Abstract
Purpose: Colorectal cancer (CRC) is one of the most frequent tumors and causes of mortality worldwide. Ubiquitin ligase was reported to regulate multiple cellular processes, including tumorigenesis. As ubiquitin E3 ligases, RING-finger proteins play a key role in physiological and pathophysiological processes. Methods: We compared the expression levels of RNF128 in CRC tissues by western-blotting and qRT-PCR. Knockdown and overexpression of RNF128 were performed to examine its effect on proliferation and metastasis of CRC cells. Using western blot and co-immunoprecipitation assays, we explored the possible mechanisms underlying the effect of RNF128 in CRC cells. Results: We found that the expression level of RNF128 was correlated with the CRC tumorigenicity. Overexpression or knockdown of RNF128 suppressed or elevated CRC cell proliferation, migration and invasion, respectively. We further determined that RNF128 regulated β-catenin ubiquitination and thus inhibited Wnt/β-catenin signaling in CRC cells. Conclusion: Our research demonstrated that RNF128 inhibited cell proliferation and metastasis of CRC cells via Wnt/β-catenin signaling-mediated deubiquitination.
Keywords: RNF128, Wnt/β-catenin signaling, ubiquitination, colorectal cancer
Introduction
Colorectal cancer (CRC) is the third frequent malignant tumor and a major cause of mortality globally. The International Cancer Research Agency (IARC) estimated that new CRC cases are about 1.8 million, and about 0.9 million people die from CRC [1]. CRC arises through the multi-factor, multi-stage and multi-step processes [2]. The underlying mechanism of colorectal cancer remains largely unknown [3].
Ubiquitination is an essential function of proteins in all eukaryotic species, involving a sequence of enzymatic reactions [4,5]. Protein ubiquitination includes ubiquitin-activating enzyme, ubiquitin-conjugating enzyme, and ubiquitin ligase. Ubiquitin ligase (E3 ligase) recognizes substrates and then attaches ubiquitin (a small protein in all eukaryotes) to target proteins [6-8]. Depending on the proteasome (the ubiquitin-proteasome system), the substrates will be degraded or altered in localization and function. Ubiquitination was reported to regulate multiple cellular processes and fundamental mechanisms of cell signaling and regulation [9]. Aberrant ubiquitination causes abnormal metabolic physiology and several diseases and even results in tumorigenesis [10-12].
The activation of Wnt/β-catenin pathway, which participates in diverse cellular functions, is a hallmark of cancer [13]. β-catenin is a transcriptional co-activation factor and plays a significant role in Wnt/β-catenin pathway. As a core component, β-catenin was involved in commanding growth and development in cancer [14]. Furthermore, accumulation of β-catenin was associated with proto-oncogene, such as cyclin D1 [15]. Studies have reported that the ubiquitin-proteasome pathway was a major regulatory mode of β-catenin [16].
RING-finger proteins belong to the E3 ubiquitin ligase family, whose members all contain RING finger motifs. Ubiquitin E3 ligases mainly consist of RING-finger proteins with more than 200 different members [17]. RING-finger proteins may play a widespread role in physiological and pathophysiological processes in organisms [18-20]. RNF128 (also called gene related to anergy in lymphocytes; Grail) is a 428 amino acid protein with extracellular protease-associated domains, transmembrane single-subunit, and an intracellular zinc-finger domain [21-23]. The up-regulation of RNF128 in CD4+ T-cell correlated with suppressing T cell activation [22,24,25]. Although it is widely expressed in various human tissues, studies of RNF128 mainly focused on immunity and inflammation, and the function of the RNF128 is largely unknown. Several recent studies showed that RNF128 might play an essential role in tumor occurrence and development [26-28].
RNF128 associates with the N-terminal domain of p53 and regulates its transactivation activity [26,27]. Overexpression of Grail prevents p53-dependent apoptosis. Some researchers analyzed the clinical samples and found that downregulation of RNF128 predicts poor prognosis in patients with urothelial carcinoma of the upper tract and urinary bladder [29]. An increased expression of RNF128 was detected in tumor-infiltrating CD8+ T cells in transplanted lymphoma tumors [30]. Recently, it has also been reported in esophageal squamous cell carcinoma and melanoma [28,31]. Nevertheless, available studies are limited. Whether RNF128 exerts an important role in colorectal cancer cells growth, invasion and metastasis is not clearly defined.
Materials and methods
Clinical tissue samples and animals
Tissue samples were collected from the clinical biological sample bank of Nanjing Medical University Affiliated Cancer Hospital. All six patients were diagnosed with CRC, and their fresh-frozen tissues were stored in liquid nitrogen before used. This study was approved by Jiangsu Institute of Cancer Research Ethics Committee.
Six-week-old female BALB/c nude mice were randomly divided into 2 groups (n=4). The use of animals was approved by the Ethics Committee of Nanjing Medical University.
Cell culture and transfection
Colorectal cancer cell lines were bought from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Colorectal cancer cells were cultured in DMEM medium containing 10% fetal bovine serum at 37°C along with 5% CO2. TransIT-X2 (mirus) was used for siRNA transfection and plasmid construction.
Stable knockdown and overexpression of RNF128
The pLV3ltr-ZsGreen-puro-U6 shRNA vector was used to create the RNF128 shRNA expression vector. The pLV3ltr-ZsGreen-puro-CMV overexpression vector system was used. Colorectal cancer cells were infected with lentiviruses and selected by puromycin. Infection efficiency was assessed by Western blot and RT-PCR.
RNA isolation and quantitative reverse-transcriptase PCR
Total RNAs were isolated using the MiniBEST Plant RNA Extration Kit (Takara, Japan). The RNA concentration was measured by NanoDrop spectrophotometer. The extracted RNA was reverse transcribed with PrimeScript RT reagent Kit (Takara, Japan). We then carried out PCR amplification with the SYBR Green (Thermofisher scientific, USA) in 7500HT Fast Real-Time PCR System (Applied Biosystems, USA).
Western blotting assay and antibodies
Total proteins were extracted with RIPA lysis buffer, protease and phosphatase inhibitors (Thermofisher Scientific, USA). After cell lysis for 20 minutes, samples were centrifuged at 12000 g for fifteen minutes at four degrees. Supernatants were collected for measuring protein concentration by BCA assay (Thermofisher Scientific, USA).
β-tubulin Rabbit mAb, Lamin B1 Rabbit mAb and β-actin Rabbit mAb were purchased from Proteintech (china); β-Catenin Rabbit mAb and CCND1 Rabbit mAb were from Cell Signaling Technology (USA); Ub and RNF128 Rat mAb were from Santa Cruz Biotechnology (USA); goat anti-rabbit and goat anti-rat were from Cell Signaling Technology (USA).
Cell proliferation assays
To assess cell proliferation, the CCK8 assay was used. Stock solution was diluted in culture medium in one-tenth of final treatment volume. After cells were attached, the medium was replaced by dilutions. Absorbance (450 nm) was measured after incubating for an hour.
Scratch wound assay
Cells were cultured in six-well plates. When 90% of confluence was reached, the cells were scratched with sterile pipette tips. The width of the scratch was observed under a microscope at 0 h and 24 h. The change of wound width was analyzed using ImageJ.
Clonogenicity assay
Stable knockdown and overexpression of RNF128 cells were plated at a low density and medium was changed every 2-3 days. After 10 days in culture, colonies were scored. Cells were fixed and stained, and the number of colony-forming-units was thencounted.
Transwell migration and invasion assays
In transwell invasion assay, the upper chambers were coated with matrigel, and the lower chambers were added with DMEM medium containing 20% FBS. After incubatation at 37°C for 48 h, cells were fixed with 4% paraformaldehyde for 1 h and stained with crystal violet for 30 min. Then the transwell chambers were washed and photographed under the optical microscope. The migration assay was the same as described above without matrigel coating.
Cell nucleus/cytoplasm fraction isolation
For cell nucleus/cytoplasm fraction isolation, nuclear and cytoplasmic protein extraction kit (KeyGen Biotech, China) was used according to the manufacturer’s instructions.
Cycloheximide (CHX) chase assay and MG132 treatment assay
The protein half-life was estimated by treating cell with cycloheximide (Merck, Germany, 25 μg/ml) to block de novo protein synthesis at different time points. MG132 (Merck, 25 μg/ml), a proteasome inhibitor, was added to the experimental group at a concentration of 10 nmol/l for 6 hours.
Statistical analyses
qRT-PCR and cell proliferation assays were repeated 3 times. The measurement data were expressed as means and standard deviations. The differences between two groups were analyzed using student’s t-test. P value of less than 0.05 was defined as significant differences.
Results
Expression levels of RNF128 in colorectal cancer tissues
We measured the expression of RNF128 in six colorectal cancer patients with their colon cancer and para-cancer tissues. Western-blotting results indicated that RNF128 expression was lower in the CRC tissues compared with the para-carcinoma tissues (n=6, P<0.05, Figure 1A). Results of the quantitative real-time PCR (qRT-PCR) analysis demonstrated that RNF128 expression was higher in CRC tissues than that in adjacent normal tissues (n=6, P<0.05, Figure 1B). The low expression of RNF128 in CRC tissues may predict its close relation with cancer progression. Statistical results identified that RNF128 had a higher expression level in CRC cells. We also counted RNF128 expression of colon adenocarcinoma in TCGA datasets (Figure 1C). The result suggests that RNF128 may be associated with the development of CRC. The differential expressions with RNF128 are likely related to malignant phenotypes in CRC.
Figure 1.
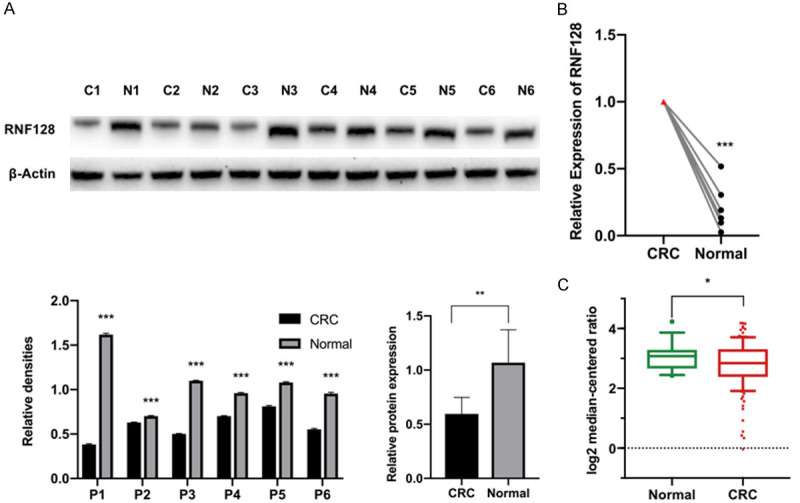
RNF128 is downregulated in colorectal tumors. (A and B) Expression levels of RNF128 in 6 clinical CRC tissues (C) compared to para-carcinoma normal tissue samples (N) were determined by Western blotting (A) and qRT-PCR (B). (C) RNF128 expression in TCGA datasets (Grouped by Normal and Cancer type). Normal (n=19); CRC (n=123). *P<0.05, **P<0.01 and ***P<0.001. Data represented as mean ± SD.
RNF128 participates in the regulation of proliferation, invasion and migration
After confirmed the differences in the RNF128 expression levels of CRC tissues, we next verified whether RNF128 affected CRC cell viability as well as cell invasion and migration ability.
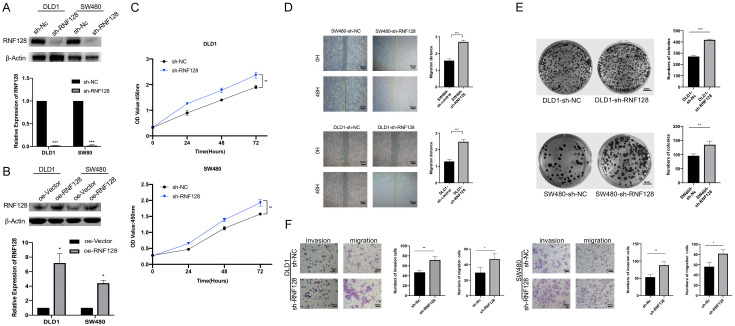
To ensure that our results were not restricted to single cell line, we chose two CRC cell lines (DLD1 and SW480) in the present study. To unravel the physiological role of RNF128, we generated a lentivirus expressing RNF128 shRNA to infect CRC cells. The qRT-PCR result indicated that cells with reduced RNF128 expression (DLD1-sh-RNF128 cells and SW480-sh-RNF128 cells) were successfully constructed (Figure 2A). We also constructed the gene expression vector and packaged the lentivirus in vitro (Figure 2B). By comparing the knockdown and overexpressing efficiency of RNF128, we used the constructed cell models for the following experiments.
Figure 2.
Knockdown of RNF128 promotes colorectal cancer cell growth, migration and invasion. (A) Expression levels of RNF128 in DLD1 and SW480 cells transfected with shRNA targeting RNF128 (sh-RNF128) or a control shRNA (sh-NC). (B) Expression levels of RNF128 in DLD1 and SW480 cells transfected with RNF128 plasmids (oe-RNF128) or an empty vector plasmid (oe-Vector). (C) CCK8 proliferation assays performed in DLD1 and SW480 cells transfected with sh-RNF128 and sh-control. (D) The migration ability of the RNF128 knockdown cells was examined using scratch assay. Relative quantitative comparison of scratch width was determined. (E) Colony formation assays performed in DLD1 and SW480 cells transfected with shRNA targeting RNF128 (sh-RNF128) or a control shRNA (sh-NC). The numbers of clones formed in each group were counted and compared. (F) Cell invasion and migration assays were performed and quantified in RNF128 knockdown and control of DLD1 and SW480 cells, respectively. *P<0.05, **P<0.01 and ***P<0.001. Data represented as mean ± SD. Scratch wound assay for (D): ×40, scale bar: 500 μm. Clonogenicity assay (E) for scale bar: 5 mm. Transwell migration and invasion assays for (F): ×100, scale bar: 100 μm.
Cell viability was detected by CCK-8 assay at different time intervals. CCK-8 assay showed that stable inhibition of RNF128 expression promoted cell proliferation (Figure 2C). Scratch wound assay was used to analyze the migration of sh-RNF128 transfected cells (Figure 2D). Scratch repair for 48 h showed that the sh-RNF128 cells’ scratch width was narrower compared to that of the control group (Figure 2D). The changes of scratch areas were calculated visually from the pictures (Figure 2D). We further used plate cloning assay to detect the proliferative ability of single CRC cell transfected with shRNA targeting RNF128 in vitro (Figure 2E). Compared to the control group, the number and size of colonies formed were marked increased in SW480-sh-RNF128 cells and DLD1-sh-RNF128 cells (Figure 2E). The difference was statistically significant. With decreasing levels of RNF128 expression, colony formation capability was visibly enhanced.
Another representative experiment to assess cellar migration and invasion ability was transwell assay. Changes in cell invasiveness were evaluated using transwell chambers with matrigel. After incubation for 48 h in incubator in transwell chambers, we counted the number of cells that migrated through the chamber under an inverted microscope (Figure 2F). Compared with the control group, the number of cells penetrating the membranes in the sh-RNF128 group was significantly increased. Differences between the two groups were statistically significant (Figure 2F). These results indicated that RNF128 had significant effect on the migration and invasion of CRC cells.
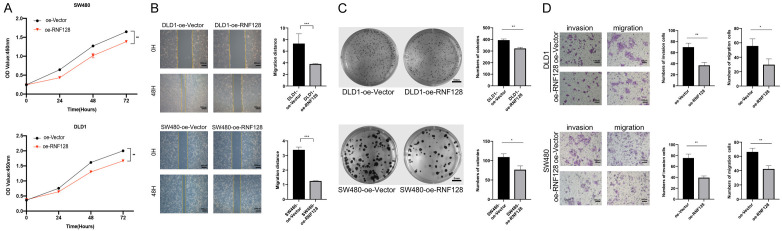
After verifying altered phenotype of RNF128 knockdown cells, we then determined whether this phenomenon was held in the converse situation. The above experiments were repeated in oe-RNF128 cell models (Figure 3). It was not difficult to find that oe-RNF128 cells could impair cell viability, hinder the repair of scratches and suppress the colony-forming ability. The overexpression group demonstrated a marked decrease in CRC tumor growth, migration and invasion.
Figure 3.
Overexpression of RNF128 promotes CRC cell migration and invasion. (A) CCK8 proliferation assays performed in DLD1 and SW480 cells transfected with RNF128 plasmid (oe-RNF128) and an empty vector plasmid (oe-Vector). (B-D) The Migration, invasion and colony formation capability of RNF128 were performed in DLD1 and SW480 cells transfected with the RNF128 plasmid (oe-RNF128) or an empty vector plasmid (oe-Vector). *P<0.05, **P<0.01 and ***P<0.001. Data represented as mean ± SD. Scratch wound assay for (B): ×40, scale bar: 500 μm. Clonogenicity assay (C) for scale bar: 5 mm. Transwell migration and invasion assays for (D): ×100, scale bar: 100 μm.
In short, RNF128 participates in the regulation of proliferation, invasion and migration.
Knockdown of RNF128 promotes CRC development and metastasis through Wnt/β-catenin signaling
We have revealed that overexpression of RNF128 can inhibit the proliferation, invasion and metastasis of CRC cells. However, the mechanism of RNF128 acting on CRC has not been thoroughly elucidated. Because RNF128 altered CRC cell proliferation, we assumed that RNF128 might affect the cell cycle. We examined the mRNA expression of several key cell cycle regulators (Figure 4A). Alterations in relative levels of CCND1 and CCNE1 indicated that the cell cycle-associated pathways would be activated or deactivated under RNF128 regulation.
Figure 4.
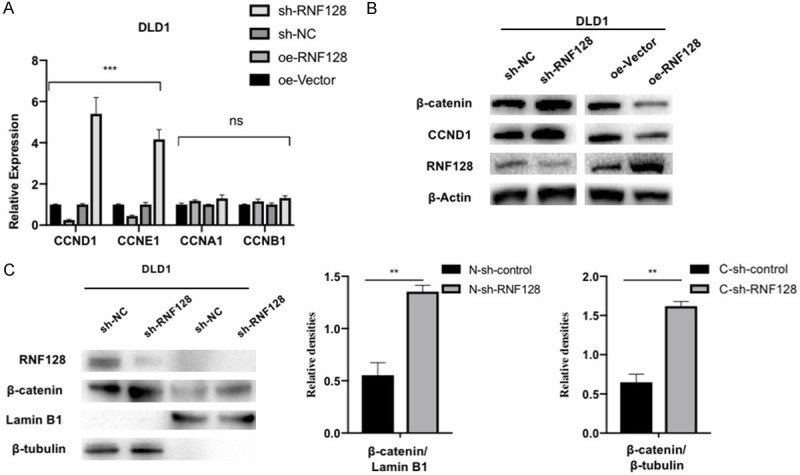
Loss of RNF128 increases β-catenin protein level and its target genes in colorectal cancer. (A) The mRNA expressions of cyclin D1, cyclin E1, cyclin A1 and cyclin B1 were quantified in RNF128 knockdown or overexpression cells compared to their respective controls in DLD1. (B) β-catenin, CCND1, and RNF128 expressions were confirmed using Western blotting. β-actin was used as a loading control. (C) Expression of β-catenin and RNF128 in nuclear and cytoplasmic fractions in RNF128-knockdown cells and control cells of DLD1. β-tubulin was used as the cytoplasmic reference protein. LaminB1 was used as the nuclear reference protein. *P<0.05, **P<0.01 and ***P<0.001. No significant difference (P>0.05, ns). Data represented as mean ± SD.
The previous studies showed that human colorectal cancer progression was closely associated with the Wnt/β-catenin signaling pathway [16]. Activation of this signaling pathway promoted the migration and invasion of CRC cells. CCND1 is an essential target gene of Wnt/β-catenin signaling. By chance, we found correlations between expression levels of β-catenin and RNF128 (Figure 4B). By isolating nuclei and evaluating nuclear levels of RNF128 and β-catenin, we found that β-catenin in the nucleus also showed an elevated expression (Figure 4C). We then measured the downstream target genes of Wnt/β-catenin pathway. The results implied that protein levels of CCND1 in CRC cells were reduced by knocking down RNF128 (Figure 4B). It suggested that Wnt/β-catenin pathway may be involved in RNF128-mediated inhibition of the proliferative and migration potential of colon tumor cells [32].
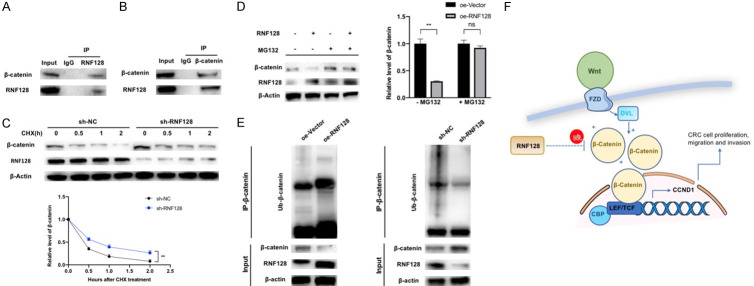
RNF128 directly interacts with β-catenin and promotes β-catenin ubiquitination
We speculated that, as an ubiquitin ligase, RNF128 acted on the Wnt/β-catenin pathway’s key protein via ubiquitination. Next, we assayed the interaction possibility among RNF128 and several critical proteins of Wnt/β-catenin pathway by co-immunoprecipitation (co-IP) and Western blotting. The interaction between RNF128 and β-catenin was verified (Figure 5A, 5B). To determine whether RNF128 regulated β-catenin protein stability, the half-life of β-catenin was analyzed at 0, 0.5, 1 and 2 h in CHX (25 μg/ml) treated cells. Our data indicated that RNF128 knockdown increased the stability of the β-catenin protein (Figure 5C). Moreover, MG132 rescued RNF128-mediated β-catenin downregulation (Figure 5D). Consistently, we also performed co-IP to measure the level of β-catenin. As expected, overexpression of RNF128 significantly increased the ubiquitination of β-catenin and RNF128 knockdown significantly decreased it (Figure 5E).
Figure 5.
RNF128 directly interacts with and ubiquitinates β-catenin for degradation. (A and B) co-immunoprecipitation (co-IP) was performed with anti-RNF128 antibody (A) or β-catenin antibody (B) and anti-lgG antibody in DLD1 cells. (C) DLD1 cells were treated with cycloheximide at the indicated time and β-catenin levels were examined by Western blotting. (D) MG132 (10 μM) was added to RNF128-overexpression cells and overexpression control cells of DLD1 for 6 h. The relative changes of β-catenin were analyzed. (E) Ubiquitination of β-catenin was analyzed by the co-IP assay and immunoblotting with anti-ubiquitin antibody in DLD1. (F) Model of the suppressive role of RNF128 in Wnt/β-catenin/CCND1 pathway. The binding of Wnt to Frizzled (FZD) receptors activates DVL. Phosphorylated DVL stabilizes cytoplasmic β-catenin to promote nuclear translocation of β-catenin. The entry of β-catenin into the nucleus interacts with TCF/LEF and enhances transcription of downstream target (CCND1). RNF128 increases β-catenin ubiquitination in this process. *P<0.05, **P<0.01 and ***P<0.001. No significant difference (P>0.05, ns). Data represented as mean ± SD.
Taken together, RNF128 facilitated the ubiquitination process of β-catenin and consequently inhibited Wnt/β-catenin signaling (Figure 5F).
Effects of RNF128 on proliferation of colorectal cancer cells in vivo
We next conducted the in vivo experiment in tumor-bearing nude mice (Figure 4A). Without affecting the weight of the mice, RNF128 inhibited colorectal cancer cells proliferation in vivo (Figure 6B, 6C). We verified the expression of RNF128 after treatment of tumor tissues in mice (Figure 6D, 6E). In addition, the correlation between the expression of RNF128 and β-catenin was evaluated in tumor tissues in vivo (Figure 6D). The results showed that expression of β-catenin in mice was increased when RNF128 was decreased. We also showed that this phenomenon was related to the increased ubiquitination level of β-catenin when the expression of RNF128 was decreased (Figure 6D).
Figure 6.
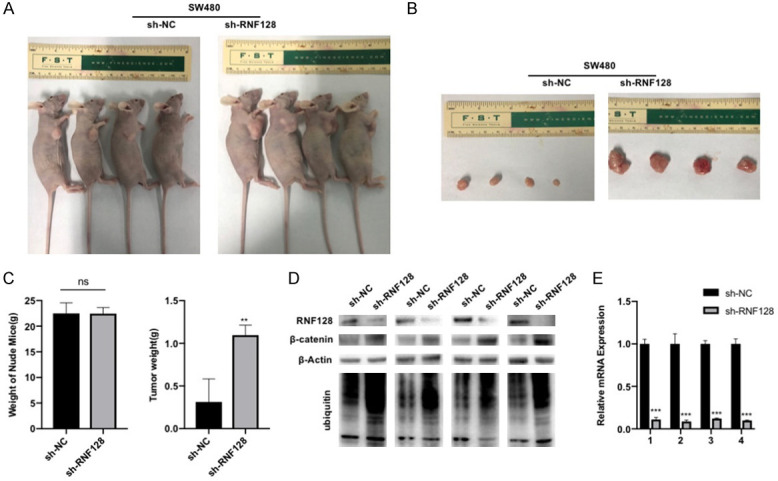
RNF128 inhibits colorectal cancer cell proliferation in vivo. (A) Construction and evaluation of implanted model of SW480-sh-RNF128 cells or their control cells in nude nice (n=4). (B and C) The volumes and weights of tumor. (D) β-catenin, RNF128 expression and the ubiquitin level of b-catenin in the tumorigenic tissue of the mouse were confirmed using Western blotting. (E) RNF128 expression in the tumorigenic tissues of the mouse was determined by qRT-PCR. **P<0.01. No significant difference (P>0.05, ns). Data represented as mean ± SD.
This finding reveals that RNF128 affects proliferation of colorectal cancer cells by promoting β-catenin ubiquitination in vivo.
Discussion
Although RNF128 was found to play a role in various cancers’ occurrence and progression, its potential role in CRC has not been examined. In this study, we focused our research on the effect of RNF128 in CRC and its mechanism. We first examined expression levels of RNF128 in CRC clinical samples. Our results demonstrated a substantially reduced level expression of RNF128 in CRC samples.
In vitro, we found that overexpression of RNF128 suppressed CRC cell proliferation, migration and invasion ability. In vitro and vivo, knocking down of RNF128 promoted proliferation, migration and invasion ability, respectively. We next demonstrated that phenotypic changes acted by RNF128 were associated with cell cycle regulation in colorectal cancer.
The Wnt/β-catenin pathway acts as a classical pathway in tumorigenesis. It has been reported that the Wnt/β-catenin pathway regulates a number of cancer types [33-35]. It is also involved in the development and progression of colorectal cancer [36]. Our analyses confirmed that RNF128 regulated ubiquitination of β-catenin to inhibit its targets transcription. Knockdown of RNF128 led to β-catenin deubiquitination and promoted β-catenin traveling to the nucleus to activate the Wnt signaling pathway. Interacted with TCF/LEF, β-catenin enhanced transcription of downstream targets. With the transcriptional activation of target gene CCND1, pro-tumorigenic environment was formed.
Our study proved that RNF128 regulates occurrence and progression of CRC through Wnt/β-catenin signaling directly. Moreover, other mechanisms are also likely to regulate CRC cell growth due to RNF128. Further investigation of the role of RNF128 in CRC tumorigenicity is needed.
Conclusions
Ultimately, the expression of RNF128 is gradually decreased in CRC tissues. Via the deubiquitination of Wnt/β-catenin signaling, low expression level of RNF128 promotes CRC cell growth. Thus, RNF128 may be considered as a therapeutic target for colorectal cancer.
Acknowledgements
This study was supported by national Natural Science Foundation of China (81902489).
Disclosure of conflict of interest
None.
Abbreviations
- RNF128
RING-finger protein 128
- Grail
gene related to anergy in lymphocytes
- CRC
colorectal cancer
- RT-qPCR
reverse transcription-real time quantitative polymerase chain reaction
- CCK-8
cell counting kit-8
- siRNA
small interfering RNAs
- si-NC
small interfering RNAs-negative control
- OD
optical density
- sh-RNA
short hairpin RNA
- CHX
cycloheximide
- co-IP
co-immunoprecipitation
- FZD
Frizzled
- IARC
The International Cancer Research Agency
References
- 1.Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 2.Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60:116–129. doi: 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sveen A, Kopetz S, Lothe R. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat Rev Clin Oncol. 2020;17:11–32. doi: 10.1038/s41571-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao T, Ndoja A. Regulation of gene expression by the ubiquitin-proteasome system. Semin Cell Dev Biol. 2012;23:523–529. doi: 10.1016/j.semcdb.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang F, Zheng C, Huang L, Lin C, Wang J. USP18 directly regulates Snail1 protein through ubiquitination pathway in colorectal cancer. Cancer Cell Int. 2020;20:346. doi: 10.1186/s12935-020-01442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrows JF, Johnston JA. Regulation of cellular responses by deubiquitinating enzymes: an update. Front Biosci (Landmark Ed) 2012;17:1184–1200. doi: 10.2741/3980. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Ma L, Wang B, Liu J, Wei W. E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 2017;36:683–702. doi: 10.1007/s10555-017-9703-z. [DOI] [PubMed] [Google Scholar]
- 8.Zheng N, Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 9.Varshavsky A. The ubiquitin system, autophagy, and regulated protein degradation. Annu Rev Biochem. 2017;86:123–128. doi: 10.1146/annurev-biochem-061516-044859. [DOI] [PubMed] [Google Scholar]
- 10.Lin Z, Li S, Feng C, Yang S, Wang H, Ma D, Zhang J, Gou M, Bu D, Zhang T, Kong X, Wang X, Sarig O, Ren Y, Dai L, Liu H, Zhang J, Li F, Hu Y, Padalon-Brauch G, Vodo D, Zhou F, Chen T, Deng H, Sprecher E, Yang Y, Tan X. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat Genet. 2016;48:1508–1516. doi: 10.1038/ng.3701. [DOI] [PubMed] [Google Scholar]
- 11.Hong L, Huang HC, Jiang ZF. Relationship between amyloid-beta and the ubiquitin-proteasome system in Alzheimer’s disease. Neurol Res. 2014;36:276–282. doi: 10.1179/1743132813Y.0000000288. [DOI] [PubMed] [Google Scholar]
- 12.Yu L, Dong L, Wang Y, Liu L, Long H, Li H, Li J, Yang X, Liu Z, Duan G, Dai X, Lin Z. Reversible regulation of SATB1 ubiquitination by USP47 and SMURF2 mediates colon cancer cell proliferation and tumor progression. Cancer Lett. 2019;448:40–51. doi: 10.1016/j.canlet.2019.01.039. [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Liang Y, Chai X, Chen S, Ye Z, Li R, Li X, Kong G, Li Y, Zhang X, Che Z, You Y, Ye S, Li L, Lin B, Huang J, Huang M, Zhang X, Qiu X, Zeng J. Ectodysplasin A receptor (EDAR) promotes colorectal cancer cell proliferation via regulation of the Wnt/β-catenin signaling pathway. Exp Cell Res. 2020;395:112170. doi: 10.1016/j.yexcr.2020.112170. [DOI] [PubMed] [Google Scholar]
- 14.Pérez-Plasencia C, López-Urrutia E, García-Castillo V, Trujano-Camacho S, López-Camarillo C, Campos-Parra AD. Interplay between autophagy and Wnt/β-catenin signaling in cancer: therapeutic potential through drug repositioning. Front Oncol. 2020;10:1037. doi: 10.3389/fonc.2020.01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borgal L, Rinschen MM, Dafinger C, Hoff S, Reinert MJ, Lamkemeyer T, Lienkamp SS, Benzing T, Schermer B. Casein kinase 1 α phosphorylates the Wnt regulator Jade-1 and modulates its activity. J Biol Chem. 2014;289:26344–26356. doi: 10.1074/jbc.M114.562165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 17.Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626–642. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horn EJ, Albor A, Liu Y, El-Hizawi S, Vanderbeek GE, Babcock M, Bowden GT, Hennings H, Lozano G, Weinberg WC, Kulesz-Martin M. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25:157–167. doi: 10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- 20.Migita K, Matsumoto S, Wakatsuki K, Kunishige T, Nakade H, Miyao S, Sho M. RNF126 as a marker of prognosis and proliferation of gastric cancer. Anticancer Res. 2020;40:1367–1374. doi: 10.21873/anticanres.14078. [DOI] [PubMed] [Google Scholar]
- 21.Kostianovsky AM, Maier LM, Baecher-Allan C, Anderson AC, Anderson DE. Up-regulation of gene related to anergy in lymphocytes is associated with notch-mediated human T cell suppression. J Immunol. 2007;178:6158–6163. doi: 10.4049/jimmunol.178.10.6158. [DOI] [PubMed] [Google Scholar]
- 22.Lineberry NB, Su LL, Lin JT, Coffey GP, Seroogy CM, Fathman CG. Cutting edge: the transmembrane E3 ligase GRAIL ubiquitinates the costimulatory molecule CD40 ligand during the induction of T cell anergy. J Immunol. 2008;181:1622–1626. doi: 10.4049/jimmunol.181.3.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lineberry N, Su L, Soares L, Fathman CG. The single subunit transmembrane E3 ligase gene related to anergy in lymphocytes (GRAIL) captures and then ubiquitinates transmembrane proteins across the cell membrane. J Biol Chem. 2008;283:28497–28505. doi: 10.1074/jbc.M805092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor JJ, Krawczyk CM, Mohrs M, Pearce EJ. Th2 cell hyporesponsiveness during chronic murine schistosomiasis is cell intrinsic and linked to GRAIL expression. J Clin Invest. 2009;119:1019–1028. doi: 10.1172/JCI36534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nurieva RI, Zheng S, Jin W, Chung Y, Zhang Y, Martinez GJ, Reynolds JM, Wang SL, Lin X, Sun SC, Lozano G, Dong C. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity. 2010;32:670–680. doi: 10.1016/j.immuni.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YC, Chan JY, Chiu YL, Liu ST, Lozano G, Wang SL, Ho CL, Huang SM. Grail as a molecular determinant for the functions of the tumor suppressor p53 in tumorigenesis. Cell Death Differ. 2013;20:732–743. doi: 10.1038/cdd.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray D, Ray P, Ferrer-Torres D, Wang Z, Nancarrow D, Yoon HW, San Martinho M, Hinton T, Owens S, Thomas D, Jiang H, Lawrence TS, Lin J, Lagisetty K, Chang AC, Beer DG. Isoforms of RNF128 regulate the stability of mutant P53 in Barrett’s esophageal cells. Gastroenterology. 2020;158:583–597. e581. doi: 10.1053/j.gastro.2019.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei CY, Zhu MX, Yang YW, Zhang PF, Yang X, Peng R, Gao C, Lu JC, Wang L, Deng XY, Lu NH, Qi FZ, Gu JY. Downregulation of RNF128 activates Wnt/β-catenin signaling to induce cellular EMT and stemness via CD44 and CTTN ubiquitination in melanoma. J Hematol Oncol. 2019;12:21. doi: 10.1186/s13045-019-0711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YY, Wang CT, Huang SK, Wu WJ, Huang CN, Li CC, Chan TC, Liang PI, Hsing CH, Li CF. Downregulation of RNF128 predicts progression and poor prognosis in patients with urothelial carcinoma of the upper tract and urinary bladder. J Cancer. 2016;7:2187–2196. doi: 10.7150/jca.16798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haymaker C, Yang Y, Wang J, Zou Q, Sahoo A, Alekseev A, Singh D, Ritthipichai K, Hailemichael Y, Hoang ON, Qin H, Schluns KS, Wang T, Overwijk WW, Sun SC, Bernatchez C, Kwak LW, Neelapu SS, Nurieva R. Absence of Grail promotes CD8(+) T cell anti-tumour activity. Nat Commun. 2017;8:239. doi: 10.1038/s41467-017-00252-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Wang Y, Yang J, Zhang W, Meng K, Sun Y, Li Y, He QY. RNF128 promotes invasion and metastasis via the EGFR/MAPK/MMP-2 pathway in esophageal squamous cell carcinoma. Cancers (Basel) 2019;11:840. doi: 10.3390/cancers11060840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Li B, Zhou L, Yu S, Su Z, Song J, Sun Q, Sha O, Wang X, Jiang W, Willert K, Wei L, Carson DA, Lu D. Prodigiosin inhibits Wnt/β-catenin signaling and exerts anticancer activity in breast cancer cells. Proc Natl Acad Sci U S A. 2016;113:13150–13155. doi: 10.1073/pnas.1616336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilchez V, Turcios L, Marti F, Gedaly R. Targeting Wnt/β-catenin pathway in hepatocellular carcinoma treatment. World J Gastroenterol. 2016;22:823–832. doi: 10.3748/wjg.v22.i2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473–481. doi: 10.1016/j.biopha.2018.11.082. [DOI] [PubMed] [Google Scholar]