Abstract
Osteosarcoma (OS) is a primary and highly malignant mesenchymal tissue tumor. The specific pathological mechanism underlying disease initiation or progression remains unclear. Circular RNAs (circRNAs) are a type of covalently circular RNA with a head‐to‐tail junction site. In this study, we aimed to investigate the sponging mechanism between circRNAs and microRNAs (miRNAs) in OS. Based on the inhibited effect of miR‐16‐5p reported on OS, circUSP34 was analyzed as a sponge of miR‐16‐5p via Starbase. We found that circUSP34 promoted the proliferation, migration, and invasion of OS in vitro and in vivo. circUSP34 increased but miR‐16‐5p decreased in OS by qRT‐PCR. Function assays showed that the malignancy of OS cells, including proliferation, migration, and invasion, was inhibited after knocking out circUSP34. Western blotting results showed that the expression level of vimentin and Ki‐67 decreased. Similarly, miR‐16‐5p mimic compromised the proliferation, migration, and invasion of OS cells. FISH assay results indicated that circUSP34 and miR‐16‐5p were colocalized in the cytoplasm. The sponging mechanism of circUSP34 and miR‐16‐5p was verified by dual‐luciferase reporter assay, RNA immunoprecipitation (RIP), and RNA pull down assays. Interestingly, the miR‐16‐5p inhibitor partly reversed the inhibitory effect of sh‐circUSP34 on the malignancy of OS cells. Further, mice tumors for IHC indicated that vimentin, N‐cadherin, and Ki‐67 protein expression decreased, but E‐cadherin protein expression increased. Collectively, circUSP34 promoted OS malignancy, including proliferation, migration, and invasion, by sponging miR‐16‐5p. It can serve as a potential therapeutic target and biomarker.
Keywords: biomarker, circRNA, miRNA, noncoding RNA, osteosarcoma
This article makes a significant contribution to our understanding of the regulation mechanisms of osteosarcoma, also considering a novel biomarker and potential therapeutic target, which would help the progression of therapeutic protocols.
Abbreviations
- circRNA
circular RNA
- miRNA
microRNA
- FBS
fetal bovine serum
- qRT‐PCR
quantitative Real‐time PCR
- cDNA
complementary DNA
- BCA
bicinchoninic acid
- EMT
epithelial‐mesenchymal transition
- PVDF
polyvinylidene difluoride membranes
- FISH
fluorescence in situ hybridization
- RBP
RNA binding protein
- RIP
RNA immonoprecipate
- OD
optical density
1. INTRODUCTION
OS is a primary mesenchymal malignant tumor that commonly occurs in adolescents and young adults. 1 , 2 , 3 , 4 Under estimation, the OS incidence in teenagers is approximately 5.6/1 000 000. 5 Long tubular bones, such as tibia, fibula, and femur, suffer from OS lesions most commonly. 6 Studies have shown that the combination of surgery and chemotherapy significantly improves patients' prognosis. 7 However, due to its high malignancy and frequent distant metastases, most patients with advanced‐stage disease have a terrible prognosis. 8 And more notably, it is still a tough problem to tackle lung metastasis, which remarkably declines patients’ survival. Obviously, the positive treatment of metastasis can improve the OS prognosis significantly. 5 The initial cause of OS remains unclear, hindering its diagnosis and early treatment. Therefore, investigating its pathophysiologic mechanisms and exploring novel therapeutic targets are urgently needed.
CircRNA is a kind of head‐to‐tail closed circular RNA without 5′‐ and 3′‐terminals. It is formed by back‐splicing pre‐mRNA and is usually over 200 nucleotides long. 9 , 10 CircRNA can regulate a variety of physiological and pathophysiological activities by interacting with transcript factors, miRNA, and RNA‐binding protein (RBP). As the covalent combination, circRNA can be detected in body fluid. Therefore, it can be used as a diagnostic and prognostic biomarker. 11 Increasingly, studies have reported that abnormally expressed circRNAs are associated with tumorigenesis, proliferation, metastasis, and invasion. 12 , 13 They are often aberrantly expressed in a variety of tumors, such as non–small cell lung cancer, colorectal cancer, and hepatocellular cancer. 14 , 15 , 16 Some circRNAs have oncogenic function, whereas others have anti‐oncogenic function. 17 It is well known that circRNA sponging miRNA is a classic interaction in tumors. For example, circ_0000285 promotes the proliferation, migration, and invasion but inhibits the apoptosis of osteosarcoma by sponging miR‐409‐3p. 18 Upregulated circ_0028171 in cells competes endogenously with miR‐218‐5p to promote osteosarcoma progression. 19 Based on above literatures, we speculate that circUSP34 plays an oncogenic role in OS by sponging miR‐16‐5p. It was reported that miR‐16‐5p inhibited OS malignancy. Herein, circUSP34 was predicted as the sponge of miR‐16‐5p by Starbase. Previously, circUSP34 was not involved in any tumors. The regulatory mechanisms of circUSP34 in OS by sponging miR‐16‐5p need to be further explored.
2. MATERIALS AND METHODS
2.1. Cell culture
Normal human umbilical vein endothelial cells (HUVEC) and OS cells (KHOS and 143B cell lines) were purchased from the American Type Culture Collection (ATCC). All of them were stored in CELLSAVING solution (New Cell & Molecular Biotech) at −80°C. KHOS cells were cultured in RPMI‐1640 (Gibco), and 143B cells were cultured in DMEM (Gibco); both were supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin and streptomycin (Gibco). Both cell lines were cultured in a humidified incubator with 5% CO2 at 37°C.
2.2. RNase R treatment
Total RNA extracted from 143B cells was treated with RNase R (Epicenter) under 3 U/μg. Then, the mix was incubated at 37℃ for 20 minutes. The product was examined by quantitative real time PCR (qRT‐PCR) and nucleic acid electrophoresis.
2.3. RNA extraction and qRT‐PCR
Total RNA was isolated from OS cells according to the manufacturer's protocol using RNeasy Plus Mini (Qiagen, Germany) and dissolved in 15 μL of RNase‐free water. The concentration and purification of RNA were assessed using a NanoPhotometer Pearl (IMPLEN) by measuring the A260/280 absorbance. Complementary DNA (cDNA) was synthesized using PrimeScript RT Master Mix (TaKaRa Biotechnology). CircUSP34 and miR‐16‐5p were quantified using iTaq™ Universal SYBR® Green (Bio‐Rad) with GAPDH and U6 as internal references, respectively. The sequences of primers are listed in Table 1.
TABLE 1.
Sequences of primers
| 5′→3′ | |
|---|---|
| circUSP34 | |
| Forward | GCCAAAGAAAGAAAAACTAAAGTGG |
| Reverse | AGGTCTCAGCCTCTGAATCAC |
| USP34 | |
| Forward | CGACTTAGATGCCTTGGCAAGAC |
| Reverse | GGAGTCCTGTAAGCCCATCATC |
| miR‐16‐5p | |
| Forward | TAGCAGCACGTAAATATTGGCG |
| Reverse | TGCGTGTCGTGGAGTC |
| GAPDH | |
| Forward | GTCTCCTCTGACTTCAACAGCG |
| Reverse | ACCACCCTGTTGCTGTAGCCAA |
| U6 | |
| Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT |
2.4. Lentiviruses preparation and oligonucleotide transfection
KHOS and 143B cells were cultured in six‐well plates (8 × 105 cells per well) to 70%‐80% confluence before transfection. miR‐16‐5p mimic and negative control were synthesized by GenePharma. Lentivirus, the sh‐circUSP34 vector #1 and #2, was constructed targeting the specific head‐to‐tail junction site purchased from Hanbio Biotechnology Company. Sequences of mimic, inhibitor, and vectors are listed in Table 2. Lipofectamine 3000 (Invitrogen) and Polybrene (Hanbio Biotechnology Company) were applied to facilitate miR‐16‐5p mimic and sh‐circUSP34 complex formation, respectively.
TABLE 2.
Sequences of sh‐RNA vector and miRNA mimic or inhibitor
| sh‐circUSP34 #1 | Sense | AAAGAAAAACUAAAGUGGUTT |
| Antisense | ACCACUUUAGUUUUUCUUUTT | |
| sh‐circUSP34 #2 | Sense | CUAAAGUGGUUUUUAGAUCTT |
| Antisense | GAUCUAAAAACCACUUUAGTT | |
| miR‐16‐5p mimic | Sense | UAGCAGCACGUAAAUAUUGGCG |
| Antisense | CCAAUAUUUACGUGCUGCUAUU | |
| miR‐16‐5p mimic NC | Sense | UUCUCCGAACGUGUCACGUTT |
| Antisense | ACGUGACACGUUCGGAGAATT | |
| miR‐16‐5p inhibitor | CGCCAAUAUUUACGUGCUGCUA | |
| miR‐16‐5p inhibitor NC | CAGUACUUUUGUGUAUACAA |
2.5. CCK‐8 assay
Three thousand cells per well were seeded in a 96‐well plate in triplicate. Subsequently, 10 μL of CCK‐8 reagent (Dojindo Crop) were added to 100 μL of culture medium for 24, 48, 72, and 96 hours, according to the manufacturer's instructions. The cells were incubated for 2 hours in incubator, and the optical density (OD) was measured at 450 nm using a microplate reader (Bio‐Rad).
2.6. EdU assay
The proliferation ability of cells was evaluated by EdU assay using a BeyoClick EdU Cell Proliferation Kit with Alexa Fluor 555 (Beyotime Biotechnology, China) following the manufacturer's instructions. Approximately, 5 × 104 OS cells per well were seeded in 12‐well plates. Both cell lines were incubated with 10 μmol/L EdU solution for 2 hours and were fixed using 4% paraformaldehyde. Next, EdU was examined using Click Additive Solution, and cellular nuclei were stained with Hoechst 33342. EdU‐treated cells in five different areas were photographed using a Leica DMi1 microscope.
2.7. Wound‐healing assay
KHOS and 143B cells transfected with sh‐circUSP34 and sh‐NC were seeded in six‐well plates at 8 × 105 cells per well. When the cell confluency reached ~80%, a wound line was scratched using a P‐200 pipette tip. Next, images were taken using a microscope (Leica) at 0 hour and after 24 hours. The distance between the injury lines was measured and analyzed using Image J.
2.8. Transwell assay
Briefly, 200 μL serum‐free medium containing 5 × 105 cells was added to the upper chamber (Corning) in triplicate. The bottom chamber was filled with 600 μL of medium having 20% FBS. All chambers were incubated at 37°C for 24 hours. The next day, the chambers were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Cells in the upper membrane of the chamber were slightly wiped using a swab. Finally, the migrating cells in five different areas were counted under a microscope.
2.9. Dual luciferase reporter assay
Luciferase reporter vector, named circUSP34‐WT and circUSP34‐MUT, was synthesized by GenePharma. Then, miR‐16‐5p mimic and miR‐16‐5p mimic NC were transfected into KHOS cells. The relative luciferase activity was examined by Dual Luciferase Assay Kit (Beyotime).
2.10. RNA immunoprecipitation (RIP)
Using the Magna RIP™ RNA‐binding protein immunoprecipitation kit (Millipore,USA), KHOS cells transfected by miR‐16‐5p mimic and miR‐16‐5p mimic NC were lysed. The magnetic beads combined with anti‐AGO2 and anti‐IgG were incubated with cell lysate overnight at 4°C. Finally, total RNA was isolated and purified by TRIzol Reagent (Invitrogen), and qRT‐PCR analysis and agarose gel electrophoresis were performed.
2.11. RNA pull down
Biotinylated miR‐16‐5p was synthesized by GenePharma. Briefly, KHOS cells were collected and then lysed. M280 streptavidin dynabeads (Invitrogen) were incubated with the probe. Then the probe‐dynabeads complex was incubated with cell lysate overnight at 4℃. RNA conjugated with dynabeads was eluted and purified using TRIzol (Invitrogen) for further qRT‐PCR analysis.
2.12. Protein extraction and Western blot
Cells were lysed using radioimmunoprecipitation assay lysis buffer (Beyotime). Protein concentration was quantified using the BCA Protein Assay kit (Solarbio), and equal proteins collected from different types of cell lysates were loaded onto 10% SDS‐PAGE gels (Invitrogen) and then transferred onto polyvinylidene difluoride membranes (PVDF). All membranes were blocked with a final concentration of 1× quick‐blocking liquid at room temperature and then incubated with primary antibodies anti‐vimentin (1:1000, Proteintech), anti‐E‐cadherin (1:1000, Proteintech), anti‐N‐cadherin (1:1000, Proteintech), anti‐Ki‐67 (1:1000, Proteintech), and anti‐GAPDH (1:1000, OriGene) at 4°C overnight. Membranes were then washed in TBS‐T and incubated with secondary antibodies for 1 hour at 37°C. Proteins were visualized using the Image Lab Software (Bio‐Rad).
2.13. Fluorescence in situ hybridization (FISH) assay
FAM‐labeled circUSP34 probes and Cy3‐labeled miR‐16‐5p probes were designed and synthesized by GenePharma. Hybridization was performed overnight at 4°C with circUSP34 and miR‐16‐5p probes according to the manufacturer's instructions. Images were acquired on a Nikon Eclipse Ti scanning confocal microscope.
2.14. Immunohistochemistry
Tissue samples were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned. The tissue sections were incubated with anti‐vimentin (1:5000), anti‐N‐cadherin (1:1000), anti‐E‐cadherin (1:1000), and anti‐Ki‐67(1:8000) primary antibodies at 4°C overnight and then incubated with an HRP‐conjugated secondary antibody. The results were photographed under a microscope.
2.15. Animal experimentation
Four‐week‐old female BALB/c nude mice were prepared for the subcutaneous tumor experiments. The stably transfected 143B cells with a knockdown for circUSP34 or negative control were injected subcutaneously (5 × 106, 150 μL). After 21 days, tumors removed from euthanized mice were subjected to weight comparison, IHC staining, and Western blot assay. The animal experiments were approved by the Ethics Committee of Peking University People's Hospital.
2.16. Statistical analyses
SPSS software (version 22.0) and GraphPad Prism 7 were used for statistical analyses. The data were analyzed via Student's t‐test or one‐way ANOVA, and the results are presented as the mean ± SD. No significance is indicated by ns. Significant data are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
3. RESULTS
3.1. The characteristics and expression level of circUSP34 in OS cells and tissues
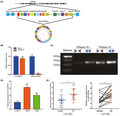
To better understand the characteristics of circUSP34, its location on the chromosome was identified. Data from circBank (http://www.circbank.cn/index.html) and UCSC (http://genome.ucsc.edu/) showed that circUSP34, which is present on chr.2p14, exhibited a circular structure via post‐transcriptional head‐to‐tail splicing (Figure 1A). Reportedly, circRNAs, including circUSP34 which is relatively stable, are resistant to RNase R digestion. 20 Subsequently, divergent and convergent primers were designed to verify the stability of circUSP34 with and without RNase R treatment using qRT‐PCR. The result showed that circUSP34 was not digested, but USP34 mRNA was digested (Figure 1B). Agarose gel electrophoresis was performed to examine the product of qRT‐PCR, and we confirmed that circUSP34 was resistant to RNase R digestion (Figure 1C), as opposed to USP34 mRNA. qRT‐PCR results revealed that circUSP34 had upregulated expression in both 143B and KHOS cells and tissues (Figure 1D,E). Therefore, we verified the circular structure and stability of circUSP34 and found that it was upregulated in OS.
FIGURE 1.
The characteristics and expression level of circUSP34 in OS cells and tissues. A, The head‐to‐tail junction site was identified via UCSC and circBank. B and C, The stability and existence of circUSP34 were detected by qRT‐PCR and nucleic acid electrophoresis. D and E, CircUSP34 expression level in OS cells and tissues was confirmed by qRT‐PCR. Data are shown as mean ± SD. ns, no significance; *P < 0.05, **P < 0 .01
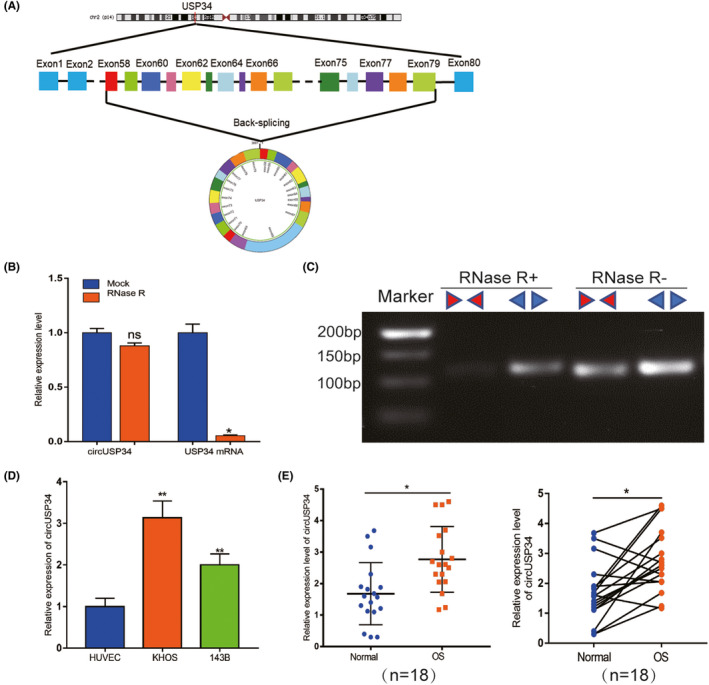
3.2. CircUSP34 promotes OS cells’ malignancy, especially proliferation and migration
143B and KHOS cells were transfected with sh‐circUSP34 #1 and #2 to knock down circUSP34. qRT‐PCR results showed that the expression level of circUSP34 significantly decreased in both cell lines (Figure 2A and Figure S1A). To determine the role of circUSP34 in proliferation viability in vitro, CCK8 and EdU assays were performed. All results revealed that interfering circUSP34 compromised the proliferation ability of 143B and KHOS cells (Figure 2B,C and Figure S1B,C). Moreover, the colony formation assay also indicated that interfering circUSP34 suppressed the proliferation of OS cells, which was consistent with the above results (Figure 2D and Figure S1D). Overall, these results suggested that circUSP34 can promote the proliferation of OS cells.
FIGURE 2.
CircUSP34 promotes OS cells' malignancy, especially proliferation and migration. A, Scenes of both KHOS and 143B cells transfected by sh‐circUSP34#1 and sh‐NC vector were photographed, and knockout efficacy was confirmed by qRT‐PCR. B‐D, CCK8, EdU, and colony formation assay results showed that proliferation was inhibited in KHOS and 143B cells transfected by sh‐circUSP34#1 or sh‐NC. E and F, OS cells’ migration and invasion were evaluated by wound‐healing and transwell assays. G, The expression level of Vimentin and Ki‐67 protein was assessed by western blot in KHOS cell. Data are shown as mean ± SD. *P < 0.05, **P < 0.01
Transwell migration as well as invasion and wound‐healing assays were performed to further explore the effect of circUSP34 on the migration and invasion of OS cells. A wound‐healing assay showed that migration capacity was inhibited in 143B and KHOS cells transfected with sh‐circUSP34 compared with that in the NC group (Figure 2E and Figure S1E). Transwell migration and invasion assays demonstrated that both were inhibited (Figure 2F). Therefore, these results demonstrated that circUSP34 can advance both migration and invasion ability.
Furthermore, western blot results indicated that vimentin expression decreased after knocking out circUSP34 in KHOS cell. Additionally, Ki‐67 expression level was also downregulated in KHOS cell(Figure 2G and Figure S1F). Taken together, our data indicated that circUSP34 can promote the progression of OS in vitro.
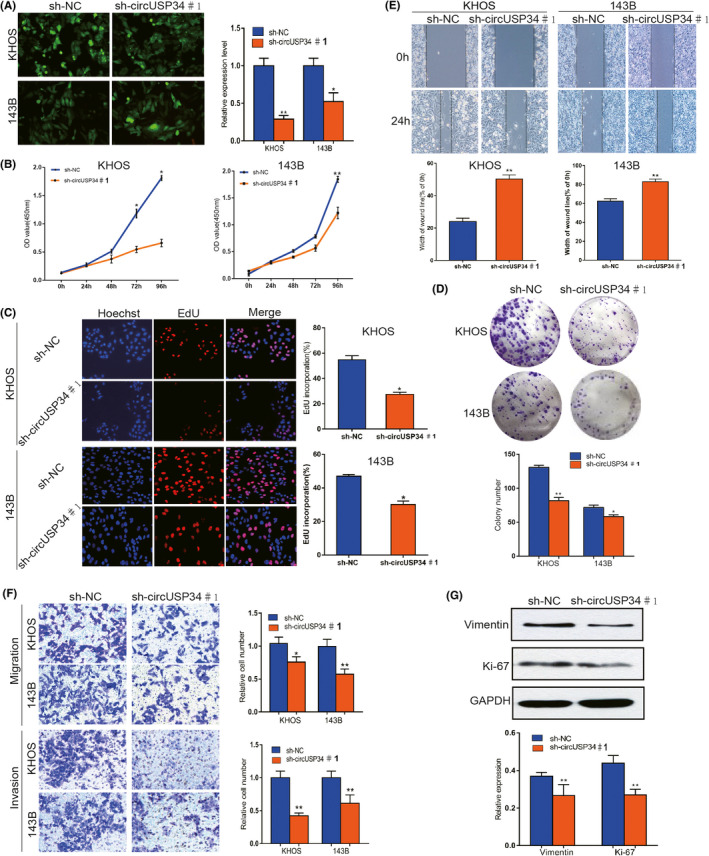
3.3. CircUSP34 acts as a sponge to regulate miR‐16‐5p
Given the interaction of circUSP34 and miR‐16‐5p, the target site was predicted using Starbase (http://starbase.sysu.edu.cn/starbase2) as shown in Figure 3A. Then, we performed FISH assay to colocalize circUSP34 (green) and miR‐16‐5p (red) in OS cells, both colocalized in the cytoplasm (Figure 3B). The relative expression level of miR‐16‐5p was detected by qRT‐PCR in OS cells transfected with sh‐circUSP34 and obviously increased (Figure 3C). Further, a dual‐luciferase reporter assay was performed in KHOS cells. The result showed that the luciferase activity of the circUSP34‐WT group with the miR‐16‐5p mimic group decreased compared with the miR‐16‐5p mimic NC group (Figure 3D). Subsequently, RIP assay by anti‐AGO2 was performed in KHOS cells. circUSP34 and miR‐16‐5p complex binding to AGO2 was pulled down and detected by qRT‐PCR. Results of qRT‐PCR and nucleus acid electrophoresis revealed that circUSP34 can sponge miR‐16‐5p (Figure 3E). The enrichment of circUSP34 pulled down by biotinylated miR‐16‐5p was measured by qRT‐PCR. It showed that miR‐16‐5p was a target of circUSP34 (Figure 3F). Given these results, circUSP34 acts as a sponge of miR‐16‐5p.
FIGURE 3.
CircUSP34 acts as a sponge to regulate miR‐16‐5p. A, The potential target site between circUSP34 and miR‐16‐5p was predicted by Starbase. B, The cellular location of circUSP34 (green) and miR‐16‐5p (red) in cells was observed by performing FISH (magnification: 400×, scale bar: 100 μm). C, miR‐16‐5p expression level was confirmed by qRT‐PCR after transfecting sh‐circUSP34. D, The relative luciferase activity was detected in the KHOS cells after cotransfecting with circUSP34‐WT or circUSP34‐MUT and mimics or NC, respectively. E, RNA immunoprecipitation (RIP) assay was performed in KHOS cells after transfecting with miR‐16‐5p mimic or NC respectively, with qRT‐PCR and nucleic acid electrophoresis. F, The enrichment of circTLK1 was measured by qRT‐PCR in RNA pull down assay. Data are shown as mean ± SD. ns, no significance;*P < 0.05, **P < 0.01
3.4. miR‐16‐5p inhibits the malignancy of OS in vitro
We next explored miR‐16‐5p expression in OS cells as well as tissues. qRT‐PCR data showed that miR‐16‐5p expression was significantly downregulated in OS cells compared with HUVEC and in OS tissues compared with non‐cancerous tissues(Figure 4A). To confirm the potential anti‐tumorous effect of miR‐16‐5p, gain‐of‐function assays were performed. CCK‐8 assay results showed that miR‐16‐5p mimic suppressed the proliferation of KHOS and 143B cells (Figure 4B). For additional verification, the EdU assay was used, and the result showed increased miR‐16‐5p inhibited OS cells in vitro (Figure 4C). Similarly, the colony formation assay showed that the proliferation ability was inhibited in vitro (Figure 4F). miR‐16‐5p mimic comprised the migration ability of KHOS and 143B cells, as confirmed by the wound‐healing assay (Figure 4D). Transwell assay results revealed that migration and invasion abilities were inhibited by miR‐16‐5p mimic (Figure 4E). Our results showed that miR‐16‐5p plays a suppressive role in malignant progression in vitro.
FIGURE 4.
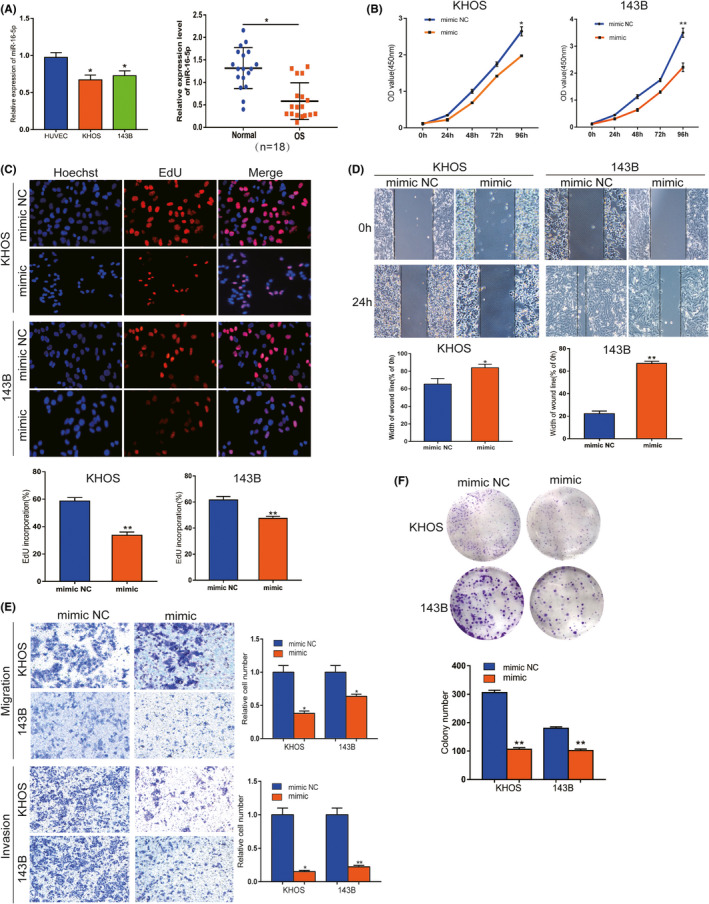
miR‐16‐5p inhibits malignancy of OS in vitro. A, The expression level of miR‐16‐5p was confirmed by qRT‐PCR in OS cells and tissues. B and C, CCK8 and EdU assays showed that proliferation viability was inhibited by mimic in both OS cells. D and E, Wound‐healing and transwell assays indicated that migration and invasion abilities were suppressed in vitro. F, Colony formation results showed that proliferation ability was suppressed. Data are shown as mean ± SD. *P < 0.05, **P < 0.01
3.5. miR‐16‐5p inhibitor partly reversed the antitumor function of sh‐circUSP34
To clarify the specific relationship between miR‐16‐5p and circUSP34, OS cells treated with sh‐circUSP34 were transfected with miR‐16‐5p inhibitor. The efficacy of transfection was quantified by qRT‐PCR. The result showed that the expression level of miR‐16‐5p decreased but that of circUSP34 increased in OS cells (Figure 5A). EdU and CCK8 results indicated that miR‐16‐5p inhibitor rescued the effect ofcircUSP34 on OS cell viability (Figure 5B,C). Herein, miR‐16‐5p inhibitor promoted the oncogenic effect of circUSP34 on proliferation. Wound‐healing assay results indicated that the function of circUSP34 after knocking out was rescued by the miR‐16‐5p inhibitor (Figure 5D). Collectively, these results showed that circUSP34 served as a sponge of miR‐16‐5p.
FIGURE 5.
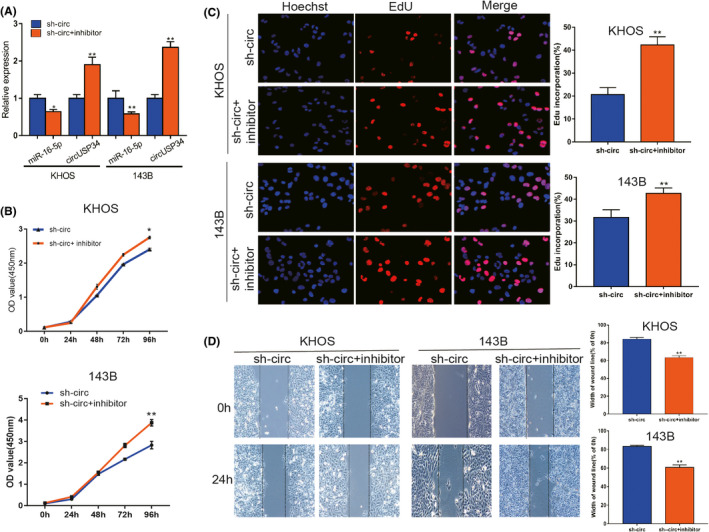
miR‐16‐5p inhibitor reversed the antitumor function of sh‐circUSP34. A, The expression level of miR‐16‐5p and circUSP34 with inhibitor and sh‐circUSP34 was confirmed by qRT‐PCR, respectively. B and C, CCK8 and EdU assays showed that the oncogenic function of circUSP34 was rescued by the miR‐16‐5p inhibitor . D, The effect of the miR‐16‐5p inhibitor on migration in OS cells transfected by sh‐circUSP34 was verified by wound‐healing assay. Data are shown as mean ± SD. *P < 0.05, **P < 0.01
3.6. CircUSP34 promotes proliferation and metastasis of OS cells in vivo
To confirm the oncogenic function of circUSP34 in vivo, we constructed five BALB/c nude mice models of every group. Every week, the tumor volume of every mouse was measured, and results showed that tumor weight and volume in the sh‐circUSP34 group were significantly inhibited compared with the sh‐NC group (Figure 6A‐C). We performed qRT‐PCR to detect miR‐16‐5p expression level in tumor tissues. The result showed an increased expression level in the sh‐circUSP34 group (Figure 6D). Subsequently, HE staining of sections indicated that these results were consistent with the pathological characteristics of OS (Figure 6E). Additionally, immunohistochemistry results showed that vimentin, N‐cadherin, and Ki‐67 protein expression decreased, but E‐cadherin protein expression increased (Figure 6F). Simultaneously, Western blot results also indicated that OS malignancy was markedly compromised. Vimentin, N‐cadherin, and Ki‐67 expression levels decreased significantly; however, E‐cadherin expression level increased (Figure 6G). These results suggest that circUSP34 acts as an oncogene to promote OS progression.
FIGURE 6.
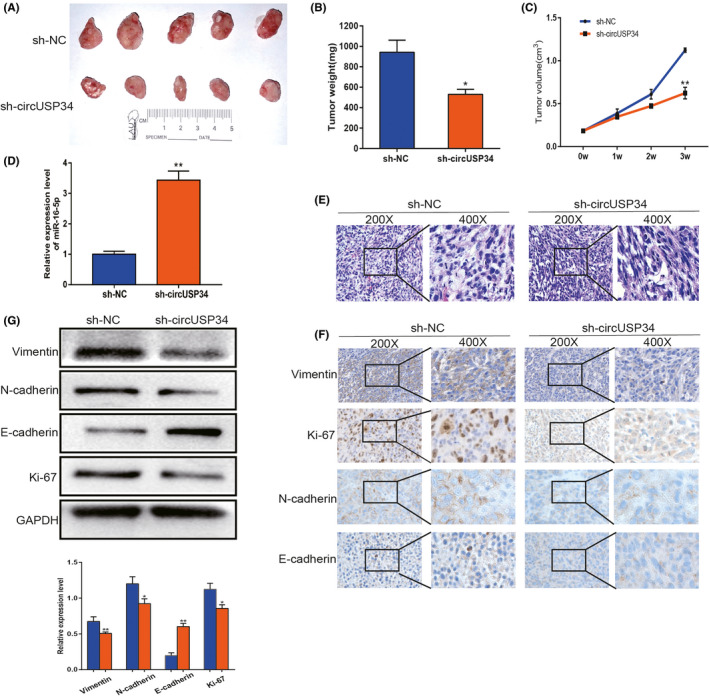
CircUSP34 promotes proliferation and metastasis of OS in vivo. A, Subcutaneous tumors of circUSP34‐knockout and NC. B, Tumor weight was measured in the sh‐circUSP34 and sh‐NC groups which were analyzed. C, Tumor volume was measured every week. D, Relative expression level of miR‐16‐5p was quantified in tumor tissues by qRT‐PCR. E, Representative images of HE staining were exhibited from the sh‐circUSP34 group and sh‐NC group. F, The expression of Vimentin, Ki‐67, N‐cadherin, and E‐cadherin was evaluated by performing IHC. G, Western blot assay showed the protein expression level of Vimentin, Ki‐67, N‐cadherin, and E‐cadherin from tumor tissue. Data are shown as mean ± SD. *P < 0.05, **P < 0.01
4. DISCUSSION
OS is a primary malignant tumor derived from mesenchymal tissue, but its high‐grade malignancy, especially distant metastasis, cannot be neglected. The combination of surgery and chemotherapy has markedly improved patients’ prognosis. 21 However, local invasion, metastasis, and drug resistance phenotypes still need to be resolved. 22 Exploring an effective therapeutic target is an urgent and critical issue. Increasingly, studies have shown that circRNAs are aberrantly expressed in various tumors including OS. 23 , 24 For instance, high expression of circRNA can promote the invasion ability of gastrointestinal cancer, lung cancer, and liver cancer. 25 , 26 , 27 The above prompted us to further investigate the potential role of circUSP34 in OS. Thus, we identified circUSP34 using Starbase as an upstream “sponge” of miR‐16‐5p, which was reported to inhibit OS. 28 In this present study, the expression level of circUSP34 was confirmed by qRT‐PCR, which was elevated in OS cells compared with HUVEC. Epithelial‐mesenchymal transition (EMT) is considered to play a significant role in malignancy progression in OS. To confirm the effect of circUSP34 on EMT‐related proteins in the context of OS, including vimentin, E‐cadherin, and N‐cadherin, their expression was quantified via Western blot. Loss‐of‐function assays to verify the oncogene circUSP34 in KHOS and 143B cells showed that knocking out circUSP34 compromised malignant behaviors of OS cells in vitro. circUSP34 after being intefered inhibited tumor growth in vivo by establishing subcutaneous tumors in mice.
It is widely accepted that miRNA function is modulated by circRNA sponging in the context of pathological and pathophysiological activities. 29 , 30 Circ_NOTCH3 competes with miR‐205‐5p to promote the progression of basal‐like breast carcinoma. 31 Circular RNA MYLK, a competing endogenous RNA, promotes bladder cancer progression. 32 Moreover, miR‐622 is sponged by circ_0119872 to promote uveal melanoma development. 33 It is worth noting that not every circRNA can sponge miRNAs to modulate their biological functions, as their activity has several requirements, such as a consistent subcellular location. Although miR‐16‐5p has been previously reported to impede tumor growth, 28 in our study we found that it has an inhibitory function. The low expression level was quantified by performing qRT‐PCR, but it significantly increased after circUSP34 knockdown. This result further supported the hypothesis advocating an interaction between circUSP34 and miR‐16‐5p. Similarly, gain‐of‐function assay results showed that overexpression of miR‐16‐5p impeded OS development. Therefore, it is probable that circUSP34 can sponge miR‐16‐5p. To better understand this activity, a rescue experiment was performed. Importantly, Zhang et al. 34 reported that Smad3 acts as a target of miR‐16‐5p in chordoma cells. Therefore, we hypothesize that miR‐16‐5p inhibits OS malignancy by targeting Smad3. Further studies are required to explore the underlying mechanisms. Our results showed that circUSP34 promotes OS proliferation, migration, and invasion by sponging miR‐16‐5p. However, the expression of circUSP34 was negatively correlated with tumor size in vivo.
In summary, our findings revealed that the highly regulated circUSP34 plays a role in oncogenes and underlying biomarkers in OS. Furthermore, circUSP34 may be a potential therapeutic target for OS. Ours is the first study to demonstrate that circUSP34 promotes OS proliferation, migration, and invasion by sponging miR‐16‐5p. However, our study requires further exploration of the mechanism of circUSP34 and its clinicopathological correlation with patients with OS.
DISCLOSURE
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
ACKNOWLEDGEMENTS
Not applicable.
Lou J, Zhang H, Xu J, et al. circUSP34 accelerates osteosarcoma malignant progression by sponging miR‐16‐5p. Cancer Sci.2022;113:120–131. 10.1111/cas.15147
Funding information
This work was supported by the National Natural Science Foundation of China (No. 81972509) and the Beijing Science and Technology Planning Project (Z171100001017097).
Contributor Information
Xiaodong Tang, Email: Tang15877@163.com.
Wei Guo, Email: 1811110346@bjmu.edu.cn.
DATA AVAILABILITY STATEMENT
Data are available on request to the authors.
REFERENCES
- 1. Chen C, Mao X, Cheng C, et al. miR‐135a reduces osteosarcoma pulmonary metastasis by targeting both BMI1 and KLF4. Front Oncol. 2021;11:620295. 10.3389/fonc.2021.620295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang Q, Liang X, Ren T, et al. The role of tumor‐associated macrophages in osteosarcoma progression – therapeutic implications. Cell Oncol. 2021;44(3):525‐539. 10.1007/s13402-021-00598-w [DOI] [PubMed] [Google Scholar]
- 3. Zhang H, Wang J, Ren T, et al. LncRNA CASC15 is upregulated in osteosarcoma plasma exosomes and CASC15 knockdown inhibits osteosarcoma progression by regulating miR‐338‐3p/RAB14 axis. Onco Targets Ther. 2020;13:12055‐12066. 10.2147/OTT.S282053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niu J, Yan T, Guo W, et al. Identification of potential therapeutic targets and immune cell infiltration characteristics in osteosarcoma using bioinformatics strategy. Front Oncol. 2020;10:1628. 10.3389/fonc.2020.01628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang J, Piao CD, Ding J, Li ZW. LncRNA MALAT1 facilitates lung metastasis of osteosarcomas through miR‐202 sponging. Sci Rep. 2020;10(1):12757. 10.1038/s41598-020-69574-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu W, Long Q, Zhang W, et al. miRNA‐221‐3p derived from M2‐polarized tumor‐associated macrophage exosomes aggravates the growth and metastasis of osteosarcoma through SOCS3/JAK2/STAT3 axis. Aging. 2021;13(15):19760‐19775. 10.18632/aging.203388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lilienthal I, Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: a review of current and future strategies. Int J Mol Sci. 2020;21(18):6885. 10.3390/ijms21186885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duffaud F. Role of TKI for metastatic osteogenic sarcoma. Curr Treat Options Oncol. 2020;21(8):65. 10.1007/s11864-020-00760-w [DOI] [PubMed] [Google Scholar]
- 9. Ragan C, Goodall GJ, Shirokikh NE, Preiss T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci Rep. 2019;9(1):2048. 10.1038/s41598-018-37037-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hou LD, Zhang J. Circular RNAs: an emerging type of RNA in cancer. Int J Immunopathol Pharmacol. 2017;30(1):1‐6. 10.1177/0394632016686985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li C, Zhang J, Yang X, et al. hsa_circ_0003222 accelerates stemness and progression of non‐small cell lung cancer by sponging miR‐527. Cell Death Dis. 2021;12(9):807. 10.1038/s41419-021-04095-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jafari GF. Circular RNA in saliva. Adv Exp Med Biol. 2018;1087:131‐139. 10.1007/978-981-13-1426-1_11 [DOI] [PubMed] [Google Scholar]
- 13. Wang S, Zhang X, Li Z, et al. Circular RNA profile identifies circOSBPL10 as an oncogenic factor and prognostic marker in gastric cancer. Oncogene. 2019;38(44):6985‐7001. 10.1038/s41388-019-0933-0 [DOI] [PubMed] [Google Scholar]
- 14. Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial‐mesenchymal transitioning and metastasis of NSCLC cells by sponging miR‐96‐5p. EBioMedicine. 2019;44:182‐193. 10.1016/j.ebiom.2019.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long F, Lin Z, Li L, et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. 2021;20(1):26. 10.1186/s12943-021-01318-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu G, Sun J, Yang Z‐F, et al. Cancer‐associated fibroblast‐derived CXCL11 modulates hepatocellular carcinoma cell migration and tumor metastasis through the circUBAP2/miR‐4756/IFIT1/3 axis. Cell Death Dis. 2021;12(3):260. 10.1038/s41419-021-03545-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qiu L, Xu H, Ji M, et al. Circular RNAs in hepatocellular carcinoma: Biomarkers, functions and mechanisms. Life Sci. 2019;231:116660. 10.1016/j.lfs.2019.116660 [DOI] [PubMed] [Google Scholar]
- 18. Long Z, Gong F, Li Y, Fan Z, Li J. Circ_0000285 regulates proliferation, migration, invasion and apoptosis of osteosarcoma by miR‐409‐3p/IGFBP3 axis. Cancer Cell Int. 2020;20:481. 10.1186/s12935-020-01557-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan F, Zhang J, Tang B, Jing L, Qiu B, Zha Z. The novel circ_0028171/miR‐218‐5p/IKBKB axis promotes osteosarcoma cancer progression. Cancer Cell Int. 2020;20:484. 10.1186/s12935-020-01562-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma S, Kong S, Wang F, Ju S. CircRNAs: biogenesis, functions, and role in drug‐resistant tumours. Mol Cancer. 2020;19(1):119. 10.1186/s12943-020-01231-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fernandes I, Melo‐Alvim C, Lopes‐Brás R, Esperança‐Martins M, Costa L. Osteosarcoma pathogenesis leads the way to new target treatments. Int J Mol Sci. 2021;22(2):813. 10.3390/ijms22020813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marchandet L, Lallier M, Charrier C, Baud’huin M, Ory B, Lamoureux F. Mechanisms of resistance to conventional therapies for osteosarcoma. Cancers. 2021;13(4):683. 10.3390/cancers13040683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen J, Yang J, Fei X, Wang X, Wang K. CircRNA ciRS‐7: a novel oncogene in multiple cancers. Int J Biol Sci. 2021;17(1):379‐389. 10.7150/ijbs.54292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z, Li X, Xu D, et al. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2021;54(1):e12936. 10.1111/cpr.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639‐644. 10.1016/j.biopha.2018.03.084 [DOI] [PubMed] [Google Scholar]
- 26. Lin QU, Ling Y‐B, Chen J‐W, et al. Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int J Oncol. 2018;53(1):246‐256. 10.3892/ijo.2018.4371 [DOI] [PubMed] [Google Scholar]
- 27. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132‐136. 10.1016/j.cca.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 28. Gu Z, Li Z, Xu R, et al. miR‐16‐5p suppresses progression and invasion of osteosarcoma via targeting at Smad3. Front Pharmacol. 2020;11:1324. 10.3389/fphar.2020.01324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nisar S, Bhat AA, Singh M, et al. Insights into the role of CircRNAs: biogenesis, characterization, functional, and clinical impact in human malignancies. Front Cell Dev Biol. 2021;9:617281. 10.3389/fcell.2021.617281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang X, Li H, Lu Y, Cheng L. Circular RNAs in human cancer. Front Oncol. 2020;10:577118. 10.3389/fonc.2020.577118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guan B, Li Q, Zhang HZ, Yang HS. circ_NOTCH3 functions as a protooncogene competing with miR‐205‐5p, modulating KLF12 expression and promoting the development and progression of basal‐like breast carcinoma. Front Oncol. 2020;10:602694. 10.3389/fonc.2020.602694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305‐317. 10.1016/j.canlet.2017.06.027 [DOI] [PubMed] [Google Scholar]
- 33. Liu S, Chen L, Chen H, Xu K, Peng X, Zhang M. Circ_0119872 promotes uveal melanoma development by regulating the miR‐622/G3BP1 axis and downstream signalling pathways. J Exp Clin Cancer Res. 2021;40(1):66. 10.1186/s13046-021-01833-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang H, Yang K, Ren T, Huang Y, Tang X, Guo W. miR‐16‐5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis. 2018;9(6):680. 10.1038/s41419-018-0738-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on request to the authors.


