Abstract
An aptamer is a short oligonucleotide chain that can specifically recognize targeting analytes. Due to its high specificity, low cost, and good biocompatibility, aptamers as the targeting elements of biosensors have been applied widely in non‐invasive tumor imaging and treatment in situ to replace traditional methods. In this review, we will summarize recent advances in using aptamer‐based biosensors in tumor diagnosis. After a brief introduction of the advantage of aptamers compared with enzyme sensors and immune sensors, the different sensing designs and mechanisms based on 3 signal transduction modes will be reviewed to cover different kinds of analytical methods, including: electrochemistry analysis, colorimetry analysis, and fluorescence analysis. Finally, the prospective advantages of aptamer‐based biosensors in tumor theranostics and post‐treatment monitoring are also evaluated in this review.
Keywords: colorimetric aptasensor, electrochemical aptasensor, fluorescent aptasensor, tumor diagnosis, tumor marker
This review introduces the advantages of aptamer sensors in disease diagnosis compared with traditional biosensors. It also shows the design strategies of the 3 most studied aptamer sensors: fluorescent aptasensors, electrochemical aptasensors, and colorimetric aptasensors, and their application in early tumor diagnosis. Moreover, taking the fluorescent aptasensor as an example, the design of aptasensors combined with nanomaterials can not only be used for tumor imaging, but also has important significance in tumor treatment and efficacy evaluation. The design provides a new idea for the integration of tumor theranostics.
Abbreviations
- AFP
alpha‐fetoprotein
- Apt
aptamer
- AuNPs
gold nanoparticles
- CEA
carcinoembryonic antigen
- CGO
carboxylated graphene oxide
- COU
coumarin‐3‐carboxylic acid
- CTCs
circulating tumor cells
- Cy5
cyanine 5
- Cyt c
cytochrome c
- DAR
1,3‐phenylenediamine resin
- DHA
dihydroartemisinin
- dsDNA
double‐stranded DNA
- EDS
energy dispersive X‐ray spectroscopy
- FAM
fluorescein amidite
- Fc
ferrocene
- FRET
fluorescence resonance energy transfer
- FTO
fluorine tin oxide
- GA
glutaraldehyde
- GCE
glassy carbon electrode
- GO
graphene oxide
- GQD
graphene quantum dots
- H1
hairpin probe 1
- H2
hairpin probe 2
- HCR
hybrid chain reaction
- HD‐CHR
hairpin DNA cascade hybridization reaction
- HER2
human epidermal growth factor receptor 2
- HRP
horseradish peroxidase
- MB
methylene blue
- MUC1
mucin 1
- NBPs
nanobipyramids
- NCL
nucleolin protein
- NCs
nanoclusters
- NG
nitrogen doped graphene
- NHS
N‐hydroxysuccinimide
- NPs
nanoparticles
- PDA
polydopamine
- PFN
polyfluorene
- POCT
point‐of‐care testing
- PSMA
prostate‐specific membrane antigen
- R101
rhodamine 101
- R6G
rhodamine 6G
- RCA
rolling circle amplification
- ROS
reactive oxygen species
- SDR
strand placement reaction
- SELEX
systematic evolution of ligands by exponential enrichment
- s‐SWCNTs
single‐walled carbon nanotubes that being excellent water solubility
- TMB
tetramethylaniline
- TPE‐TA
tertiary amine‐containing tetraphenylethene
1. INTRODUCTION
The latest statistics from the American Cancer Society found that the average number of people dying from cancer in the United States each year is as high as 600 000. 1 Fear of cancer is a common phenomenon at this stage. The limitation of effective treatment and delay in diagnosis are the main reasons for the high incidence of cancer mortality. 2 In view of this, timely and accurate diagnosis of early cancer is particularly important for improving the cure rate of cancer. Currently, common cancer diagnosis methods include tissue biopsy, proteomics, tumor imaging, and biomarker detection. 3 Compared with the first 3, biomarker detection is more common in clinical screening and diagnosis due to its characteristics of less invasive damage and lower cost. However, the detection of tumor markers at this stage has low sensitivity and cannot be used for low‐level concentration screening in the early stages of cancer, resulting in a certain missed diagnosis rate. Designing a low detection limit and high affinity detection strategy is particularly important for the detection of early cancer marker proteins. 4 Recently, due to the application of various signal amplification technologies in biosensors, such as enzyme catalysis, nucleic acid chain reaction, biotin‐streptavidin, click chemistry, cascade reaction, nanomaterials, etc., biosensors have high reproducibility and sensitivity to effectively circumvent the limitations of traditional methods. 5 With a very low detection limit, the biological signal is converted to a visual signal that can be used to measure the level of specific proteins on or secreted by tumor cells. The biosensor was defined as "an independent integrated device." 6 Usually, they mainly include 3 major components: biometric identification component, signal conversion component, and information reading component. Most of the biological recognition elements are macromolecules such as antibodies, aptamers, and enzymes, which have the characteristics of specifically recognizing target analytes to facilitate quantitative or semi‐quantitative analysis of a certain target by biosensors. 7
At present, the recognition element of the most applicable aptasensor is a short oligonucleotide chain separated from a random library in vitro by SELEX, 8 , 9 using different screening strategies and manipulation of the selection conditions to closely control the aptamer‐target binding affinity and specificity. 10 Generally, the size of the aptamer sequence can be ~30‐70 nucleotides in average length, folded into a three‐dimensional structure, and connected to specific biological elements through specificity and affinity, such as metal ions, tumor marker proteins, small molecules, or even viruses, circulating tumor cells, etc. 11 , 12 , 13 , 14 , 15 For example, Li et al used aptamers and nanomaterials to assemble fluorescent aptasensors to detect tumor‐associated proteins on exosomes derived from prostate cancer and breast cancer, and successfully used them in the clinical differentiation of healthy specimens from tumor specimens with the advantage of high sensitivity. 16 Based on the characteristics of rapid response and portability, biosensors using enzymes as identification elements have been used for immediate detection of tumor patients, such as detecting circulating tumor cells, prostate antigen, etc. 17 , 18 However, enzymes are sensitive to temperature and pH, having a short shelf life. For antibody sensors, the generation and characterization of new antibodies are time consuming and difficult, 19 , 20 such as induction by target preparations, animal immunity, antibody purification, and other operations are complicated and require time and material costs. 21 , 22 , 23 For some small molecules and proteins with low immunogenicity, it is also difficult for newly generated antibodies to control their binding properties and bind to similar structures to cause non‐target interference, 24 and are easily affected by immunosuppressive agents. 25 Different from enzyme and antibody sensors, the most notable feature of aptamers is their ease of modification and low immunogenicity. The specific aptamer sequence can be synthesized in vitro with low cost, reproducible mass production, and is easily modified by nanomaterials for tumor marker analysis and treatment. In addition, they can tolerate various pH and salt concentrations, and have good thermal stability. 9 , 26 These features make aptamers an ideal recognition element for biosensors for tumor monitoring instead of enzymes or antibodies.
According to the signal conversion elements of different types of aptasensors, biological information is usually converted to fluorescent signals, electrochemical signals or color changes, 27 , 28 , 29 which can be divided into fluorescence, electrochemistry, and colorimetric aptasensors. 8 , 9 , 10 These changed signals are read by simple instruments, and the result can be visualized. Based on portable design, good signal theory, and compatibility with biochemical components, the most traditional electrochemistry is still the most commonly used in the assembly of aptasensors. Compared with other transducers, the advantages of electrochemical aptasensors are miniaturization and automation, insensitivity to turbidity or quenching effects, and almost completely avoids complicated sample preparation. Moreover, electrochemical aptasensors have lower detection limits than other types of sensors. The detection limit (particle number/mL) of most optical aptasensors is mostly 105‐106, while the detection limit of most electrochemical aptasensors can reach 103‐104. 3 Currently, the transduction mediums of electrochemical sensors usually affect the diffusion efficiency, electron transfer, and the final detected signal. Finding suitable electroactive substances is particularly important for the design of electrochemical sensors. 30 Due to the characteristics of ease of use, accessibility, and instant detection, colorimetric aptasensors can be used to detect tumor markers by observing the color change of the solution by the naked eye. However, the colorimetric aptasensor has some limitations because it is easily affected by the color of the sample, making it difficult in the clinical diagnosis of high‐demand multi‐target detection. Different from the first 2 sensors, the most significant advantage of the fluorescent aptasensor is that it can distinguish different wavelengths of visible light, used in multiple detection in the form of a sensor array to simultaneously distinguish and even quantify a variety of tumor cells with extremely low detection limits. Based on the above 3 signal transduction modes of aptasensors, this review summarizes the latest developments and current challenges of different detection strategies for electrochemical, colorimetric and fluorescent aptasensors, and evaluates the application trend of some types of aptasensors in the future combined with nanomaterials to realize the integration of tumor diagnosis and treatment.
2. ELECTROCHEMICAL APTASENSOR
Electrochemical aptasensors are currently the most widely used biosensors in tumor imaging, which was first proposed in 2004, and they can provide low‐power and ultra‐low detection limits of target analytes. 31 , 32 Due to its high accuracy and good reproducibility, this type of sensor is often used as a minimally invasive device for POCT. 33 Generally, in the electrochemical aptasensor, the aptamer is fixed on the electrode surface as a biological recognition element. Through the specific binding of the aptamer to the target, the capacitance change caused by binding of the analyte or the current or potential response generated by the oxidation and reduction reactions on the electrode surface is evaluated. According to the type of response signal, it can be divided into ampere method, cyclic voltammetry, electrical impedance method, etc. 34 , 35 , 36 This section discusses the design schemes of several common electrochemical aptasensors for tumor marker monitoring, including direct fixation, sandwich, and immobilization free.
2.1. Direct immobilization
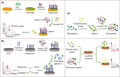
In most cases, the aptamer probe can be immobilized on the electrode surface to capture the target protein by electrostatic adsorption, covalent attachment, and affinity. 37 Several electrochemical aptasensors based on direct induction of target have been developed, with paper, ion‐exchangeable polymer membrane, and high‐load graphene oxide fixed on the surface of GCE to provide a conductive path, 38 as shown in Figure 1A. Bharti et al used electrodeposited gold‐platinum bimetallic nanoparticles on the surface of the CGO/FTO electrode for signal amplification, and used EDS‐NHS to activate CGO, and then modified with streptomycin to further deposit biotin‐labeled aptamers. After incubation with the breast cancer marker protein MUC1, the aptamer electrode binds to the target and the signal changes are measured by differential pulse voltammetry, which is concentration‐dependent with MUC1. 31
FIGURE 1.
A, Schematic illustration of aptamer direct immobilization electrochemical sensor for capturing target proteins. B, Schematic illustration of peroxidase‐labeled aptamer sandwich electrochemical sensor for capturing tumor cells and converting into electrical signals. C, Schematic illustration of immobilization‐free aptasensor based on MB‐labeled dsDNA conformation
2.2. Sandwich format
The design strategy of the sandwich electrochemical aptasensor comes from the structure of the immunosensor, including antibody‐aptamer sandwich, aptamer‐antibody sandwich, and aptamer‐aptamer sandwich sensing layer. 39 , 40 Compared with the participation of a single aptamer, the sandwich form usually has higher selectivity. Signal aptamers often bind to redox markers such as Fc, MB, or peroxidase, 30 as shown in Figure 1B. Shekari et al designed a sensor to deposit gold nanoparticles and graphene quantum dots on a graphene‐nitrogen‐modified GCE to obtain a GQD/AuNP/NG/GCE type structure. The CEA Apt I was fixed on the modified GCE. The amino‐modified CEA Apt II was connected to the heme‐G‐quadruplex via GA as a linker to produce Apt II/GA/DNAzyme. Through the sandwich mode, the Apt II /GA/DNAzyme bioconjugate was captured by the CEA aptamer on the electrode. Hemin‐G4 acts as a peroxidase to rapidly catalyze the electroreduction of hydrogen peroxide, and quantify CEA by differential pulse voltammetry. 41
In addition, the sandwich format facilitates multiple amplification strategies such as a DNA walker for signal amplification. Ji et al built a DNA walker track by self‐assembly of Fc‐labeled anchored DNA and thrombin Apt I on the surface of the gold electrode. Thrombin Apt II and walking strand DNA were introduced into the gold electrode through aptamer‐target specific recognition, thereby starting the hybridization of DNA walker with anchored DNA. The DNA walker gradually cut the hybridized anchored DNA by cutting endonuclease to release multiple Fc molecules for signal amplification. The electrochemical signal produced by Fc decreased linearly with the log value of thrombin concentration in the range 10 pM to 100 nM. 42 Similarly, Cai et al 43 built a signal amplification electrochemical aptasensor for detecting breast cancer cells through a free‐running DNA walker.
2.3. Immobilization free
For the above 2 types, the process of fixing the aptamer to the electrode surface is time consuming. The aptamer assembled on the electrode sometimes hinders the effective recognition between the target and the aptamer. Unlike the conventional strategy of fixing aptamers on electrodes, in Figure 1C, this design strategy is usually to form a dsDNA conformation by complementing the hybridized strand with MB or Fc‐labeled aptamer. The dsDNA modified with electroactive substances is easily adsorbed by the electrode modified with specific nanomaterials, and then the electrical signal is turned on. When the target marker is present, the aptamer preferentially binds to the target, the complex falls off, and the electrical signal is turned off. In the study by Wang et al, a thiolated complementary chain was used to hybridize with MB‐labeled aptamer, adsorbed by the electrode modified to trigger the electrical signal. After the MB‐aptamers recognized K562 circulating tumor cells, the Apt‐CTC complex fell off from the electrode, causing the signal to turn off, and the detection limit reached 23 cells/mL. 44 Due to the avoidance of complicated electrode modification and identification probe fixation processes, as well as expensive labeling procedures, label‐free homogeneous detection with high sensitivity and accuracy has gained popularity in the design of electrochemical aptasensor.
3. COLORIMETRIC APTASENSOR
Among various signal transduction modes, colorimetry is undoubtedly the simplest, most convenient, and intuitive detection method. 45 , 46 This method can visually distinguish the color change by visual observation or the change of absorbance value to feedback the response of the target analyte, having the application prospect of instant diagnosis at the bedside when there is no complicated instrument. 47 , 48
3.1. AuNPs
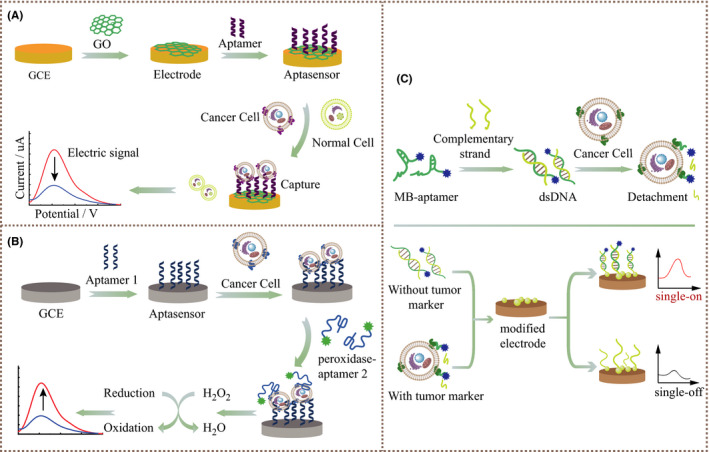
At present, the most common AuNPs are used as transducers of colorimetric aptasensors in various studies. 49 Due to the surface plasmon resonance of gold nanoparticles, they have strong distance‐dependent optical properties. 50 Once the differently modified gold nanoparticles are close to each other, their absorption spectra will shift, and the scattering profile will change, eventually resulting in a change in the color and absorption spectra of the sample. The aggregation and redispersion of AuNPs regulate the color change of the solution and indicate whether there is a certain tumor marker in the sample, 51 , 52 as shown in Figure 2. As gold nanoparticles are easily biofunctionalized and have good biological stability and spectral performance, Borghei et al used 2 sets of single‐stranded DNA probes to functionalize 2 sets of gold nanoparticles. Afterwards, the complementary aptamer AS1411 was hybridized to the specific sites of the 2 sets of DNAs to induce cross‐linking and aggregation of the 2 sets of nanoparticles, and the solution was purple. When the NCL was present, the aptamer AS1411 preferentially bound to the target protein NCL and then fell off from the DNA sequence. There were only 2 groups of unrelated DNA‐AuNPs in the solution, which were red. 52 Direct visual observation was used to distinguish MCF‐7 breast cancer cells that expressed high NCL from normal cells not expressing NCL, and the detection limit of this method was estimated to be ~10 cells/mL, which gave sufficient sensitivity and selectivity. In addition, the operation was simpler than the more costly fluorescence and electrochemical measurements. 53
FIGURE 2.
Schematic illustration of the colorimetric aptasensor identifying tumor cells by AuNPs aggregation‐inducing color change
3.2. H2O2 oxidation
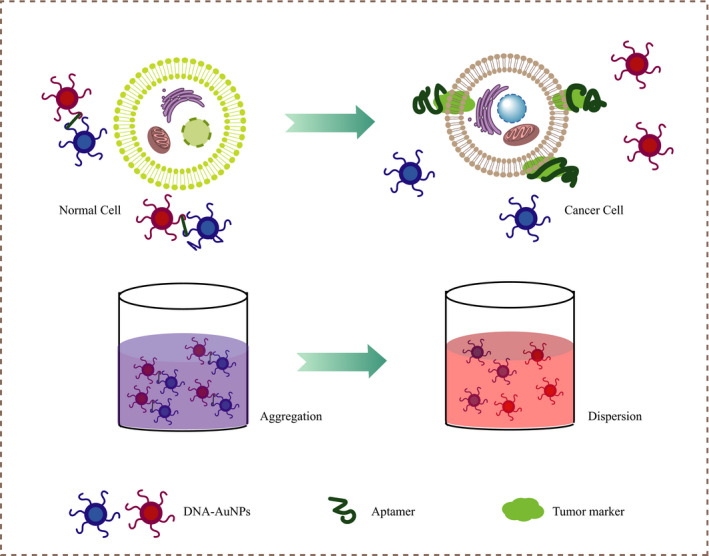
Another commonly used detection strategy is to simulate intrinsic enzymes, 54 such as modifying nanozymes with aptamers to increase the activity of peroxidase‐like enzymes, to enhance the oxidation of tetramethylaniline in the presence of H2O2 and present a strong blue color. 55 When tumor cells with high expression of target markers are present, the aptamer preferentially binds to the target protein. After the target responsive structure changes, the nanozyme returns to the original lower peroxidase‐like activity, thereby weakening the catalytic effect; the solution does not turn into blue significantly, 54 as shown in Figure 3. Compared with HRP, these nano‐artificial enzymes such as AuNPs, Au@Fe3O4 NPs, ZnFeO reduced GO, and heme/G‐quadruplex, 56 , 57 , 58 , 59 due to their low cost and tolerance to pH and temperature, have been widely used in colorimetric sensors to distinguish tumors. Xia et al used TMB and CD63 aptamer‐terminated single‐walled carbon nanotubes s‐SWCNTs as catalytic substrates and catalytic enzymes to detect serum exosomes in breast cancer patients. 60 In view of this, they can be widely used in the rapid detection and discrimination of different cancers, by just changing the aptamer. 61
FIGURE 3.
Schematic illustration of the colorimetric aptasensor identifying tumor cells by nanozyme inducing TMB oxidation to change color in the presence of H2O2
4. FLUORESCENT APTASENSOR
The design of fluorescent aptasensors mainly involves fluorophores, dyes, or fluorescent nanomaterials, aptamers, and quenchers. 62 , 63 Different from colorimetric aptasensors, they are not affected by the background color of the sample and can simultaneously design multiple aptamers in the form of a sensor array for multi‐channel rapid detection. 64 Based on the different detection mechanisms of fluorescence signal recovery, they are mainly divided into: fluorescence resonance energy transfer, fluorescence signal amplification, and fluorescence polarization. 65 Using its non‐invasiveness and visualization, combined with a variety of nanomaterials such as photosensitizers, real‐time in situ monitoring and killing of tumor cells, and treatment effect monitoring are other significant directions in the design strategy of fluorescent aptasensors.
4.1. Fluorescence resonance energy transfer
The most typical way is to modify the 3' or 5' end of the aptamer with a fluorophore or an aggregation‐inducing luminescence agent, and then introduce a fluorescence quencher through electrostatic adsorption or complementary structure. The fluorescence signal is turned off based on FRET between the luminescent agent and the quencher. 16 , 66 When the target marker is present, the conformation switch shifts, the quencher and the fluorophore are separated, and the fluorescent signal is turned on again to quickly detect the target protein 67 (Figure 4A). This method of turning off and then turning on the fluorescent signal effectively reduces the background signal. 3 Common high‐distance‐dependent fluorescence signal quenchers include graphene oxide, mesoporous carbon nanospheres, organic frameworks, magnetic nanoparticles, etc. 67 , 68 , 69 , 70 Xu et al designed a four‐color sensor based on FRET to distinguish 4 tumor markers, AFP, VEGF165, CEA and HER2, with a very low detection limit, 64 realizing the simultaneous detection and imaging of multiple tumor‐related proteins in living cells. Based on the same detection strategy, but changing different aptamers or nanoquenchers, Li et al 16 designed GO/Apt‐TPE‐TA nanocomposites that quenched the aptamer PSMA‐modified aggregation‐inducing luminescence of TPE‐TA by GO, and finally turned on the fluorescence signal in the serum of prostate cancer patients to successfully detect tumor exosomes.
FIGURE 4.
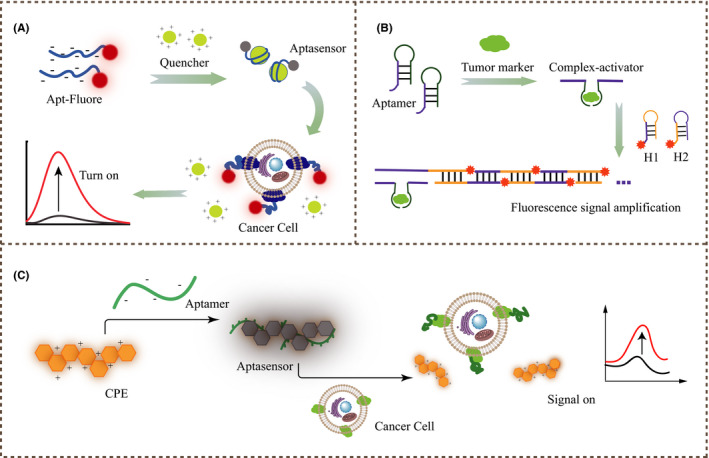
A, Schematic illustration of the fluorescent aptasensor based on FRET between fluorophores and quenchers to turn on the fluorescent signal and recognize tumor cells. B, Schematic illustration of the aptasensor based on HCR for amplifying signals by activating H1, H2. C, Schematic diagram of the aptasensor assembled by the cationic copolymer and aptamer to detect tumor marker proteins through fluorescence polarization
4.2. Fluorescence signal amplification
Common nucleic acid signal amplification reactions include RCA, HCR, HD‐CHR, and SDR. 3 As shown in Figure 4B, Huang et al used the MB‐CD63 antibody to capture leukemia‐derived exosomes. DNA probes containing AS1411 aptamers and RCA primers bound to exosomes and initiated RCA amplification. Finally, FAM was released to enhance the fluorescence signal. 71 Feng et al developed a double‐amplification detection method for adenosine based on HCR and exonuclease III‐assisted DNA circulation. In the presence of adenosine, the adenosine aptamer triggered the initiation of HCR between H1 and H2, resulting in longer double‐stranded DNA polymerization. Finally, the dye SYBR Green I was inserted into the groove of DNA, causing significant amplification of the fluorescence signal. 72 Using the same principle, Zhao et al monitored early prostate cancer through HCR to amplify the fluorescent signal mechanism. 34 Ma et al 35 also applied HCR to the detection of MUC1 model peptides; this is expected to be applied to the detection of breast cancer and other tumor cells that highly express MUC1. Figure 4B shows the principle of signal amplification in common HCR‐based sensor designs.
4.3. Fluorescence polarization
Compared with the former, the design based on fluorescence polarization is more simplified and requires no quenchers or donor‐acceptor pairs. As shown in Figure 4C, when the aptamer detaches from the polymer and forms a specific three‐dimensional structure with the target protein, the fluorescence signal is restored. 36 Bao et al reported that the Apt‐PFN+ complex was used to detect tumor markers AFP and CEA. 73 Similarly, Ho et al prepared aptamer sgc8c‐conjugated DAR NPs trapped with R6G and aptamer TD05‐conjugated DAR NPs trapped with R101 to recognize CCRF‐CEM and Ramos cells, respectively. 74 Based on this detection strategy, Lyu et al proposed the afterglow effect, bypassing real‐time luminescence monitoring, and minimizing the tissue background signal. 75
4.4. Prospects for theranostics
In addition to early screening and diagnosis, timely treatment of pre‐cancer is the top priority to reduce the mortality rate of cancer. At present, aptasensors are mature in various cancer screening and detection technologies. However, being able to report and trigger targeted killing of tumor cells at the same time is still a challenge we need to face. 76 At this stage, some researchers have reported that aptasensors can be combined with photodynamic, photothermal, and chemotherapy to diagnose and treat tumors. 77 , 78 , 79
Wang et al reported a composite material composed of Au NBPs@PDA and Au NCs. Au NCs labeled with MUC1 aptamer (probe 1) competitively bound with the target protein MUC1, and then detached from the surface of Au NBPs@PDA, turning on the red fluorescence to accurately image MCF‐7 breast cancer cells. At the same time, the hybridization of the complementary single strand (probe 2) with high expression of MicroRNA‐21 in the HepG2 cells triggered the green fluorescence of Au NCs, which realized the original dual‐type imaging of tumor biomarkers with different spatial distributions of different tumor cells. In addition, the marked photothermal properties of Au NBPs@PDA can kill cancer cells more effectively for accurate diagnosis and tumor treatment. 77 Han et al designed the combined application of Cy5‐labeled Cyt c aptamer, GO, and the anti‐tumor drug COU‐DHA targeted mitochondrial for real‐time imaging of cell death. After the tumor cells had endocytosed the composite material, COU‐DHA was released from the GO surface to target the mitochondria, inducing the production of ROS in situ to kill tumor cells. Then, Cyt c was released from mitochondria, the Cy5‐labeled aptamer preferentially bound to Cyt c and fell off from GO, emitting red fluorescence, indicating tumor cell apoptosis. 80 It is inevitable that most research on the integration of diagnosis and treatment is the complex structure of composite materials. Committed to build a multifunctional aptasensor as a new research direction, and use for imaging and killing tumor cells to achieve the dual goals of diagnosis and treatment will be the direction of the extensive research on aptasensors in the future.
5. CONCLUSION AND OUTLOOK
In this review, we focused on the detection strategies of aptamer biosensors with 3 signal transduction modes. Each type of aptamer sensing has unique advantages and corresponding limitations, which must also be considered before determining the purpose of detection. For example, the fluorescent aptasensor is easier to use for multi‐channel simultaneous detection of different tumor cells than the other 2; the colorimetric aptasensor is the most intuitive and simplest of the 3 to distinguish between cancer and normal samples; the electrochemical aptasensor is more focused on non‐invasive, small, portable, and low detection limits to screen tumor markers. In addition, the article also mentions the expanded application of aptasensors in treatment and post‐treatment monitoring. At present, many studies have been successfully applied in the clinic to detect cancer patient samples. We have reason to believe that the application prospect of aptasensors will become more available in the clinical practice in the foreseeable future.
CONFLICT OF INTEREST
The authors have no conflict of interest.
ACKNOWLEDGMENTS
This work was supported by the Scientific and Technological Innovation Major Base of Guangxi (No. 2018‐15‐Z04), the State Project for Essential Drug Research and Development (No. 2019ZX09301132), Guangxi Key Research and Development Project (No. AB20117001), the National University of Singapore Start‐up Grant (NUHSRO/2020/133/Startup/08), NUS School of Medicine Nanomedicine Translational Research Programme (NUHSRO/2021/034/TRP/09/Nanomedicine), and NUS School of Medicine Kickstart Initiative (NUHSRO/2021/044/Kickstart/09/LOA).
Mo T, Liu X, Luo Y, et al. Aptamer‐based biosensors and application in tumor theranostics. Cancer Sci.2022;113:7–16. 10.1111/cas.15194
Contributor Information
Pan Wu, Email: w_pwupan@163.com.
Xiaoyuan Chen, Email: chen.shawn@nus.edu.sg.
REFERENCES
- 1. Siegel R, Miller K, Fuchs H, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7‐33. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Jemal A, Wender R, et al. An assessment of progress in cancer control. CA Cancer J Clin. 2018;68(5):329‐339. [DOI] [PubMed] [Google Scholar]
- 3. Zhu C, Li L, Wang Z, et al. Recent advances of aptasensors for exosomes detection. Biosens Bioelectron. 2020;160:112213. [DOI] [PubMed] [Google Scholar]
- 4. Sun Z, Yang J, Li H, et al. Progress in the research of nanomaterial‐based exosome bioanalysis and exosome‐based nanomaterials tumor therapy. Biomaterials. 2021;274:120873. [DOI] [PubMed] [Google Scholar]
- 5. Huang F, Zhang Y, Lin J, et al. Biosensors coupled with signal amplification technology for the detection of pathogenic bacteria: A review. Biosensors. 2021;11(6):190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uygun ZO. State‐of‐the‐art Hepatocellular Carcinoma Biomarker Detection by Biosensor Technology‐a Review. J Gastrointest Cancer. 2021;52(3):1081‐1085. [DOI] [PubMed] [Google Scholar]
- 7. Bhalla N, Jolly P, Formisano N, et al. Introduction to biosensors. Essays Biochem. 2016;60(1):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaban SM, Kim DH. Recent advances in aptamer sensors. Sensors (Basel). 2021;21(3):979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang G, Zhong L, Yang N, et al. Screening of aptamers and their potential application in targeted diagnosis and therapy of liver cancer. World J Gastroenterol. 2019;25(26):3359‐3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen C, Feng S, Zhou M, et al. Development of a structure‐switching aptamer‐based nanosensor for salicylic acid detection. Biosens Bioelectron. 2019;140:111342. [DOI] [PubMed] [Google Scholar]
- 11. Labib M, Zamay A, Muharemagic D, et al. Aptamer‐based viability impedimetric sensor for viruses. Anal Chem. 2012;84(4):1813‐1816. [DOI] [PubMed] [Google Scholar]
- 12. Wang J, Wang Y, Hu X, et al. Dual‐Aptamer‐conjugated molecular modulator for detecting bioactive metal ions and inhibiting metal‐mediated protein aggregation. Anal Chem. 2019;91(1):823‐829. [DOI] [PubMed] [Google Scholar]
- 13. Lee K, Lee J, Ahn B. Design of refolding dna aptamer on single‐walled carbon nanotubes for enhanced optical detection of target proteins. Anal Chem. 2019;91(20):12704‐12712. [DOI] [PubMed] [Google Scholar]
- 14. Niu C, Wang C, Li F, et al. Aptamer assisted CRISPR‐Cas12a strategy for small molecule diagnostics. Biosens Bioelectron. 2021;183:113196. [DOI] [PubMed] [Google Scholar]
- 15. Safarpour H, Dehghani S, Nosrati R, et al. Optical and electrochemical‐based nano‐aptasensing approaches for the detection of circulating tumor cells (CTCs). Biosens Bioelectron. 2020;148:111833. [DOI] [PubMed] [Google Scholar]
- 16. Li B, Liu C, Pan W, et al. Facile fluorescent aptasensor using aggregation‐induced emission luminogens for exosomal proteins profiling towards liquid biopsy. Biosens Bioelectron. 2020;168:112520. [DOI] [PubMed] [Google Scholar]
- 17. Yang J, Huang X, Gan C, et al. Highly specific and sensitive point‐of‐care detection of rare circulating tumor cells in whole blood via a dual recognition strategy. Biosens Bioelectron. 2019;143:111604. [DOI] [PubMed] [Google Scholar]
- 18. Abardía‐Serrano C, Miranda‐Castro R, de‐los‐Santos‐Álvarez N, Lobo‐Castañón MJ. New uses for the personal glucose meter: detection of nucleic acid biomarkers for prostate cancer screening. Sensors. 2020;20(19):5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rossetti M, Brannetti S, Mocenigo M, et al. Harnessing effective molarity to design an electrochemical dna‐based platform for clinically relevant antibody detection. Angew Chem Int Ed Engl. 2020;59(35):14973‐14978. [DOI] [PubMed] [Google Scholar]
- 20. Verdian A, Fooladi E, Rouhbakhsh Z. Recent progress in the development of recognition bioelements for polychlorinated biphenyls detection: Antibodies and aptamers. Talanta. 2019;202:123‐135. [DOI] [PubMed] [Google Scholar]
- 21. Li W, Jing Z, Wang S, et al. P22 virus‐like particles as an effective antigen delivery nanoplatform for cancer immunotherapy. Biomaterials. 2021;271:120726. [DOI] [PubMed] [Google Scholar]
- 22. Huang D, Tran J, Olson A, et al. Vaccine elicitation of HIV broadly neutralizing antibodies from engineered B cells. Nat Commun. 2020;11(1):5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amritkar V, Adat S, Tejwani V, et al. Engineering Staphylococcal Protein A for high‐throughput affinity purification of monoclonal antibodies. Biotechnol Adv. 2020;44:107632. [DOI] [PubMed] [Google Scholar]
- 24. Yu H, Alkhamis O, Canoura J, et al. Advances and Challenges in Small‐Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew Chem Int Ed Engl. 2021;60(31):16800‐16823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu L, Bai X, Tenguria S, et al. Mammalian cell‐based immunoassay for detection of viable bacterial pathogens. Front Microbiol. 2020;11:575615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li L, Xu S, Yan H, et al. Nucleic acid aptamers for molecular diagnostics and therapeutics: advances and perspectives. Angew Chem Int Ed Engl. 2021;60(5):2221‐2231. [DOI] [PubMed] [Google Scholar]
- 27. Dinarvand M, Neubert E, Meyer D, et al. Near‐infrared imaging of serotonin release from cells with fluorescent nanosensors. Nano Lett. 2019;19(9):6604‐6611. [DOI] [PubMed] [Google Scholar]
- 28. Rauf S, Lahcen A, Aljedaibi A, et al. Gold nanostructured laser‐scribed graphene: A new electrochemical biosensing platform for potential point‐of‐care testing of disease biomarkers. Biosens Bioelectron. 2021;180:113116. [DOI] [PubMed] [Google Scholar]
- 29. Wei Y, Wang D, Zhang Y, et al. Multicolor and photothermal dual‐readout biosensor for visual detection of prostate specific antigen. Biosens Bioelectron. 2019;140:111345. [DOI] [PubMed] [Google Scholar]
- 30. Abd‐Ellatief R, Abd‐Ellatief M. Electrochemical aptasensors: current status and future perspectives. Diagnostics. 2021;11(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goud KY, Reddy KK, Khorshed A, et al. Electrochemical diagnostics of infectious viral diseases: Trends and challenges. Biosens Bioelectron. 2021;180:113112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. An Y, Jin T, Zhu Y, et al. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens Bioelectron. 2019;142:111503. [DOI] [PubMed] [Google Scholar]
- 33. Villalonga A, Perez‐Calabuig AM, Villalonga R. Electrochemical biosensors based on nucleic acid aptamers. Anal Bioanal Chem. 2020;412(1):55‐72. [DOI] [PubMed] [Google Scholar]
- 34. Zhao R, Zhao L, Feng H, et al. A label‐free fluorescent aptasensor based on HCR and G‐quadruplex DNAzymes for the detection of prostate‐specific antigen. Analyst. 2021;146(4):1340‐1345. [DOI] [PubMed] [Google Scholar]
- 35. Ma C, Liu H, Tian T, et al. A simple and rapid detection assay for peptides based on the specific recognition of aptamer and signal amplification of hybridization chain reaction. Biosens Bioelectron. 2016;83:15‐18. [DOI] [PubMed] [Google Scholar]
- 36. Zhao X, Dai X, Zhao S, et al. Aptamer‐based fluorescent sensors for the detection of cancer biomarkers. Spectrochim Acta A Mol Biomol Spectrosc. 2021;247:119038. [DOI] [PubMed] [Google Scholar]
- 37. Malecka K, Mikuła E, Ferapontova E. Design strategies for electrochemical aptasensors for cancer diagnostic devices. Sensors. 2021;21(3):736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yen YK, Chao CH, Yeh YS. A graphene‐PEDOT:PSS modified paper‐based aptasensor for electrochemical impedance spectroscopy detection of tumor marker. Sensors. 2020;20(5):1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ikebukuro K, Kiyohara C, Sode K. Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens Bioelectron. 2005;20(10):2168‐2172. [DOI] [PubMed] [Google Scholar]
- 40. Amouzadeh Tabrizi M, Shamsipur M, Saber R, et al. Flow injection amperometric sandwich‐type aptasensor for the determination of human leukemic lymphoblast cancer cells using MWCNTs‐Pd/PTCA/aptamer as labeled aptamer for the signal amplification. Anal Chim Acta. 2017;985:61‐68. [DOI] [PubMed] [Google Scholar]
- 41. Shekari Z, Zare HR, Falahati A. Electrochemical sandwich aptasensor for the carcinoembryonic antigen using graphene quantum dots, gold nanoparticles and nitrogen doped graphene modified electrode and exploiting the peroxidase‐mimicking activity of a G‐quadruplex DNAzyme. Mikrochim Acta. 2019;186(8):530. [DOI] [PubMed] [Google Scholar]
- 42. Ji Y, Zhang L, Zhu L, et al. Binding‐induced DNA walker for signal amplification in highly selective electrochemical detection of protein. Biosens Bioelectron. 2017;96:201‐205. [DOI] [PubMed] [Google Scholar]
- 43. Cai S, Chen M, Liu M, et al. A signal amplification electrochemical aptasensor for the detection of breast cancer cell via free‐running DNA walker. Biosens Bioelectron. 2016;85:184‐189. [DOI] [PubMed] [Google Scholar]
- 44. Wang Y, Zhang W, Tang X, et al. Target‐triggered "signal‐off" electrochemical aptasensor assisted by Au nanoparticle‐modified sensing platform for high‐sensitivity determination of circulating tumor cells. Anal Bioanal Chem. 2020;412(29):8107‐8115. [DOI] [PubMed] [Google Scholar]
- 45. Du Y, Dong S. Nucleic acid biosensors: recent advances and perspectives. Anal Chem. 2017;89(1):189‐215. [DOI] [PubMed] [Google Scholar]
- 46. Xu L, Chopdat R, Li D, et al. Development of a simple, sensitive and selective colorimetric aptasensor for the detection of cancer‐derived exosomes. Biosens Bioelectron. 2020;169:112576. [DOI] [PubMed] [Google Scholar]
- 47. Dong J, He L, Wang Y, et al. A highly sensitive colorimetric aptasensor for the detection of the vascular endothelial growth factor in human serum. Spectrochim Acta Part A Mol Biomol Spectrosc. 2020;226:117622. [DOI] [PubMed] [Google Scholar]
- 48. Zhang H, Peng L, Li M, et al. A label‐free colorimetric biosensor for sensitive detection of vascular endothelial growth factor‐165. Analyst. 2017;142(13):2419‐2425. [DOI] [PubMed] [Google Scholar]
- 49. Yoo S, Lee S. Optical biosensors for the detection of pathogenic microorganisms. Trends Biotechnol. 2016;34(1):7‐25. [DOI] [PubMed] [Google Scholar]
- 50. Zhang X, Xiao K, Cheng L, et al. Visual and highly sensitive detection of cancer cells by a colorimetric aptasensor based on cell‐triggered cyclic enzymatic signal amplification. Anal Chem. 2014;86(11):5567‐5572. [DOI] [PubMed] [Google Scholar]
- 51. Medley C, Smith J, Tang Z, et al. Gold nanoparticle‐based colorimetric assay for the direct detection of cancerous cells. Anal Chem. 2008;80(4):1067‐1072. [DOI] [PubMed] [Google Scholar]
- 52. Borghei YS, Hosseini M, Dadmehr M, et al. Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal Chim Acta. 2016;904:92‐97. [DOI] [PubMed] [Google Scholar]
- 53. Zhu S, Wang X, Jing C, et al. A colorimetric ATP assay based on the use of a magnesium(II)‐dependent DNAzyme. Mikrochim Acta. 2019;186(3):176. [DOI] [PubMed] [Google Scholar]
- 54. Zhou Y, Xu H, Wang H, et al. Detection of breast cancer‐derived exosomes using the horseradish peroxidase‐mimicking DNAzyme as an aptasensor. Analyst. 2019;145(1):107‐114. [DOI] [PubMed] [Google Scholar]
- 55. Dehghani Z, Hosseini M, Mohammadnejad J, et al. Colorimetric aptasensor for Campylobacter jejuni cells by exploiting the peroxidase like activity of Au@Pd nanoparticles. Mikrochim Acta. 2018;185(10):448. [DOI] [PubMed] [Google Scholar]
- 56. Liu S, Xu N, Tan C, et al. A sensitive colorimetric aptasensor based on trivalent peroxidase‐mimic DNAzyme and magnetic nanoparticles. Anal Chim Acta. 2018;1018:86‐93. [DOI] [PubMed] [Google Scholar]
- 57. Wang C, Qian J, Wang K, et al. Colorimetric aptasensing of ochratoxin A using Au@Fe3O4 nanoparticles as signal indicator and magnetic separator. Biosens Bioelectron. 2016;77:1183‐1191. [DOI] [PubMed] [Google Scholar]
- 58. Wu S, Duan N, Qiu Y, et al. Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFeO‐reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int J Food Microbiol. 2017;261:42‐48. [DOI] [PubMed] [Google Scholar]
- 59. Zhao C, Hong C, Lin Z, et al. Detection of Malachite Green using a colorimetric aptasensor based on the inhibition of the peroxidase‐like activity of gold nanoparticles by cetyltrimethylammonium ions. Mikrochim Acta. 2019;186(5):322. [DOI] [PubMed] [Google Scholar]
- 60. Xia Y, Liu M, Wang L, et al. A visible and colorimetric aptasensor based on DNA‐capped single‐walled carbon nanotubes for detection of exosomes. Biosens Bioelectron. 2017;92:8‐15. [DOI] [PubMed] [Google Scholar]
- 61. Jiang Y, Shi M, Liu Y, et al. Aptamer/AuNP biosensor for colorimetric profiling of exosomal proteins. Angew Chem Int Ed. 2017;56(39):11916‐11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zahra Q, Khan Q, Luo Z. Advances in optical aptasensors for early detection and diagnosis of various cancer types. Frontiers in Oncology. 2021;11: 632165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pei X, Wu X, Xiong J, et al. Competitive aptasensor for the ultrasensitive multiplexed detection of cancer biomarkers by fluorescent nanoparticle counting. Analyst. 2020;145(10):3612‐3619. [DOI] [PubMed] [Google Scholar]
- 64. Xu J, Chen W, Shi M, et al. An aptamer‐based four‐color fluorometic method for simultaneous determination and imaging of alpha‐fetoprotein, vascular endothelial growth factor‐165, carcinoembryonic antigen and human epidermal growth factor receptor 2 in living cells. Microchim Acta. 2019;186(3):204. [DOI] [PubMed] [Google Scholar]
- 65. Kou X, Zhang X, Shao X, et al. Recent advances in optical aptasensor technology for amplification strategies in cancer diagnostics. Anal Bioanal Chem. 2020;412(25):6691‐6705. [DOI] [PubMed] [Google Scholar]
- 66. Zhang Y, Guo S, Huang H, et al. Silicon nanodot‐based aptasensor for fluorescence turn‐on detection of mucin 1 and targeted cancer cell imaging. Anal Chim Acta. 2018;1035:154‐160. [DOI] [PubMed] [Google Scholar]
- 67. Cui J, Kan L, Li Z, et al. Porphyrin‐based covalent organic framework as bioplatfrom for detection of vascular endothelial growth factor 165 through fluorescence resonance energy transfer. Talanta. 2021;228:122060. [DOI] [PubMed] [Google Scholar]
- 68. Li C, Meng Y, Wang S, et al. Mesoporous carbon nanospheres featured fluorescent aptasensor for multiple diagnosis of cancer in vitro and in vivo. ACS Nano. 2015;9(12):12096‐12103. [DOI] [PubMed] [Google Scholar]
- 69. Ou X, Zhan S, Sun C, et al. Simultaneous detection of telomerase and miRNA with graphene oxide‐based fluorescent aptasensor in living cells and tissue samples. Biosens Bioelectron. 2019;124‐125:199‐204. [DOI] [PubMed] [Google Scholar]
- 70. Zhu N, Li G, Zhou J, et al. A light‐up fluorescence resonance energy transfer magnetic aptamer‐sensor for ultra‐sensitive lung cancer exosome detection. J Mater Chem B. 2021;9(10):2483‐2493. [DOI] [PubMed] [Google Scholar]
- 71. Huang L, Wang DB, Singh N, et al. A dual‐signal amplification platform for sensitive fluorescence biosensing of leukemia‐derived exosomes. Nanoscale. 2018;10(43):20289‐20295. [DOI] [PubMed] [Google Scholar]
- 72. Feng C, Hou Z, Jiang W, et al. Binding induced colocalization activated hybridization chain reaction on the surface of magnetic nanobead for sensitive detection of adenosine. Biosens Bioelectron. 2016;86:966‐970. [DOI] [PubMed] [Google Scholar]
- 73. Bao B, Su P, Zhu J, et al. Rapid aptasensor capable of simply detect tumor markers based on conjugated polyelectrolytes. Talanta. 2018;190:204‐209. [DOI] [PubMed] [Google Scholar]
- 74. Ho L, Wu W, Chang C, et al. Aptamer‐conjugated polymeric nanoparticles for the detection of cancer cells through "turn‐on" retro‐self‐quenched fluorescence. Anal Chem. 2015;87(9):4925‐4932. [DOI] [PubMed] [Google Scholar]
- 75. Lyu Y, Cui D, Huang J, et al. Near‐infrared afterglow semiconducting nano‐polycomplexes for the multiplex differentiation of cancer exosomes. Angew Chem Int Ed Engl. 2019;58(15):4983‐4987. [DOI] [PubMed] [Google Scholar]
- 76. Bai H, Peng R, Wang D, et al. A minireview on multiparameter‐activated nanodevices for cancer imaging and therapy. Nanoscale. 2020;12(42):21571‐21582. [DOI] [PubMed] [Google Scholar]
- 77. Wang J, Gao Y, Liu P, et al. Core‐shell multifunctional nanomaterial‐based all‐in‐one nanoplatform for simultaneous multilayer imaging of dual types of tumor biomarkers and photothermal therapy. Anal Chem. 2020;92(22):15169‐15178. [DOI] [PubMed] [Google Scholar]
- 78. Esmaeili Y, Zarrabi A, Mirahmadi‐Zare S, et al. Hierarchical multifunctional graphene oxide cancer nanotheranostics agent for synchronous switchable fluorescence imaging and chemical therapy. Mikrochim Acta. 2020;187(10):553. [DOI] [PubMed] [Google Scholar]
- 79. Liu J, Zhang Y, Liu W, et al. Tumor antigen mediated conformational changes of nanoplatform for activated photodynamic therapy. Adv Healthcare Mat. 2019;8(20):e1900791. [DOI] [PubMed] [Google Scholar]
- 80. Han C, Xu X, Zhang C, et al. Cytochrome c light‐up graphene oxide nanosensor for the targeted self‐monitoring of mitochondria‐mediated tumor cell death. Biosens Bioelectron. 2020;173:112791. [DOI] [PubMed] [Google Scholar]


