to the m6A site on HDGF mRNA and enhance HDGF mRNA stability [20]. Han and colleagues have demonstrated that METTL3 can promote the proliferation of bladder cancer via accelerating the maturation of pri-miR221/222, leading to the reduction of PTEN [21]. Chen et al. have proved that overexpression of METTL3 can obviously promote hepatocellular carcinoma (HCC) growth both in vitro and in vivo. Besides, METTL3 can lead to m6A-mediated SOCS2 mRNA degradation via the m6A reader protein YTHDF2-dependent pathway [22]. Liu et al. also showed that CNV and DNA methylation may contribute to the abnormal upregulation of METTL3 in HCC [23]. Besides, METTL3 is highly expressed in lung adenocarcinoma and facilitates proliferation and invasion of lung adenocarcinoma cells [24]. It was reported that METTL3 is remarkably overexpressed in glioblastoma, and silencing METTL3 could down-regulate glioma recombinant factors such as SOX2, POU3F2, OLIG2 and SALL2, thereby inhibiting the growth of GSCs [25]. A few studies suggested that lower levels of METTL3 promote tumor progression. Nevertheless, the role of METTL3 in the pathogenesis of HCC needs further explore.
Materials and methods
Sample selection
From October 2018 to March 2019, 50 paired HCC tissues as well as adjacent normal tissues were harvested from diagnosed patients in Peking University Shenzhen Hospital. All tissue samples were stored at -80°C in liquid nitrogen quickly after surgical resection. All tissue samples were pathologically validated. All included patients and their families signed an informed consent and the investigation received approval from the ethics committee of the hospital (approval number: 2020-336). Tumor pathological classification and staging were carried out according to the staging standards of the Union for International Cancer Control (UICC).
Cell culture
We purchased HCC cell lines (Hep3B, HCCLM3, MHCC97-L, HUH7) and human normal liver cell line (L02) from Institute of Hematology, China Academy of Traditional Chinese Medicine (Beijing, China). All cells were cultured in a complete medium containing 10% heat-inactivated fetal bovine serum (FBS; Invitrogen), streptomycin (100 U/mL)/ penicillin (100 μg/mL; Invitrogen) (RPMI-1640; Invitrogen, Grand Island, NY, USA). We maintained cells in an incubator with humidified atmosphere at 37°C with 5% CO2.
Cell transfection
In this study, the METTL3 siRNA and USP7 siRNA, as well as the corresponding siRNA negative control were obtained from GenePharma Company (Shanghai, China). The cells used in the experiment (5×105 cells/well) were placed in a 6-well plate until the cell density reached 80%. According to the manufacturer’s instructions, the transfection reagent was mixed with Lipofectamine 2000 (Invitrogen) and incubated at room temperature for 30 min. The products were added to the petri dishes and transfected for 24-48 hours, and then the transfection efficiency was examined by qRT-PCR. The sequences of siRNAs are as follows: si-METTL3 sense: 5’-GGCAAUAAUUAGUAGUCAAGU-3’; si-METTL3 anti-sense: 5’-UUGACUACUAAUUAUUGCCUG-3’; si-USP7 sense: 5’-GGAUGUCUGUAGAAUAUUAAA-3’; si-USP7 anti-sense: 5’-UAAUAUUCUACAGACAUCCUG-3’.
RNA extraction and qRT-PCR
Purified total RNA was extracted from tissues and cells using TRIzol (Invitrogen) solution. As per the manufacturer’s instruction, the cDNA was extracted by reverse transcription (RT) in a 20 μL reaction system using the PrimeScript RT Reagent Kit (TAKARA, Code No. RR037A). qRT-PCR analysis was performed using the following conditions: 92°C 10 min, followed by 40 cycles of 10 s at 92°C, 1 min at 60°C and 30 s at 72°C. Primer sequences are as follows: METTL3 (F: 5’-CTCTGGGGGTATGAACGGG-3’, R: 5’-CTCTGGGGGTATGAACGGG-3’); USP7 (F: 5’-CCCTCCGTGTTTTGTGCGA-3’, R: 5’-AGACCATGACGTGGAATCAGA-3’); GAPDH (F: 5’-GGAATCCACTGGCGTCTTCA-3’, R: 5’-GGTTCACGCCCATCACAAAC-3’).
M6A RNA methylation
The Total RNA was extracted using TRIzol (Thermo Fisher, USA) according to the product specification. The relative content of m6A in total RNA was determined using the EpiQuik m6A RNA Methylation Quantitative Kit (colorimetric method; P-9005, Epigentek, USA), according to the product instructions. The m6A level was colorimetrically analyzed with absorbance at 450 nm.
CCK-8 assay
The cells in the medium were digested with 0.25% trypsin, and the cell suspension was harvested and inoculated into 96-well plates with 6 wells in each group. Each well contained 2×103 cells and 200 μL medium at least. The cells were incubated overnight for cell adhesion and growth, and then the supernatant was washed with phosphate buffer saline (PBS). Mixture containing 90 μL pure 1640 medium and 10 μL CCK-8 solution (Beyotime Biotechnology, Shanghai, China) was added into each well. After incubation for 2 h, we read the absorbance of each well at 450 nm with a microplate analyzer.
Transwell assay
Cell migration and invasion experiments were performed using the Transwell chamber (Corning). Cells were inoculated into a serum-free medium at 1×104 cells per compartment and added to the upper compartment coated with or without 200 mg/mL Matrigel for migration or invasion assay, respectively. A medium containing 10% FBS was added to the lower chamber as a chemical attractant. After 24 h of culture, the cells in the upper chamber were removed by swabbing, and the cells migrated to the lower surface of the filter were fixed in 100% formaldehyde for 20 min and stained with 0.2% crystal violet for 5 min. The migratory and invasive cells were photographed and counted by taking five random fields of view in each chamber under a light microscope of 200× magnification.
EdU assay
Cells in each group at logarithmic growth stage were inoculated in 12-well plates at a cell density of 5×104 cells/mL. According to the instructions of EdU reagent (Ribobio, China), EdU labeling, cells fixation, Apollo staining and DNA staining were implemented respectively. Finally, the image was obtained through a fluorescence microscope. Three duplicate holes were set up in each group. We performed all experiments thrice.
Western blotting
After the transfected HCC cells were digested and resuspended, they were seeded into 6-well plates at a cell density of 1×106/well. After the cells were cultured for 24 h, the protein expression amount in the cells was examined. With RIPA lysate we extracted the total protein and measured the total protein level. In each group, 100 μg proteins were taken for electrophoresis, and PVDF membrane was used for membrane transfer. The proteins were sealed for 1 h at room temperature and incubated with primary antibody overnight at low temperature. The antibodies against METTL3 (ab195352; 1:1000; abcam), USP7 (ab108931; 1:2000; abcam) and GAPDH (ab8245; 1:1000; abcam) were purchased from Abcam (Cambridge, MA, USA). HRP-conjugated secondary goat anti-mouse and goat anti-rabbit antibodies (Proteintech, USA) were then added dropwise to membrane and incubated at room temperature for 1 h. Then, chromochrome solution was added for color development and photographic analysis.
Animal assays
Male nu/nu mice between 4 and 6 weeks of age received subcutaneous injections of equivalent Hep3B cells expressing either LV-shMETTL3 or LV-USP7 within 30 min of harvesting on the right and left flanks. The tumor was weighed after approximately 4 weeks, and the volume was measured every 5 days. Animal assays were approved by the animal research ethics committee of our hospital, and the data of each group of mice were expressed as the mean ± standard deviation (x̅±sd).
Statistical analysis
For data analysis, SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA) was used. Graph-Pad Prism (Version X; La Jolla, CA, USA) was used for photo editing. Student’s t-test was used to compare the mean of measurement data between the two groups, and analysis of variance (ANOVA) was used to compare the difference between multiple groups with Tukey honestly significant difference (HSD) post hoc test. Kaplan-Meier analysis with log-rank tests was used for overall survive analysis. If P<0.05, statistical significance was considered.
Results
METTL3 was highly expressed in HCC tissues
Through qRT-PCR, it was found that the expression level of METTL3 was remarkably increased in HCC tissues in comparison with paired adjacent tissues (Figure 1A). In comparison with human normal liver cell lines (L02), the expression level of METTL3 in HCC cell lines (Hep3B, HCCLM3, MHCC97-L, HUH7) was remarkably increased (Figure 1B). Subsequently, we further confirmed the high expression of METTL3 protein in HCC tissues by western blot (Figure 1C). We then analyzed the levels of total methylated RNA (m6A) in HCC tissues and cell lines, and found that m6A levels were remarkably increased in HCC tissues and cell lines (Figure 1D, 1E).
Figure 1.
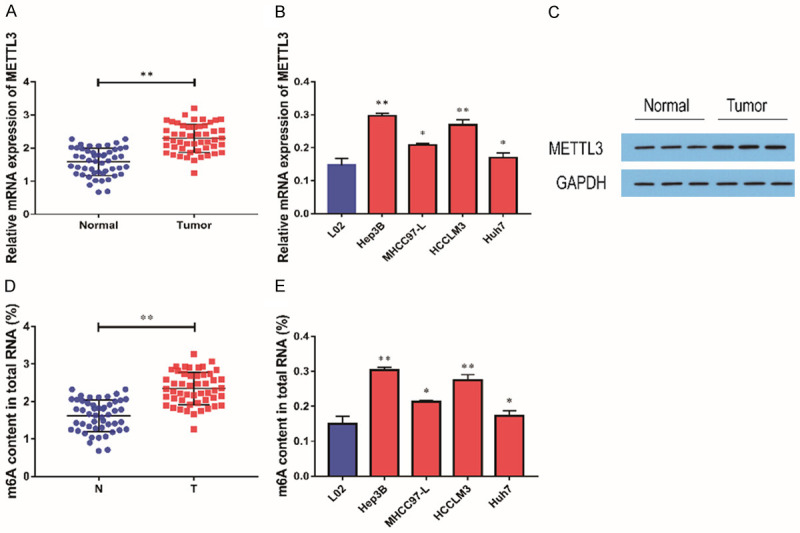
METTL3 was highly expressed in HCC tissues. A: In comparison with adjacent tissues, the expression level of METTL3 was remarkably up-regulated in HCC tissues (n=50); B: In comparison with human normal liver cell lines (L02), the expression level of METTL3 was higher in hepatocellular carcinoma cell lines (Hep3B, HCCLM3, MHCC97-L, HUH7; n=3); C: Elevated METTL3 protein levels in HCC tissues were examined by Western blot (n=3); D: The levels of m6A in HCC tissues were measured by methylation assay (n=50); E: The level of m6A in HCC cells was measured by methylation assay (n=3). Compared with LO2 cells, *P<0.05; compared with normal tissues or LO2 cells, **P<0.01.
High expression of METTL3 in HCC cells predicted a poor prognosis
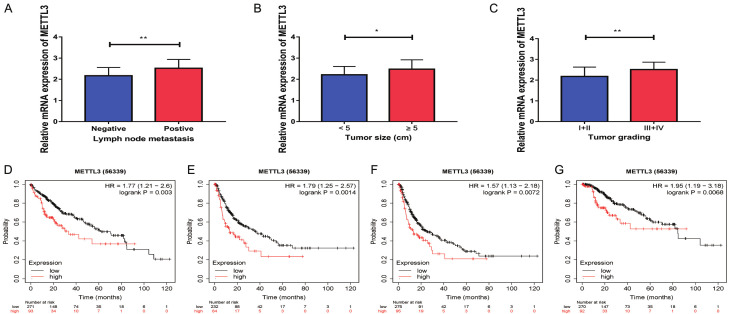
Meanwhile, through correlation analysis with clinicopathological data, it was found that the expression level of METTL3 in HCC tissues of patients with lymph node metastasis was also remarkably up-regulated in comparison with that of patients without lymph node metastasis (Figure 2A). In comparison with HCC tissues of patients with tumor diameter of <5 cm, the expression level of METTL3 in HCC tissues of patients with tumor diameter of 5 cm or above was remarkably increased (Figure 2B). In comparison with the HCC tissues of the patients with pathological stage I and stage II, the expression of METTL3 was increased in the HCC tissues of the stage III and IV patients (Figure 2C). Further analysis of METTL3 in Kaplan-Meier Plotter database found that the increase of METTL3 expression predicted a lower overall survival (OS) rate (Figure 2D), Relapse Free Survival (RFS) rate (Figure 2E), Progress Free Survival (PFS) rate (Figure 2F) and Disease Specific Survival rate (DSS) (Figure 2G). Therefore, it was found that the expression level of METTL3 was remarkably associated with the pathological stage, tumor size and lymph node metastasis of the patients (Table 1), and METTL3 could be a potential biomarker indicating poor prognosis of patients with HCC.
Figure 2.
The high expression of METTL3 indicated a poor prognosis of HCC. A: Correlation analysis of METTL3 and tumor lymph node metastasis in patients (n=50); B: Correlation analysis of METTL3 expression and tumor size in patients (n=50); C: Correlation analysis of METTL3 expression and TNM stage of patients (n=50); D: By analyzing Kaplan-Meier Plotter database, patients with high METTL3 expression had a worse overall survival prognosis (n=364); E: By analyzing Kaplan-Meier Plotter database, patients with high METTL3 expression had a worse relapse free survival prognosis (m=316); F: By analyzing Kaplan-Meier Plotter database, patients with high METTL3 expression had a worse progression free survival prognosis (n=370); G: By analyzing Kaplan-Meier Plotter database, patients with high METTL3 expression had a worse disease-specific survival prognosis (n=362). Compared with <5 cm tumors, *P<0.05; compared with negative lymph node metastasis or I+II tumors, **P<0.01.
Table 1.
Correlation between METTL3 expression and clinicopathological factors
| Factors | Cases | Expression of METTL3 | P | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| Overall | 50 | 25 | 25 | |
| Gender | ||||
| Male | 40 | 21 | 19 | 0.7252 |
| Female | 10 | 4 | 6 | |
| Age | ||||
| ≤40 | 11 | 6 | 5 | >0.9999 |
| >40 | 39 | 19 | 20 | |
| TNM Stage | ||||
| I+II | 25 | 5 | 20 | <0.0001 |
| III+IV | 25 | 20 | 5 | |
| Lymph node | ||||
| Negative | 15 | 3 | 12 | 0.0121 |
| Positive | 35 | 22 | 13 | |
| Tumor Size | ||||
| <5 cm | 20 | 6 | 14 | 0.0421 |
| ≥5 cm | 30 | 19 | 11 | |
Silencing of METTL3 decreased the invasion, migration and proliferation of HCC cells
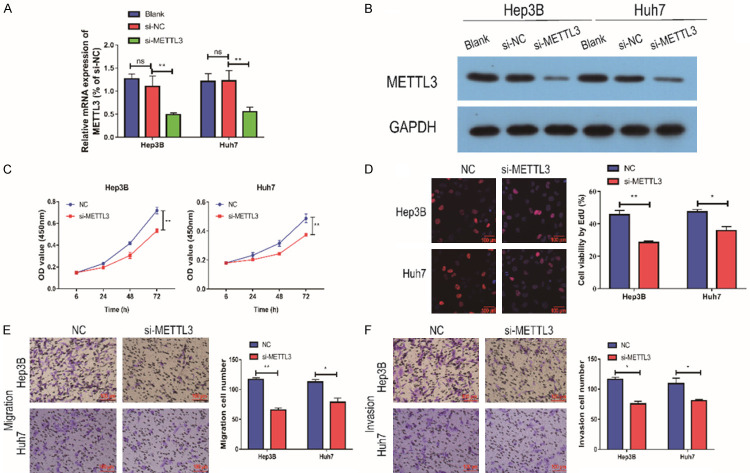
In order to further investigate the mechanism of METTL3 involvement in the development of HCC, we used qRT-PCR assay. Hep3B and HUH7 cells were selected for subsequent experiments. We transfected si-METTL3 in Hep3B and HUH7 cells. The transfection efficiency was examined by qRT-PCR and Western blot assay, and the results showed that the expression of METTL3 was remarkably decreased (Figure 3A, 3B). In order to detect the effect of METTL3 on cells, CCK-8 assay was performed. The results showed that cell proliferation ability was remarkably attenuated after the down-regulation of METTL3 in Hep3B and HUH7 cells (Figure 3C). The EdU assay also found the same result (Figure 3D). Transwell assay was performed to detect cell migration ability, and it showed that cell migration ability was remarkably reduced after the down-regulation of METTL3, and cell invasion ability was also remarkably attenuated (Figure 3E, 3F). Taken together, it was found that down-regulation of METTL3 can remarkably inhibit the invasion, migration and proliferation of HCC cells.
Figure 3.
Silencing of METTL3 inhibited the proliferation and migration of HCC cells. A: After si-METTL3 transfection in Hep3B and HUH7 cells, METTL3 expression was remarkably down-regulated (n=3); B: CCK-8 assay showed that down-regulation of METTL3 expression in Hep3B cells resulted in reduced cell proliferation (n=3); C: CCK-8 assay showed that METTL3 expression was down-regulated in HUH7 cells, and cell proliferation was reduced (n=3); D: EdU assay showed that down-regulation of METTL3 expression in Hep3B and HUH7 cells resulted in reduced cell proliferation (Magnification: 200×; n=3); E: Transwell assay showed that down-regulation of METTL3 expression in Hep3B and HUH7 cells resulted in reduced cell migration (Magnification: 200×; n=3); F: Transwell assay showed that down-regulation of METTL3 expression in Hep3B and HUH7 cells resulted in reduced cell invasion (Magnification: 200×; n=3). Compared with NC group cells, *P<0.05; compared with NC group cells, **P<0.01; compared with blank group cells, ns, no significant difference.
Silencing of METTL3 inhibited the tumor growth of HCC
To explore the effect of METTL3 on the growth of HCC tumor, the tumor size of mice after 4 weeks of HCC cell injection was detected by tumor loading experiment in nude mice. The xenograft tumor volume and weight of nude mice injected with Lv-shMETTL3 was significantly smaller than that of Lv-NC (Figure 4).
Figure 4.
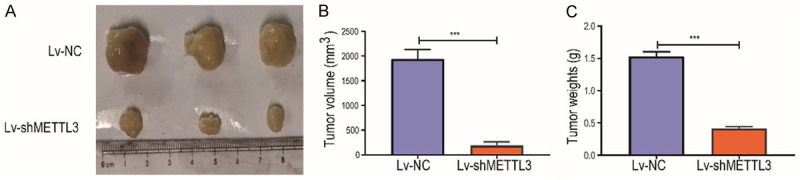
Silencing of METTL3 inhibited the tumor growth of HCC. A: After Lv-shMETTL3 transfection in HCC cells, the HCC tumor formed in nude mice four weeks after injection was removed (n=3); B: The tumor volume in nude mice (n=3); C: The tumor weight in nude mice (n=3). Compared with LV-Nc group, ***P<0.001.
METTL3 could regulate USP7 expression
To further study the role of METTL3 in the development of HCC, we found that USP7 could undergo methylation modification through the prediction website (Figure 5A). Therefore, we speculated that METTL3 could regulate the expression level of USP7. We further detected the expression level of USP7 in HCC tissues, and the results showed that the level of USP7 in HCC tissues was remarkably higher than that in paracancerous tissues, and it was positively correlated with the level of METTL3 (Figure 5B, 5C). In order to further study the regulatory relationship between the two, qRT-PCR test was performed and it showed that the expression level of USP7 was remarkably reduced after down-regulating the expression of METTL3, and the same result was also found by the Western blot assay (Figure 5E). Through methylation detection, it was found that the m6A level of USP7 was remarkably reduced after the down-regulation of METTL3 expression (Figure 5F). In addition, si-USP7 was transfected into cells to inhibit its expression. qRT-PCR detection indicated that the mRNA and protein expression levels of USP7 were remarkably reduced after transfection (Figure 5G, 5H). Taken together, METTL3 could regulate the expression of USP7 via m6A methylation modification in HCC cells.
Figure 5.
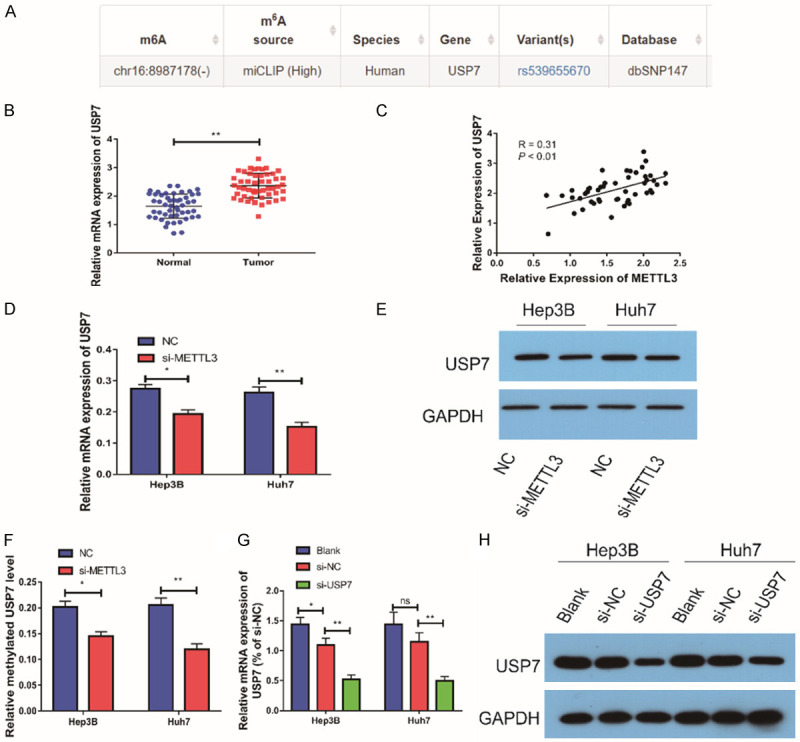
METTL3 could regulate USP7 expression. A: Methylation of USP7 was predicted by the predictive site; B: In comparison with adjacent tissues, the expression of USP7 was remarkably up-regulated in HCC tissues (n=50); C: Statistical analysis results showed that the expression of USP7 was positively correlated with the expression of METTL3 (n=50); D: USP7 expression was down-regulated after downregulation of METTL3 expression in Hep3B and HUH7 cells (n=3); E: USP7 protein levels were down-regulated after downregulation of METTL3 expression in Hep3B and HUH7 cells (n=3); F: m6A level of USP7 in HCC cells after down-regulation of METTL3 expression by methylation detection (n=3); G: The expression of USP7 was down-regulated after si-USP7 transfection in Hep3B and HUH7 cells (n=3); H: After USP7 expression was down-regulated in Hep3B and HUH7 cells, METTL3 expression was down-regulated (n=3). Compared with NC group cells, *P<0.05; compared with NC group cells or normal group, **P<0.01; compared with blank group cells, *P<0.05. ns, no significant difference.
Silencing of USP7 inhibited the invasion, migration and proliferation of HCC cells
In order to further explore the specific role of USP7 in the development of HCC, we tested the effect of USP7 on cell proliferation by CCK-8 assay. It showed that cell proliferation was remarkably attenuated when USP7 expression was down-regulated in Hep3B and HUH7 cells (Figure 6A). The EdU experiment also showed the same result (Figure 6B). We used Transwell assay to detect the cell migration ability, and it showed that the cell migration ability was remarkably reduced after the down-regulation of USP7 expression, and the cell invasion ability was also remarkably attenuated (Figure 6C, 6D). In conclusion, down-regulation of USP7 expression could remarkably inhibit the invasion, migration and proliferation of HCC cells.
Figure 6.
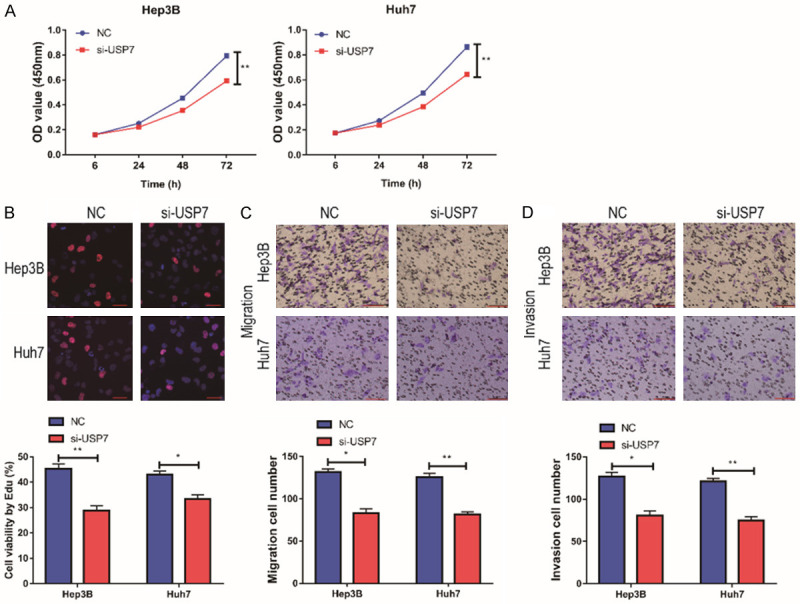
Silencing of USP7 inhibited the proliferation and migration of HCC cells. A: CCK-8 assay showed that USP7 expression was down-regulated in Hep3B cells, and cell proliferation was reduced (n=3); CCK-8 assay showed that USP7 expression was down-regulated in HUH7 cells, and cell proliferation was reduced (n=3); B: EdU assay showed that USP7 expression was down-regulated in Hep3B and HUH7 cells, and cell proliferation was reduced (Magnification: 200×; n=3); C: Transwell assay showed that USP7 expression was down-regulated in Hep3B and HUH7 cells, and cell migration was reduced (Magnification: 200×; n=3); D: Transwell assay showed that USP7 expression was down-regulated in Hep3B and HUH7 cells, and cell invasion ability was weakened (Magnification: 200×; n=3). Compared with NC group cells, *P<0.05; compared with NC group cells, **P<0.01.
Downregulation of METTL3 partially reversed the effect of USP7 on the malignant phenotype of HCC
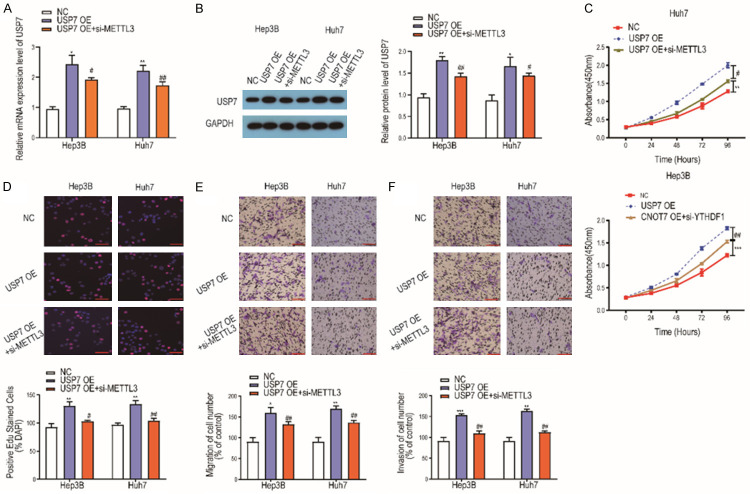
We investigated the interaction of USP7 and METTL3 in HCC by torsion test. We first co-transfected si-METTL3 and USP7 OE in HCC cells and detected the efficiency of transfection by qRT-PCR and Western blot, the results showed that the mRNA or protein level of USP7 in HCC cells co-transfected with si-METTL3 and USP7 OE was significantly higher than that in NC, but lower than that in USP7 OE transfection group (Figure 7A, 7B). Then we found that the OD value of HCC cells at 450 nm co-transfected with si-METTL3 and USP7 OE was significantly higher than that of NC, but lower than that of USP7 OE (Figure 7C). EdU assay showed that the positive rate of EdU in HCC cells co-transfected with si-METTL3 and USP7 OE was significantly higher than that in NC cells, but lower than that in USP7 cells (Figure 7D). The results of Transwell migration assay showed that the migration rate of HCC cells co-transfected with si-METTL3 and USP7 OE was significantly higher than that of NC, but lower than that of USP7 OE (Figure 7E). Meanwhile the results of Transwell invasion assay showed that the invasion rate of HCC cells co-transfected with si-METTL3 and USP7 OE was significantly higher than that of NC, but lower than that of USP7 OE (Figure 7F).
Figure 7.
Downregulation of METTL3 partially reversed the effect of USP7 on the malignant phenotype of HCC. A: The mRNA expression of USP7 in HCC cells co-transfected with si-METTL3 and USP7 OE was detected by qRT-PCR (n=3); B: The protein level of USP7 in HCC cells co-transfected with si-METTL3 and USP7 OE was detected by western blot (n=3); C: Detection of 450 nm OD value of HCC cells co-transfected with si-METTL3 and USP7 OE by CCK8 experiment (n=3). D: EdU assay was used to detect the positive rate of EdU in HCC cells co-transfected with si-METTL3 and USP7 OE (Magnification: 200×; n=3); E: Transwell migration assay was used to detect the migration rate of HCC cells co-transfected with si-METTL3 and USP7 OE (Magnification: 200×; n=3); F: Transwell invasion assay was used to detect the invasion rate of HCC cells co-transfected with si-METTL3 and USP7 OE (Magnification: 200×; n=3). Compared with NC group cells, *P<0.05; **P<0.01; ***P<0.001; compared with USP7 OE group cells, #P<0.05; ##P<0.01.
Overexpression of USP7 promoted the tumor growth of HCC
To explore the effect of USP7 on the growth of HCC tumor, the tumor size of mice after 4 weeks of HCC cells injection was detected by tumor loading experiment in nude mice. The xenograft tumor volume and weight of nude mice injected with LV-USP7 were obviously larger than those of LV-NC (Figure 8).
Figure 8.
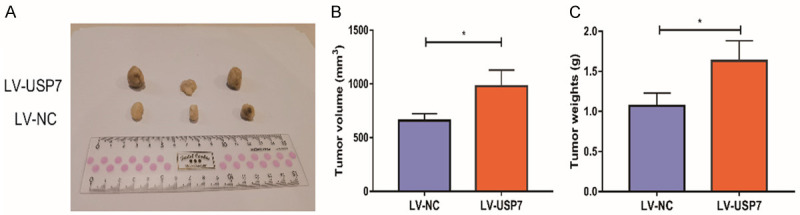
Overexpression of USP7 promoted the tumor growth of HCC. A: After LV-USP7 transfection in HCC cells, the HCC tumor formed in nude mice four weeks after injection was removed (n=3); B: The tumor volume in nude mice (n=3); C: The tumor weight in nude mice (n=3). Compared with LV-NC group, *P<0.05.
Discussion
Primary HCC is one of the common malignant tumors, and has become the third cause of cancer death in the world. The onset of HCC is covert, and early diagnosis is difficult. Due to the limitations including low resection rate, high postoperative recurrence rate, insensitivity to radiotherapy and chemotherapy, and narrow source of liver transplantation, in recent years the research focus in the field of HCC treatment has turned to molecular targeted therapy [26]. Sorafenib is currently the only molecular targeted drug approved by FDA for the treatment of advanced HCC, but its action target is in endothelial cells and exerts an influence of anti-angiogenesis, rather than directly acting on tumor cells, so its anti-tumor effect is still limited to some extent [27]. Thus, it is very significant to explore the occurrence and development mechanism of HCC and find new drug targets for the treatment of HCC.
METTL3, as the most important methylase of m6A, plays a vital role in cancers, including influencing methylation levels, cancer-related gene mRNA stability, the expression of oncogenes in cancer cells, cancer cell signaling pathways and cancer cell apoptosis and other regulatory mechanisms [28-30]. Changes in the expression levels of METTL3 and related genes may be a potential target for the use of molecular therapies to treat cancer. Some current studies have shown some key steps in the regulation of cancer by METTL3, but it is still necessary to further study the complex regulatory network of cancer driver gene transcription, post-transcriptional modification and translation, and gain insight into the role of METTL3 in it. Therefore, in-depth elucidation of the regulatory mechanism of METTL3 in different tumors and perfecting the regulatory network of cancer occurrence and development have important theoretical and practical significance for cancer treatment. Although some studies are promising in identifying targets for molecular diagnosis and treatment, a large number of clinical trials are still needed to determine the potential diagnostic and therapeutic functions of METTL3 in cancers.
In our previous studies, we found that the level of METTL3 was remarkably up-regulated in HCC tissues, and the expression of METTL3 in HCC cell lines was remarkably increased in comparison with that in L02. Previously, METTL3 has been reported to have the potential to be a novel biomarker in tumors. Bi et al. showed that METTL3 exerts hypomethylation and high expression in ovarian cancer tissues and cells. Hypomethylation of METTL3 was pronounced in ovarian cancer samples, which was negatively associated with patient survival [31]. Chen et al. found that METTL3 expression is higher in ESCC tissues and markedly associated with depth of invasion and poor prognosis [32]. Jiang and colleagues demonstrated that METTL3 expression is positively associated with a higher malignant grade and poorer prognosis of IDH-wild type gliomas but not IDH-mutant gliomas [33]. We found that the expression level of METTL3 was remarkably correlated with the pathological stage, tumor size and lymph node metastasis of the patients. It meant that METTL3 might be a latent biomarker in HCC. Further research results showed that cell invasion, migration and proliferation were remarkably attenuated after down-regulation of the expression of METTL3 in cells.
At present, deubiquitination enzymes mainly consist of 6 families: USP, UCH, OUT, MJDs, JAMMs and MCPIPs. Among them, the USP family has the largest number of members and is also the most studied family at present. In the USP family, the function of USP7 (Ubiquitin Specific Peptidase 7) has also been discovered by more and more researchers. USP7 is an evolutionarily conserved protein that was originally isolated as a binding partner of the herpes simplex virus protein Vmw110. Meanwhile, USP7 is also necessary for cell cycle regulation, cell growth control, stress response and receptor functional development [34]. More and more studies have also attempted to clarify the antiviral pathway of USP7, but it is still not fully understood [35]. Studies at home and abroad have shown that ubiquitination and deubiquitination play a very significant role in the occurrence of liver diseases. The largest subfamily of deubiquitinases is the ubiquitin-specific protease (USPs) family, a deubiquitinase that removes ubiquitin from specific protein substrates to prevent their degradation by targeted proteasomes [36]. USP7 is an evolutionarily conserved protein, which is responsible for modulating a large number of cellular processes [37]. USP7 is a potential target for drug development due to its substrate specificity. Many studies have shown that USP7 participates in the initiation and development of several tumors. For instances, USP7 acts as an oncogene involved in melanoma cell proliferation and metastasis via decreasing the Wnt/β-catenin signaling pathway [38]. USP7 can increase YAP stability under increased serine conditions by regulating deubiquitination, so as to regulate cell proliferation and tumor growth in colon cancer [39]. Besides, USP7 is also involved in the progression of prostate cancer [40], gastric cancer [41], esophageal carcinoma [42] and T-lineage acute lymphoblastic leukemia [43]. Attempts to inhibit USP7 activity from a therapeutic point of view have led to the development of several USP7 inhibitors that primarily induce apoptosis in cancer cells. Recent studies showed that USP7 is highly expressed in HBV-associated HCC, which provides a new idea for the study of the mechanism of HCC [44]. Through bioinformatics website, we predicted that USP7 could undergo methylation. Through qRT-PCR assay, we found that the level of USP7 was remarkably up-regulated in HCC tissues and was positively correlated with the level of METTL3. Meanwhile, METTL3 could regulate the expression of USP7 through m6A methylation. In further studies, it was found that down-regulation of USP7 reduced cell invasion, migration and proliferation. Besides, the animal assay also showed that USP7 could promote cell growth in HCC. The rescue experiment results confirmed that the METL3/USP7 regulatory axis played an important regulatory role in the occurrence and development of HCC.
However, this study still has many shortcomings. First, which downstream m6A recognition protein can recognize the USP7 methylation site and regulate expression is still unknown. Second, whether the m6A recognition site of USP7 can be recognized and regulated by other RNA methylases still needs further study.
In conclusion, we found that METTL3 might be involved in the development of HCC by modulating the level of USP7 through the modification of m6A methylation.
METTL3 could regulate the expression of USP7 via the m6A methylation modification to promote the invasion, migration and proliferation of HCC cells. METTL3 might be used as a potential biomarker for a poor prognosis of HCC.
Acknowledgements
This work was supported by Sanming Project of Medicine in Shenzhen (SZSM201612071), Shenzhen Key Medical Discipline Construction Fund (SZXK078), the Cell Technology Center and Transformation Base, Innovation Center of Guangdong-Hong Kong-Macao Greater Bay Area, Ministry of Science and Technology of China (Grant No. YCZYPT (2018) 03-1).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu L, He J. Report of incidence and mortality in China cancer registries, 2009. Chin J Cancer Res. 2013;25:10–21. doi: 10.3978/j.issn.1000-9604.2012.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaub MR, McGlaughlin VG. A knowledge assessment of combat stress reaction in army family practice residents. Mil Med. 1990;155:539–542. [PubMed] [Google Scholar]
- 4.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(Suppl 1):S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 6.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 8.Roignant JY, Soller M. m(6)A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Niemi M, Mustakallio KK. A model for NADH-tetrazolium reductase. Histochemical implications of light-induced electron transfer in NADH-riboflavin-tetrazolium complexes. Histochemie. 1965;4:451–458. doi: 10.1007/BF00306255. [DOI] [PubMed] [Google Scholar]
- 10.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berulava T, Buchholz E, Elerdashvili V, Pena T, Islam MR, Lbik D, Mohamed BA, Renner A, von Lewinski D, Sacherer M, Bohnsack KE, Bohnsack MT, Jain G, Capece V, Cleve N, Burkhardt S, Hasenfuss G, Fischer A, Toischer K. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail. 2020;22:54–66. doi: 10.1002/ejhf.1672. [DOI] [PubMed] [Google Scholar]
- 13.Theler D, Dominguez C, Blatter M, Boudet J, Allain FH. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014;42:13911–13919. doi: 10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, Hong GM, Huang H, Wang X, Chen P, Gurbuxani S, Arnovitz S, Li Y, Li S, Strong J, Neilly MB, Larson RA, Jiang X, Zhang P, Jin J, He C, Chen J. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Huang J, Zou T, Yin P. Human m(6)A writers: two subunits, 2 roles. RNA Biol. 2017;14:300–304. doi: 10.1080/15476286.2017.1282025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613. doi: 10.1016/j.biopha.2019.108613. [DOI] [PubMed] [Google Scholar]
- 19.Yun S, He X, Zhang W, Chu D, Feng C. Alleviation effect of grape seed proanthocyanidins on neuronal apoptosis in rats with iron overload. Biol Trace Elem Res. 2020;194:210–220. doi: 10.1007/s12011-019-01766-8. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, Zhou J, Sun B, Zou X, Wang S. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, Wei JF, Yang H. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 23.Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of METTL3 as an adverse prognostic biomarker in hepatocellular carcinoma. Dig Dis Sci. 2021;66:1110–1126. doi: 10.1007/s10620-020-06260-z. [DOI] [PubMed] [Google Scholar]
- 24.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 26.Kokudo N, Hasegawa K, Akahane M, Igaki H, Izumi N, Ichida T, Uemoto S, Kaneko S, Kawasaki S, Ku Y, Kudo M, Kubo S, Takayama T, Tateishi R, Fukuda T, Matsui O, Matsuyama Y, Murakami T, Arii S, Okazaki M, Makuuchi M. Evidence-based clinical practice guidelines for hepatocellular carcinoma: the Japan society of hepatology 2013 update (3rd JSH-HCC guidelines) Hepatol Res. 2015;45 doi: 10.1111/hepr.12464. [DOI] [PubMed] [Google Scholar]
- 27.Balboula AZ, Stein P, Schultz RM, Schindler K. RBBP4 regulates histone deacetylation and bipolar spindle assembly during oocyte maturation in the mouse. Biol Reprod. 2015;92:105. doi: 10.1095/biolreprod.115.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou H, Yin K, Zhang Y, Tian J, Wang S. The RNA m6A writer METTL14 in cancers: roles, structures, and applications. Biochim Biophys Acta Rev Cancer. 2021;1876:188609. doi: 10.1016/j.bbcan.2021.188609. [DOI] [PubMed] [Google Scholar]
- 29.Lan Q, Liu PY, Bell JL, Wang JY, Huttelmaier S, Zhang XD, Zhang L, Liu T. The emerging roles of RNA m(6)A methylation and demethylation as critical regulators of tumorigenesis, drug sensitivity, and resistance. Cancer Res. 2021;81:3431–3440. doi: 10.1158/0008-5472.CAN-20-4107. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Ge YZ, Xu L, Xu Z, Dou Q, Jia R. The potential roles of RNA N6-methyladenosine in urological tumors. Front Cell Dev Biol. 2020;8:579919. doi: 10.3389/fcell.2020.579919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, Liang X, Yang Y. METTL3 promotes the initiation and metastasis of ovarian cancer by inhibiting CCNG2 expression via promoting the maturation of pri-microRNA-1246. Cell Death Discov. 2021;7:237. doi: 10.1038/s41420-021-00600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Huang L, Yang T, Xu J, Zhang C, Deng Z, Yang X, Liu N, Chen S, Lin S. METTL3 promotes esophageal squamous cell carcinoma metastasis through enhancing GLS2 expression. Front Oncol. 2021;11:667451. doi: 10.3389/fonc.2021.667451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang YZ, Chai RC, Pang B, Chang X, An SY, Zhang KN, Jiang T, Wang YZ. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-kappaB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett. 2021;511:36–46. doi: 10.1016/j.canlet.2021.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 35.Kessler BM, Fortunati E, Melis M, Pals CE, Clevers H, Maurice MM. Proteome changes induced by knock-down of the deubiquitylating enzyme HAUSP/USP7. J Proteome Res. 2007;6:4163–4172. doi: 10.1021/pr0702161. [DOI] [PubMed] [Google Scholar]
- 36.Pozhidaeva AK, Mohni KN, Dhe-Paganon S, Arrowsmith CH, Weller SK, Korzhnev DM, Bezsonova I. Structural characterization of interaction between human ubiquitin-specific protease 7 and immediate-early protein ICP0 of herpes simplex virus-1. J Biol Chem. 2015;290:22907–22918. doi: 10.1074/jbc.M115.664805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng CD, Dong YF, Niu WX, Niu CS. HAUSP promoted the growth of glioma cells in vitro and in vivo via stabilizing NANOG. Pathol Res Pract. 2020;216:152883. doi: 10.1016/j.prp.2020.152883. [DOI] [PubMed] [Google Scholar]
- 38.Xiang M, Liang L, Kuang X, Xie Z, Liu J, Zhao S, Su J, Chen X, Liu H. Pharmacological inhibition of USP7 suppresses growth and metastasis of melanoma cells in vitro and in vivo. J Cell Mol Med. 2021;25:9228–9240. doi: 10.1111/jcmm.16834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Fu J, Hu B, Chen L, Wang J, Fang J, Ge C, Lin H, Pan K, Fu L, Wang L, Du J, Xu W. Serine metabolism regulates YAP activity through USP7 in colon cancer. Front Cell Dev Biol. 2021;9:639111. doi: 10.3389/fcell.2021.639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park SH, Fong KW, Kim J, Wang F, Lu X, Lee Y, Brea LT, Wadosky K, Guo C, Abdulkadir SA, Crispino JD, Fang D, Ntziachristos P, Liu X, Li X, Wan Y, Goodrich DW, Zhao JC, Yu J. Posttranslational regulation of FOXA1 by polycomb and BUB3/USP7 deubiquitin complex in prostate cancer. Sci Adv. 2021;7:eabe2261. doi: 10.1126/sciadv.abe2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Kang W, Li O, Qi F, Wang J, You Y, He P, Suo Z, Zheng Y, Liu HM. Abrogation of USP7 is an alternative strategy to downregulate PD-L1 and sensitize gastric cancer cells to T cells killing. Acta Pharm Sin B. 2021;11:694–707. doi: 10.1016/j.apsb.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N, Zhao Z, Liu P, Zheng Y, Cai S, Sun Y, Wang B. Upregulation of deubiquitinase USP7 by transcription factor FOXO6 promotes EC progression via targeting the JMJD3/CLU axis. Mol Ther Oncolytics. 2021;20:583–595. doi: 10.1016/j.omto.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaw TI, Dong L, Tian L, Qian C, Liu Y, Ju B, High A, Kavdia K, Pagala VR, Shaner B, Pei D, Easton J, Janke LJ, Porter SN, Ma X, Cheng C, Pruett-Miller SM, Choi J, Yu J, Peng J, Gu W, Look AT, Downing JR, Zhang J. Integrative network analysis reveals USP7 haploinsufficiency inhibits E-protein activity in pediatric T-lineage acute lymphoblastic leukemia (T-ALL) Sci Rep. 2021;11:5154. doi: 10.1038/s41598-021-84647-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rawat R, Starczynowski DT, Ntziachristos P. Nuclear deubiquitination in the spotlight: the multifaceted nature of USP7 biology in disease. Curr Opin Cell Biol. 2019;58:85–94. doi: 10.1016/j.ceb.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]