Abstract
Tumor‐associated macrophages (TAMs) are associated with a poor prognosis of diffuse large B‐cell lymphoma (DLBCL). As macrophages are heterogeneous, the immune polarization and their pathological role warrant further study. We characterized the microenvironment of DLBCL by immunohistochemistry in a training set of 132 cases, which included 10 Epstein–Barr virus‐encoded small RNA (EBER)‐positive and five high‐grade B‐cell lymphomas, with gene expression profiling in a representative subset of 37 cases. Diffuse large B‐cell lymphoma had a differential infiltration of TAMs. The high infiltration of CD68 (pan‐macrophages), CD16 (M1‐like), CD163, pentraxin 3 (PTX3), and interleukin (IL)‐10‐positive macrophages (M2c‐like) and low infiltration of FOXP3‐positive regulatory T lymphocytes (Tregs) correlated with poor survival. Activated B cell‐like DLBCL was associated with high CD16, CD163, PTX3, and IL‐10, and EBER‐positive DLBCL with high CD163 and PTX3. Programmed cell death‐ligand 1 positively correlated with CD16, CD163, IL‐10, and RGS1. In a multivariate analysis of overall survival, PTX3 and International Prognostic Index were identified as the most relevant variables. The gene expression analysis showed upregulation of genes involved in innate and adaptive immune responses and macrophage and Toll‐like receptor pathways in high PTX3 cases. The prognostic relevance of PTX3 was confirmed in a validation set of 159 cases. Finally, in a series from Europe and North America (GSE10846, R‐CHOP‐like treatment, n = 233) high gene expression of PTX3 correlated with poor survival, and moderately with CSF1R, CD16, MITF, CD163, MYC, and RGS1. Therefore, the high infiltration of M2c‐like immune regulatory macrophages and low infiltration of FOXP3‐positive Tregs is associated with a poor prognosis in DLBCL, for which PTX3 is a new prognostic biomarker.
Keywords: diffuse large B‐cell lymphoma; IL‐10; PD‐L1, CD163; PTX3
This research focused on the analysis of several macrophage and regulatory T lymphocyte markers in diffuse large b‐cell lymphoma. We found that high PTX3 expression correlated with poor prognosis of the patients.
Abbreviations
- ABC/non‐GCB
activated B cell‐like
- CSF1R
colony stimulating factor 1 receptor
- DLBCL
diffuse large B‐cell lymphoma
- EBER
Epstein–Barr virus‐encoded small RNA
- EBV
Epstein–Barr virus
- FFPE
formalin‐fixed paraffin‐embedded
- FL
follicular lymphoma
- GCB
germinal center B cell‐like
- HGBCL
high‐grade B‐cell lymphoma
- IHC
immunohistochemistry
- IL‐10
interleukin‐10
- IPI
International Prognostic Index
- LDH
lactate dehydrogenase
- MITF
microphthalmia transcription factor
- NK
natural killer
- NF‐κB
nuclear factor‐κB
- NOS
not otherwise specified
- OS
overall survival
- PD‐L1
programmed death‐ligand 1
- PFS
progression‐free survival
- PTX3
pentraxin 3
- R‐CHOP
rituximab, cyclophosphamide, hydroxydaunorubicin (doxorubicin), oncovin (vincristine), prednisone
- RGS1
regulator of G‐protein signaling 1
- sIL‐2R
soluble interleukin‐2 receptor
- TAM
tumor‐associated macrophage
- TNF
tumor necrosis factor
- Treg
regulatory T lymphocyte
1. INTRODUCTION
Diffuse large B‐cell lymphoma accounts for 30%‐40% of newly diagnosed non‐Hodgkin lymphomas. 1 Gene expression profiling has classified DLBCL into the GCB subtype, which is associated with a better prognosis, and ABC/non‐GCB subtype, which has a more aggressive clinical evolution. 1 The poorer prognosis of patients with the ABC/non‐GCB type is partly associated with the high infiltration of macrophages and the constitutive activation of the NF‐κB pathway. 2 , 3 , 4
The macrophage lineage is heterogeneous. 5 Classical or M1 macrophages are potent effector cells with pro‐inflammatory and antitumoral functions. 6 However, M2 macrophages are immunosuppressive and protumoral and are stratified into the M2a, M2b, and M2c subtypes. 7 M2c macrophages have immune regulatory functions, express CD163, PTX3, and IL‐10 markers, and induce Tregs. 5 , 6 , 7 , 8 Tumor‐associated macrophages are also named M2d and are abundant in solid cancers, such as gastric, ovarian, breast, and lung adenocarcinomas. 5 , 6 , 7 , 8 Although M1‐like TAMs enhance antitumoral host immune responses, M2‐like TAMs have been implicated in tumor progression, metastasis, resistance to therapy, angiogenesis, and immune suppression. 5 , 6 , 7 , 8
Pentraxin 3, also known as TNF‐inducible gene 14 protein (TSG‐14), is a protein that contributes to the regulation of innate resistance to pathogens, inflammatory reactions, and the clearance of self‐components. Pentraxin 3 regulates the inflammatory activity of macrophages and it is expressed by macrophages with M2‐like polarization, namely, the M2c‐like subtype. 5 , 6 , 7 , 8 In the context of IL‐10 stimulation, B lymphocytes acquire regulatory properties. 9 , 10 Pentraxin 3 makes a crucial contribution to tumor inflammation and is highly expressed in liposarcomas 11 and lung 12 and pancreatic carcinoma 13 , 14 ; however, its role in hematolymphoid neoplasia remains unclear.
Due to the importance of TAMs in the pathogenesis of solid and hematolymphoid neoplasia, the role of the IL‐10 molecule in the regulation of host immune responses and immune checkpoints, the targeting of TAMs and the IL‐10 regulatory pathway is an important strategy for DLBCL therapy. 15 , 16 , 17
In this study, we undertook IHC and a gene expression analysis of DLBCL samples collected from patients receiving R‐CHOP therapy to investigate the role of macrophages and Tregs in the pathogenesis of DLBCL and elucidate their impact on clinical outcomes. The results obtained revealed that PTX3 is a powerful marker that identifies patients with different survival outcomes.
2. MATERIALS AND METHODS
2.1. Patients and samples
The training set comprised 132 cases of DLBCL diagnosed according to the 2016 WHO criteria. 17 Cases were collected from Tokai University Hospital between 2004 and 2011. This study was carried out according to the Declaration of Helsinki of Ethical Principles for Medical Research Involving Human Subjects (IRB 14R‐080).
This series of patients had conventional DLBCL characteristics. Detailed information is shown in Tables 1, 2, and S1. Staging maneuvers and the assessment of treatment responses were standard. The median age of patients was 69 years (range, 14‐97 years) and the male / female ratio was 1.54. The IPI was retrospectively assessed in 106 patients (80%): low risk (37, 34.9%), low‐intermediate risk (36, 34%), high‐intermediate risk (17, 16%), and high risk (16, 12.1%). Treatments were R‐CHOP in 73.2% of cases and R‐CHOP‐like (mainly RCOP) in 22.0%; only 4.9% had other treatments (such as RT, CHOP, and THP‐COP). Among the 118 patients with assessable responses, 89 (75.4%) achieved a complete response, 12 (10.2%) progressive disease, 16 (13.6%) a partial response, and 1 (0.8%) stable disease. One‐, 3‐, 5‐, and 10‐year OS rates were 85%, 68%, 59%, and 46%, respectively.
TABLE 1.
Main pathological characteristics in patients with diffuse large B‐cell lymphoma (training set)
| Characteristic | Frequency (%) | |
|---|---|---|
| Histological | ||
| CD3+ | 0s | (1.6) |
| CD5+ | 12/129 | (9.3) |
| CD20+ | 129/131 | (98.5) |
| CD10+ | 39/130 | (30.0) |
| BCL6+ | 88/129 | (68.2) |
| MUM1+ | 89/129 | (69.0) |
| Non‐GCB (Hans classifier) | 82/129 | (63.6) |
| BCL2+ | 89/129 | (69.0) |
| EBV EBER+ | 10/125 | (7.6) |
| RGS1‐high | 64/108 | (59.3) |
| MYC‐high | 62/119 | (52.1) |
| Molecular | ||
| BCL2 split (FISH) | 13/121 | (10.7) |
| MYC split (FISH) | 13/124 | (10.5) |
| BCL6 split (FISH) | 18/106 | (17.0) |
| BCL2 and MYC split (FISH) | 2/119 | (1.7) |
| BCL2 IHC+ and MYC‐IHC‐high (double expressor) | 39/123 | (31.7) |
| MYC t+ and/or MYC‐IHC‐high | 67/132 | (50.8) |
| High grade B‐cell lymphoma (MYC t+ and BCL2 t+ and/or BCL6 t+) | 5/125 | (4.0) |
| MYD88 L265P mutation | 12/121 | (9.9) |
The cut‐off for positivity for the immunohistochemical (IHC) markers for B lymphocytes of DLBCL using the Hans algorithm (CD10, BCL6, IRF4/MUM1) was set at the conventional 30%. BCL2 was considered positive if more than 50% of the tumoral cells were positive. High regulator of G‐protein signaling 1 (RGS1) expression associates with poor prognosis in DLBCL. RGS1‐high corresponds to expression 2‐3+ as we have previously described. 21 MYC translocation positive cases positively correlated with higher IHC expression for MYC protein (Fisher’s exact test, P = .003); MYC‐high cut‐off was 40%, and after digital image quantification was set at 22%.
Abbreviations: EBER, EBV‐encoded small RNA; EBV, Epstein–Barr virus; GCB, germinal center B cell‐like.
TABLE 2.
Main clinical characteristics in patients with diffuse large B‐cell lymphoma (training set)
| Clinical features | Frequency (%) | |
|---|---|---|
| Age >60 y | 96/132 | (72.7) |
| Sex, male | 80/132 | (60.6) |
| Location | ||
| Nodal (+spleen) | 47/132 | (35.6) |
| Waldeyer’s ring | 22/132 | (16.7) |
| Gastrointestinal | 18/132 | (13.6) |
| Other extranodal | 45/132 | (34.1) |
| LDH high | 87/130 | (66.9) |
| High sIL‐2R (>530) | 107/124 | (86.3) |
| High performance status | 11/85 | (12.9) |
| Extranodal >1 site | 23/69 | (33.3) |
| Stage III‐IV | 50/122 | (41.0) |
| B symptoms | 21/101 | (20.8) |
| IPI high intermediate + high | 33/106 | (31.1) |
| Treatment | ||
| R‐CHOP | 90/123 | (73.2) |
| R‐CHOP‐like | 27/123 | (22.0) |
| Other | 6/123 | (4.9) |
| Clinical response | 89/118 | (75.4) |
Abbreviations: IPI, International Prognostic Index; LDH, lactate dehydrogenase; R‐CHOP, rituximab, cyclophosphamide, hydroxydaunorubicin (doxorubicin), oncovin (vincristine), prednisone.
The validation set included 159 cases of DLBCL diagnosed between 2009 and 2012. The clinicopathologic characteristics of these cases are shown in Table S2. In summary, this series was compatible with a conventional series of DLBCL NOS, with 73% of cases being older than 60 years, 37.7% nodal, 44.9% with IPI high‐intermediate and high, and 96.6% treated with rituximab. One‐, 3‐, 5‐, and 10‐year OS rates were 75%, 59%, 53%, and 41%, respectively.
Validation of the PTX3 gene marker in an independent series of DLBCL from Europe and North America was undertaken in GSE10846, which is publicly available in the NCBI database.
2.2. Phenotypic characterization of DLBCL samples
Immunohistochemical analysis was carried out on FFPE whole tissue sections using automatic equipment (Leica Bond‐Max system and reagents; Leica Biosystems). Immunophenotypes included the markers of CD3, CD5, CD20, CD10, MUM1 (IRF4), BCL2, BCL6, and Ki‐67 (Novocastra/Leica Biosystems), RGS1 (rabbit polyclonal; Thermo Fisher Scientific), and MITF (C5/D5/MAB10775; Abnova). We used the standard 30% cut‐off for the positivity of the Hans classifiers CD10, BCL6, and IRF4/MUM1. BCL2 was considered positive if 50% or more of the tumor cells were positive, and MYC if 40% or more of the tumor nuclei were positive. The macrophagic signature included the pan‐macrophage markers of CD68 (514H12), M1‐like CD16 (2H7), M2‐like CD163 (10D6), PTX3 (PPZ1228; Perseus Proteomics), and IL‐10 (LS‐B7432; Lifespan Bioscience). We used FOXP3 to identify Tregs (236A; CNIO). The expression of these markers was also examined in reactive tonsils. Staining was initially evaluated using an ordinal variable as 0, +1, +2, and +3 based on the density of cells positive for each marker. Digital image quantification using Fiji software was undertaken to assess the total percentage of positive cells in the microenvironment as previously described. 19 , 20 In summary, a large representative area was digitalized and the number of DAB‐positive pixels was identified in the blue stack. The percentage of positive cells was calculated as follows: percentage = ([positive pixels / all pixels] × 100).
The EBER in situ hybridization to detect EBV (Leica), FISH with the BCL2 split signal probe, MYC split signal probe, and BCL6 split signal probe (#Y5407, #Y5410, and #5408; Dako/Agilent Technologies) 21 , 22 to detect BCL2, MYC, and BCL6 rearrangements, respectively, and PCR/Sanger sequencing to detect the MYD88 (L265P) mutation 23 were also carried out.
Programmed cell death‐ligand 1 (#E1J2J; Cell Signaling Technology) and CSF1R (#FER216; CNIO) IHC were recovered from our recent previous publications (https://doi.org/10.3390/ai2010008 and https://doi.org/10.3390/hemato2020011, respectively).
2.3. Gene expression analysis
Gene expression profiling of a representative set of 37 cases was undertaken using RNA extracted from FFPE samples. The nCounter Immuno‐oncology and Lymph2Cx assay panels were used (NanoString Technology). Housekeeping gene normalization was calculated using the log2((normalData[,i]/hkGeomMeans[i])) formula.
2.4. Statistical analysis
All statistical analyses were undertaken using SPSS software (version 26; IBM). The χ2 and/or Fisher’s exact tests and the Mann–Whitney U test were used for group comparisons, and the Kaplan–Meier and log‐rank tests and Cox’s regression analysis for survival analyses. Overall survival was defined from the date of diagnosis to the last contact date. Progression‐free survival was defined from the date of diagnosis to disease progression. Bivariate correlation was carried out using Pearson and Spearman’s tests. The significance level was set at .05. 24 , 25 , 26 , 27 , 28 , 29
3. RESULTS
3.1. Clinical and histological features of patients in the training set
Detailed information is shown in Tables 1, 2, and S1. The most relevant histological features of this series were as follows: DLBCL was positive for CD5 in 9.3% of cases, BCL2 in 69.0%, and EBER in 7.6%, and RGS1 was highly expressed in 59.3% of cases. The cell‐of‐origin analysis based on the Hans classifier showed non‐GCB in 63.6% of cases. Molecular analyses revealed the MYC FISH split signal in 10.5% of cases and the MYD88 L265P mutation in 9.9%. Clinical variables that were associated with unfavorable OS were a patient age older than 60 years, high LDH, high sIL‐2R, more than one extranodal site, stage III‐IV, and IPI high‐intermediate and high. Similar results were obtained for PFS.
According to the revised 4th Edition of the WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, the HGBL with MYC and BCL2 and/or BCL6 rearrangements category therefore includes: (a) double‐hit cases previously classified as B‐cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma; (b) blastoid cases with a double hit; and (c) cases with a DLBCL NOS morphology that, upon evaluation, have rearrangements of MYC and BCL2 and/or BCL6. In the training series, we identified five cases of DLBCL NOS morphology that, following FISH evaluation, were reclassified into HGBCL (Table 1). Also, in the training set, we identified 10 cases that were positive for EBV by in situ hybridization. Those cases fell within the WHO category of EBV‐positive DLBCL NOS.
3.2. Distribution of markers in DLBCL and relationships with histopathological features in the training set
The frequencies of all markers are shown in Table 3, including means ±SD and medians. Characteristic images for each marker are shown in Figure 1. The expression of the markers was low, with a mean percentage of less than 20% of all cells, except CD163, which had a frequency of 22.7% ± 19.4%. The following correlations were observed with pathological features (Table 4): (a) the non‐GCB subtype expressed high CD16, CD163, PTX3, and IL‐10; (b) EBER‐positive cases expressed high CD163 and low PTX3; (c) HGBCL had higher CD163 but lower PTX3; and (d) double‐expressor (ie, BCL2‐positive, MYC above 22% by digital quantification) by high CD16 (Table S3).
TABLE 3.
Distribution of markers in the series of patients with diffuse large B‐cell lymphoma (training set)
| Marker | Frequency | Best OS cut‐off | Low | High | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | ±SD | Median | No | % | No | % | ||
| CD68 | 15.2 | 8.4 | 13.5 | 28.0 | 120.0 | 92.3 | 10.0 | 7.7 |
| CD16 | 6.2 | 9.6 | 0.9 | 0.7 | 56.0 | 45.9 | 66.0 | 54.1 |
| MITF | 2.9 | 2.8 | 2.1 | 3.1 | 81.0 | 66.4 | 41.0 | 33.6 |
| CD163 | 22.7 | 19.4 | 17.9 | 20.5 | 75.0 | 57.3 | 56.0 | 42.7 |
| PTX3 | 11.0 | 17.0 | 1.5 | 49.0 | 125.0 | 95.4 | 6.0 | 4.6 |
| IL‐10 | 8.3 | 9.9 | 4.9 | 9.6 | 39.0 | 66.1 | 20.0 | 33.9 |
| FOXP3 | 2.2 | 2.8 | 1.0 | 4.5 | 111.0 | 84.1 | 21.0 | 15.9 |
| RGS1 | 9.3 | 11.7 | 4.8 | 3.0 | 44.0 | 40.7 | 64.0 | 59.3 |
The immune microenvironment markers were initially evaluated as an ordinal variable a 0, 1+, 2+, and 3+ as <1%, 1%‐5%, 5%‐20%, and >20%, respectively, under the optical microscope. After digitalization the percentage of positive cells was quantified using Fiji software. Then, the best cut‐off for the overall survival (OS) was found from the quantitative data (ie, the most significant P value). Figure 1 shows immunohistochemical images of the different immune markers with the evaluation reference under the microscope and a characteristic image of high values for each marker that were associated with poor OS.
Abbreviations: IL‐10, interleukin‐10; MITF, microphthalmia transcription factor; PTX3, pentraxin 3; RGS1, regulator of G‐protein signaling 1.
FIGURE 1.
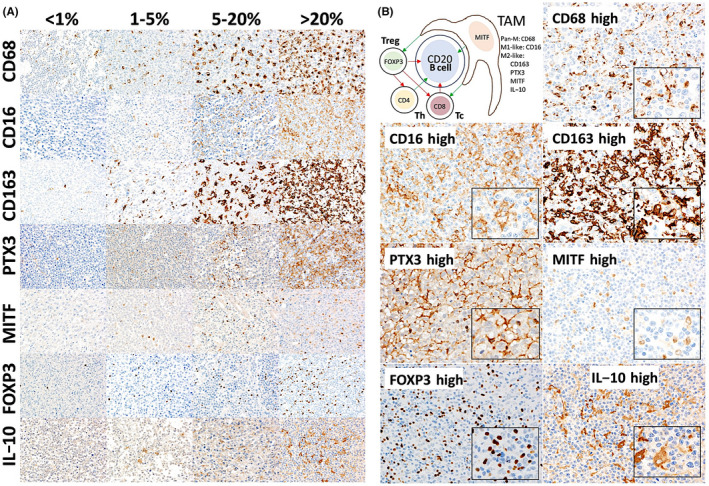
A, Distribution of tumor‐associated macrophages (TAMs) and FOXP3‐positive regulatory T lymphocytes (Tregs) in diffuse large B‐cell lymphoma (DLBCL). The frequencies of different markers of macrophages (TAMs) and FOXP3+ Tregs varied between samples. The following markers were examined: CD68 (pan‐macrophages), CD16 (pro‐inflammatory, M1‐like, with cytotoxic and antitumoral properties), CD163, pentraxin 3 (PTX3), microphthalmia transcription factor (MITF), and interleukin‐10 (IL‐10) (protumoral M2‐like; PTX3 and IL‐10 for the immune regulatory M2c‐like subtype). Original magnification, ×200 (Olympus BX63). B, Histological features of TAMs and FOXP3‐positive Tregs in DLBCL. This figure shows the immunohistochemical expression of different markers and histological characteristics of TAMs and Tregs, focusing on DLBCL cases with a high infiltration of these cells in the immune microenvironment. The effects of and relationship between the different cells in the microenvironment in DLBCL are also shown; red indicates inhibition, green indicates activation. Original magnification, ×200 (Olympus BX53)
TABLE 4.
Correlation between markers with the cell‐of‐origin classification, Epstein–Barr virus‐encoded small RNA (EBER), and high‐grade B‐cell lymphoma (HGBCL)
| Marker | Cell‐of‐origin (Hans classifier) a | ||||
|---|---|---|---|---|---|
| P value | GCB | Non‐GCB | |||
| Mean | ±SD | Mean | ±SD | ||
| CD68 | .079 | 14.4 | 7.1 | 16.4 | 8.4 |
| CD16 | .00002 | 0.7 | 1.5 | 9.2 | 11.5 |
| MITF | .288 | 3.1 | 3.7 | 3.3 | 2.9 |
| CD163 | .003 | 19.2 | 19.9 | 30.6 | 19.6 |
| PTX3 | .026 | 4.7 | 11.8 | 18.4 | 20.7 |
| IL‐10 | .020 | 3.1 | 4.0 | 11.4 | 11.0 |
| FOXP3 | .645 | 1.8 | 2.6 | 2.2 | 2.9 |
| RGS1 | .126 | 6.6 | 6.6 | 15.2 | 14.8 |
| Marker | EBER | ||||
|---|---|---|---|---|---|
| P value | Positive | Negative | |||
| Mean | ±SD | Mean | ±SD | ||
| CD68 | .875 | 16.5 | 8.0 | 15.5 | 8.2 |
| CD16 | .362 | 6.6 | 10.5 | 7.1 | 10.8 |
| MITF | .482 | 3.5 | 3.0 | 3.1 | 3.2 |
| CD163 | .035 | 38.5 | 24.1 | 25.7 | 18.6 |
| PTX3 | .017 | 24.7 | 23.7 | 13.5 | 18.5 |
| IL‐10 | .160 | 5.8 | 7.5 | 9.3 | 10.8 |
| FOXP3 | .554 | 4.6 | 4.1 | 1.8 | 2.4 |
| RGS1 | .179 | 21.7 | 19.8 | 11.6 | 11.8 |
| Marker | HGBCL genotype by FISH | ||||
|---|---|---|---|---|---|
| P value | Positive | Negative | |||
| Mean | ±SD | Mean | ±SD | ||
| CD68 | .369 | 12.1 | 9.5 | 15.1 | 8.1 |
| CD16 | .890 | 2.1 | 9.8 | 2.1 | 2.3 |
| MITF | .748 | 2.1 | 2.5 | 3.1 | 2.8 |
| CD163 | .045 | 49.6 | 29.4 | 22.1 | 18.4 |
| PTX3 | .003 | 0.2 | 0.2 | 11.9 | 17.5 |
| IL‐10 | .464 | 4.1 | 3.6 | 8.8 | 10.2 |
| FOXP3 | .084 | 0.6 | 0.9 | 2.3 | 2.9 |
| RGS1 | .905 | 18.1 | 26.6 | 8.7 | 11 |
Abbreviations: GCB, germinal center B cell‐like; IL‐10, interleukin‐10; MITF, microphthalmia transcription factor; PTX3, pentraxin 3; RGS1, regulator of G‐protein signaling 1.
The same results were found when analyzing the diffuse large B‐cell lymphoma, not otherwise specified, only group.
The markers were also correlated with the double‐expressor, MYD88 L265P mutation, and cell‐of‐origin in DLBCL NOS (Tables S3‐S5).
The immunohistochemical analysis of PD‐L1 showed a positive correlation with CD16, CD163, IL‐10, and RGS1 (Table S6).
3.3. Relationships with OS and PFS in the training set
The best (ie, optimal) cut‐off for each marker was defined based on the P value in the log‐rank test for OS. On average, between all markers, 31.1% of cases were in the poor prognosis group (Table 3).
Detailed information is shown in Table 3. Overall survival figures are shown in Figure 2. In the univariate analysis, the markers associated with poor OS were macrophages with high expression of CD68, CD16, MITF, CD163, PTX3, and IL‐10, and low infiltration of FOXP3+ Tregs. In the multivariate analysis of all of these markers and IPI, only PTX3 and IPI retained their significance (Table 5). Therefore, PTX3 was the most relevant marker in this model in addition to IPI.
FIGURE 2.
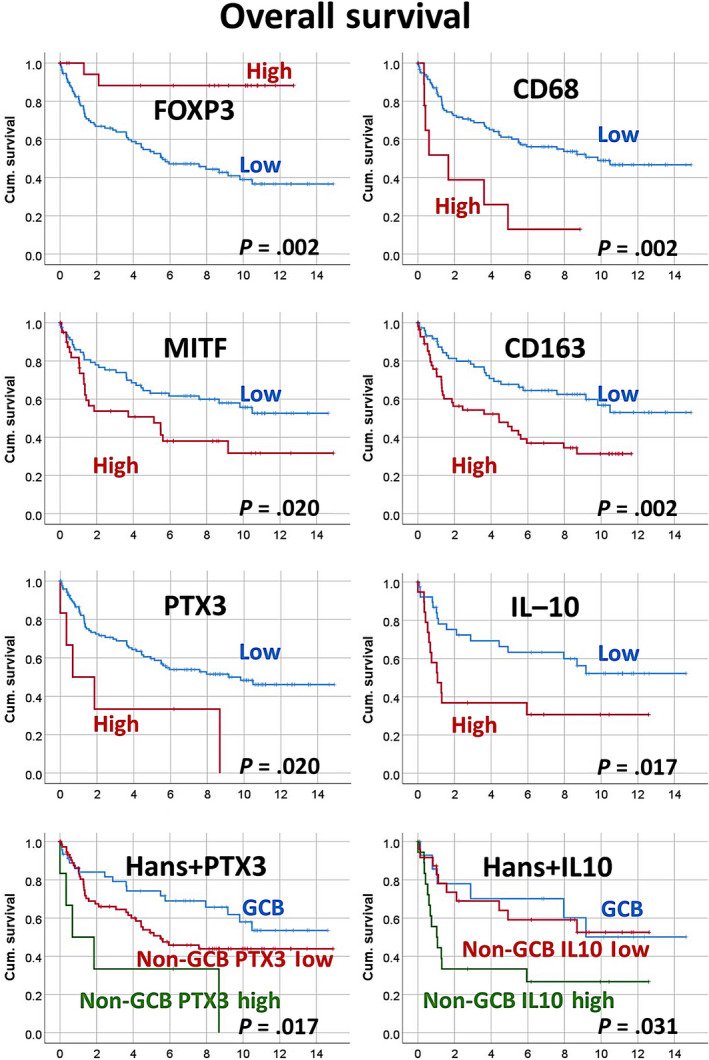
Relationship between markers and overall survival (OS) in the training set of patients with diffuse large B‐cell lymphoma (DLBCL). High expression of macrophage markers was associated with poor prognosis, including OS and progression‐free survival (PFS). High infiltration of regulatory T lymphocytes was associated with a favorable prognosis (OS). Of note, the analysis was repeated stratifying for DLBCL not otherwise specified (NOS), Epstein–Barr virus‐encoded small RNA+ DLBCL, and high‐grade B‐cell lymphoma, both for OS and PFS. In the DLBCL NOS group, the same results were found. Cum., cumulative; GCB, germinal center B cell‐like
TABLE 5.
Correlation between markers and outcome in patients with diffuse large B‐cell lymphoma
| Marker | OS | PFS | ||||||
|---|---|---|---|---|---|---|---|---|
| P value | HR | 95% CI | P value | HR | 95% CI | |||
| Univariate analysis | ||||||||
| CD68 | .002 | 3.2 | 1.5 | 7.2 | .0001 | 4.4 | 1.9 | 10.0 |
| CD16 | .049 a | 1.5 | 0.9 | 2.6 | .022 | 2.0 | 1.1 | 3.5 |
| MITF | .020 | 1.9 | 1.1 | 3.2 | .249 | 1.4 | 0.8 | 2.5 |
| CD163 | .002 | 2.1 | 1.3 | 3.5 | .002 | 2.4 | 1.4 | 4.1 |
| PTX3 | .020 | 2.8 | 1.1 | 7.1 | .015 | 3.3 | 1.2 | 9.2 |
| IL‐10 | .017 | 2.4 | 1.1 | 5.0 | .016 | 2.6 | 1.2 | 5.9 |
| FOXP3 | .002 | 0.1 | 0.0 | 0.6 | .123 | 0.5 | 0.2 | 1.2 |
| Multivariate analysis b | ||||||||
| CD68 | .457 | 2.7 | 0.2 | 37.4 | .026 | 24.1 | 1.5 | 394.1 |
| CD16 | .604 | 0.7 | 0.2 | 2.8 | .536 | 0.7 | 0.2 | 2.4 |
| MITF | .386 | 1.8 | 0.5 | 6.5 | .94 | 1.0 | 0.3 | 3.2 |
| CD163 | .829 | 1.2 | 0.3 | 5.3 | .906 | 0.9 | 0.2 | 3.5 |
| PTX3 | .033 | 6.1 | 1.2 | 31.9 | .238 | 2.7 | 0.5 | 14.1 |
| IL‐10 | .287 | 2.1 | 0.5 | 8.0 | .269 | 2.1 | 0.6 | 7.6 |
| FOXP3 | .146 | 0.3 | 0.1 | 1.5 | .826 | 1.1 | 0.3 | 3.9 |
| IPI | .035 | 2.0 | 1.1 | 3.9 | .359 | 1.3 | 0.8 | 2.2 |
| Multivariate analysis c | ||||||||
| PTX3 | .020 | 4.5 | 1.3 | 18.2 | – | – | – | – |
| IL‐10 | .009 | 7.3 | 1.7 | 15.6 | .065 | 4 | 0.9 | 17.6 |
| FOXP3 | .111 | 0.3 | 0.1 | 1.3 | – | – | – | – |
| EBER | .023 | 4.8 | 4.8 | 1.2 | – | – | – | – |
| CD68 | – | – | – | – | .014 | 26.1 | 1.9 | 349 |
Abbreviations: –, no data; CI, confidence interval; HR, hazard ratio; OS, overall survival; PFS, progression‐free survival.
Log‐rank (Mantel‐Cox) P = .136; Breslow (generalized Wilcoxon) P = .049.
Multivariate analysis, method = enter.
Multivariate analysis, Cox regression, method = backward conditional, predictors = CD68, CD16, microphthalmia transcription factor (MITF), CD163, pentraxin 3 (PTX3), interleukin‐10 (IL‐10), FOXP3, International Prognostic Index (IPI), cell‐of‐origin (Hans classifier), Epstein–Barr virus‐encoded small RNA (EBER), and high‐grade B‐cell lymphoma genotype.
Similar results were obtained for PFS (Table 5 and Figure S1). In the univariate analysis, the high expression of CD68, CD16, CD163, PTX3, and IL‐10 correlated with unfavorable PFS.
The analysis was repeated for all markers stratifying for the DLBCL NOS, EBER+ DLBCL, and HGBCL subtypes (Figure S1 and Table 6). The results showed that the previous findings in DLBCL were also confirmed in the DLBCL NOS group.
TABLE 6.
Prognostic value of markers when stratifying for diffuse large B‐cell lymphoma (DLBCL) subtypes
| Marker | P value (Kaplan–Meier with log‐rank test) | |||||
|---|---|---|---|---|---|---|
| OS | PFS | |||||
| DLBCL NOS | EBER+DLBCL | HGBCL | DLBCL NOS | EBER+DLBCL | HGBCL | |
| CD68 | 0.011 | 0.538 | NA | 0.001 | 0.705 | NA |
| CD16 | 0.142 | 0.673 | 0.317 | 0.033 | 0.736 | 0.317 |
| MITF | 0.007 | 0.283 | 0.317 | 0.136 | 0.050 | 0.317 |
| CD163 | 0.001 | 0.314 | 0.515 | 0.002 | 0.610 | 0.918 |
| PTX3 | 0.007 | 0.003 | NA | 0.012 | NA | NA |
| IL‐10 | <0.001 | NA | NA | <0.001 | NA | NA |
| FOXP3 | 0.002 | 0.110 | NA | 0.061 | 0.920 | NA |
| RGS1 | 0.140 | 0.275 | 0.157 | 0.658 | 0.307 | 0.157 |
Abbreviations: EBER, Epstein–Barr virus‐encoded small RNA; HGBCL, high‐grade B‐cell lymphoma; IL‐10, interleukin‐10; MITF, microphthalmia transcription factor; NA, nonassessable; NOS, not otherwise specified; OS, overall survival; PFS, progression‐free survival; PTX3, pentraxin 3; RGS1, regulator of G‐protein signaling 1.
3.4. Gene expression analysis in the training set
The results of the gene expression analysis are shown in Figure 3 and Table S7. The 38 genes in the Lymph2Cx assay panel were merged with the 770 genes in the immuno‐oncology panel. In 37 representative cases, gene expression was compared between two groups based on the IHC expression of PTX3: high vs low, using a cut‐off of 10%. In this analysis, we identified which genes and pathways differed between the two groups to elucidate the role of PTX3 in the pathogenesis of DLBCL. Of the 37 cases, 4 were EBER+ DLBCL and 33 DLBCL NOS.
FIGURE 3.
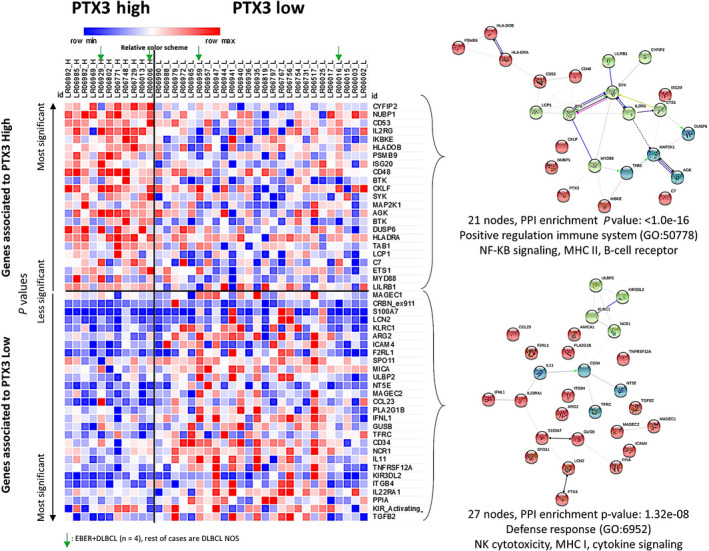
Gene expression analysis of the training set of patients with diffuse large B‐cell lymphoma (DLBCL). A set of 37 cases of DLBCL from Tokai University Hospital was selected and a gene expression analysis was carried out using Lymph2Cx and immuno‐oncology NanoString panels. Nonvariant genes between the two groups were excluded and significant genes were ranked according to P values and group associations. The heatmap visualization shows how gene expression between high and low pentraxin 3 (PTX3) groups differed, indicating a different pathogenic mechanism. A relative color scheme uses the minimum and maximum values in each row to convert values to colors and allows for the easier visualization of differences between the two groups. Genes involved in innate and adaptive immune responses and macrophage and Toll‐like receptor pathways, including CYFIP2, NUBP1, CD53, IL2RG, IKBKE, BTK, SYK, MAP2K1, and MYD88, were upregulated in the high PTX3 group. Genes involved in regulation, interleukins, adhesion, and natural killer (NK) cell function pathways, including TFFB2, KIR2DS1, PPIA, IL22RA1, and ITGB4, were upregulated in the low PTX3 group. Using the STRING database, the 21 genes associated with the high PTX3 group (top right panel) corresponded to the pathways positively regulating the immune system, nuclear factor‐κB signaling, MHC II, and B‐cell receptors. In contrast, the 27 genes associated with the low PTX3 group (bottom right panel) corresponded to the defense response pathway, NK cytotoxicity, MHC I, and cytokine signaling
Nonvariant genes were excluded from the analysis, and significant genes were ranked according to P values and group associations. Additionally, genes were annotated by the following 20 immune response categories: adhesion, antigen processing, B‐cell functions, cell cycle, cell functions, chemokines, complement, cytokines, cytotoxicity, interleukins, leukocyte functions, macrophage functions, microglial functions, NK cell functions, pathogen defense regulation, senescence, T‐cell functions, Toll‐like receptor, TNF superfamily, and transporter functions (Table S7). Genes were also annotated using the STRING database of known and predicted protein–protein interactions. Interactions included direct (physical) and indirect (functional) associations.
In Figure 3, the gene expression is shown using a relative color scheme. The relative color scheme used the minimum and maximum values in each row to convert values to colors and allowed for the easier visualization of differences between the two groups.
Eighteen genes were identified in the low PTX3 group. The most significantly expressed genes were TGFB2 (interleukins, P = .009), KIR2DS1 (regulation and NK cell function, P = .009), PPIA (signaling, apoptosis, HK, P = .012), IL22RA1 (chemokine, P = .016), and ITGB4 (adhesion, P = .017). This group had a higher number of genes involved in regulation, interleukin, adhesion, and NK cell functions.
Twenty‐one genes were identified in the high PTX3 group. The most significantly expressed genes were CYFIP2 (innate immune response, P = .002), NUBP1 (signaling, apoptosis, HK, P = .002), CD53 (adaptive immune response, P = .008), IL2RG (chemokines, P = .008), and IKBKE (innate immune response, P = .011). Other genes of interest were BTK (adaptive immune response, P = .021), SYK (macrophage function, P = .023), and MAP2K1 (innate immune response, P = .028). MYD88 (innate immune response, Toll‐like receptor) was slightly overexpressed in this group (P = .128). This group had higher numbers of genes involved in antigen processing and adaptive and innate immune responses. IKBKE, BTK, SYK, and MYD88 formed a cluster with the biological processes Gene Ontology (GO) of innate immune responses and the Toll‐like receptor signaling pathway. Using the STRING database, the 21 genes associated with the high PTX3 group corresponded to the pathway for the positive regulation of the immune system, NF‐κB signaling, MHC II, and B‐cell receptors. In contrast, the 27 genes associated with the low PTX3 group were involved in the defense response pathway, NK cytotoxicity, MHC I, and cytokine signaling (Figure 3).
3.5. Evaluation of the PTX3 marker in the validation set
The correlation between PTX3 and the survival outcomes of patients was validated in an independent series from Tokai University Hospital (Figure 4). The clinicopathologic characteristics of this validation series are shown in Table S2. The immunohistochemical analysis of PTX3 showed that high expression correlated with poor OS (high [n = 28] vs low expression [n = 120], hazard risk = 2.4, P = .003; 95% CI, 1.4‐4.2). Additionally, PTX3 stratified the non‐GCB subtype, as shown in Figure S2 (hazard risk = 1.8, P = .004; 95% CI, 1.2‐2.8). In the Figure S2A‐B, the analysis also included the stratification for EBER‐ and EBER+ DLBCL cases, and OS (A) and PFS (B).
FIGURE 4.
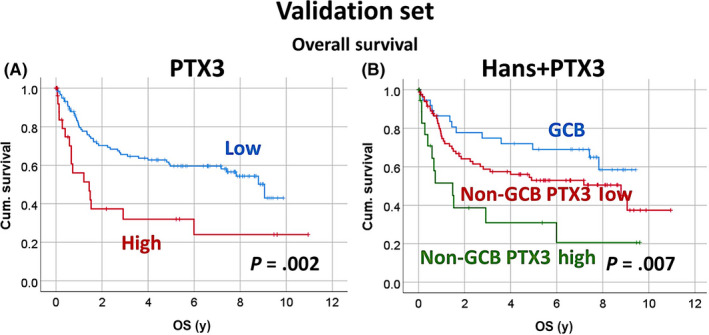
Relationship between pentraxin 3 (PTX3) and overall survival (OS) in the validation set of patients with diffuse large B‐cell lymphoma. The immunohistochemical expression of PTX3 was analyzed in a validation set. The analysis of OS confirmed that the high expression of PTX3 correlated with poor OS. PTX3 expression also stratified the non–germinal center B cell‐like (GCB) subtype. In the supplementary Figure 2A‐B, the analysis also included the stratification for EBER‐ and EBER+ DLBCL cases, and OS (A) and PFS (B). Cum., cumulative
3.6. Validation of PTX3 marker in an independent series
The publicly available GSE10846 DLBCL gene expression dataset was used to validate the association of PTX3 and poor OS of patients. This series comprises 414 cases; 181 (44%) were treated with CHOP‐like therapy, and 233 (56%) with R‐CHOP‐like therapy. The R‐CHOP‐like cases were comparable to the Tokai series. In concurrence, we found high expression of PTX3 in the R‐CHOP‐like therapy group was associated with poor OS of patients: 5‐year OS, 76% (95% CI, 85%‐67%) vs 55% (95% CI, 68%‐42%). Kaplan–Meier with log‐rank test, P = .001; hazard risk = 2.3 (95% CI, 1.4‐3.8, P = .001) (Figure 5).
FIGURE 5.
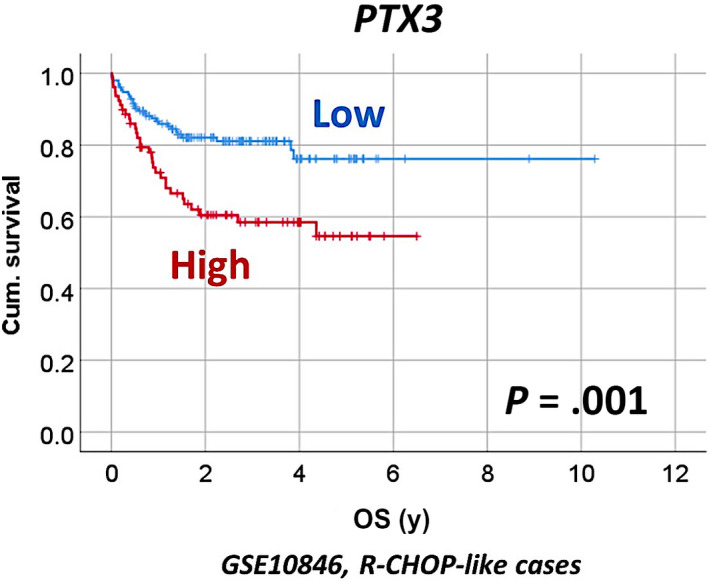
Correlation between the gene expression of PTX3 and overall survival (OS) in the GSE10846 series of patients with diffuse large B‐cell lymphoma (DLBCL). The results were validated in an independent series of DLBCL from Europe and North America. High PTX3 expression correlated with poor prognosis of the patients. Cum., cumulative; R‐CHOP, rituximab, cyclophosphamide, hydroxydaunorubicin (doxorubicin), oncovin (vincristine), prednisone
High PTX3 expression also correlated with other clinical variables including ECOG performance status of 2 or higher, LDH ratio greater than 1, and National Comprehensive Cancer Network‐IPI high‐intermediate/high (all P values ≤0.01) (Table 7).
TABLE 7.
Correlation between clinicopathological characteristics and PTX3 gene expression (GSE10846 dataset) in diffuse large B‐cell lymphoma (DLBCL)
| Variable | PTX3 | P value | |
|---|---|---|---|
| High | Low | ||
| Diagnosis | DLBCL | − | |
| Tissue | Lymph node | − | |
| Treatment | R‐CHOP‐like | − | |
| No. | 79/233 (34) | 154/233 (66) | − |
| Sex | |||
| Female | 69/99 (70) | 30/99 (30) | .318 |
| Male | 85/134 (63) | 49/134 (37) | |
| Age, y | |||
| ≤60 | 72/113 (64) | 41/113 (36) | .457 |
| >60 | 82/120 (68) | 38/120 (32) | |
| Ann Arbor stage | |||
| I‐II | 78/105 (74) | 27/105 (26) | .014 |
| III‐IV | 71/121 (59) | 50/121 (41) | |
| ECOG performance status | |||
| <2 | 116/158 (73) | 42/158 (27) | <.001 |
| ≥2 | 21/52 (40) | 31/52 (60) | |
| LDH ratio | |||
| <1 | 77 /99 (78) | 22/99 (22) | <.001 |
| >1 | 48/93 (52) | 45/93 (48) | |
| >1 extranodal site | |||
| 0‐1 | 117/173 (67) | 56/173 (32) | .062 |
| >1 | 15/30 (50) | 15/30 (50) | |
| NCCN‐IPI | |||
| Low+low‐intermediate risk | 73/101 (72) | 28/101 (28) | .010 |
| High+high‐intermediate risk | 33/63 (52) | 30/63 (48) | |
| Cell of origin | |||
| GCB | 71/107 (66) | 36/107 (34) | .938 |
| ABC+ unclassified | 83/126 (66) | 43/126 (34) | |
Data are shown n/N (%).
Abbreviations: −, no data; ABC, activated B‐cell‐like; GCB, germinal center B‐cell‐like; LDH, lactate dehydrogenase; NCCN‐IPI, National Comprehensive Cancer Network‐International Prognostic Index; R‐CHOP, rituximab, cyclophosphamide, hydroxydaunorubicin (doxorubicin), oncovin (vincristine), prednisone.
Interestingly, at RNA levels, PTX3 positively correlated with CD16, MITF, CD163, CSF1R, and RGS1 (Table 8).
TABLE 8.
Correlation between PTX3 and other relevant genes in diffuse large B‐cell lymphoma (GSE10846 series)
| Bivariate correlations | PTX3 | |
|---|---|---|
| Correlation coefficient | P value | |
| CD16 | 0.193 | .005 |
| MITF | 0.149 | .023 |
| CD163 | 0.237 | <.001 |
| IL‐10 | 0.037 | .573 |
| CSF1R | 0.18 | .006 |
| CD274 (PD‐L1) | 0.003 | .959 |
| TNFAIP8 | −0.121 | .065 |
| CASP8 | −0.116 | .078 |
| FOXP3 | −0.051 | .435 |
| RGS1 | 0.212 | .001 |
| MYC | 0.143 | .029 |
4. DISCUSSION
In this study, we showed that: (a) the tumor infiltration of macrophages in DLBCL differed and the expression of macrophage markers varied; (b) the high expression of macrophage markers and low infiltration of FOXP3+ Tregs were associated with a poor prognosis in DLBCL; (c) PTX3 and IL‐10 both played an important role in the pathogenesis of DLBCL and were associated with a poor prognosis; and (iv) gene expression differed between the high vs low PTX3 groups.
We examined various factors in a large series of DLBCL from Japan. IPI including age, LDH, clinical stage, and extranodal sites as well as sIL‐2R and the lack of a clinical response correlated with shorter OS. The biological factors associated with a poor prognosis were MUM1 positivity, the non‐GCB phenotype (Hans classifier), and EBER positivity (Table S1). The present results are consistent with the previously reported findings from Europe and North America 30 , 31 ; therefore, our series corresponds to a conventional series of DLBCL.
Tumor‐associated macrophages are abundant in solid cancers 32 , 33 , 34 ; M1‐like TAMs enhance antitumoral host immune responses, whereas M2‐like TAMs have been implicated in tumor progression. 35 Survival outcomes correlate with TAMs in hematological malignancies. A high infiltration of CD68‐positive TAMs has been associated with a poor prognosis in patients with FL. 36 CD68‐positive TAMs, specifically CD163‐positive M2‐like TAMs, have been associated with a poor prognosis in classical Hodgkin lymphomas 37 and the progression of adult T‐cell leukemia/lymphoma, 38 cutaneous T‐cell lymphoma, 39 and T‐cell/histiocyte‐rich large B‐cell lymphoma. 40 Gene expression profiling showed that the ABC subtype of DLBCL was characterized by the activation of the NF‐κB pathway and by markers of macrophages. 2 , 3 , 4 This study confirmed the prognostic relevance of TAMs in de novo DLBCL, including pan‐macrophages (CD68), M1‐like (CD16), and M2‐like (CD163, MITF, PTX3, and IL‐10) TAMs, and FOXP3+ Tregs. The high expression of CD68, CD16, MITF, CD163, PTX3, and IL‐10, but low expression of FOXP3 correlated with poor OS. The high expression of CD16 also correlated with poor OS, but only during the first years of observation (Kaplan–Meier and Breslow statistical tests, P =.049). The cell‐of‐origin and presence of EBV also influenced the prognosis of DLBCL patients. The non‐GCB type had higher expression of CD16, CD163, PTX3, and IL‐10. The EBER‐positive cases also more highly expressed CD163 and PTX3. Regulator of G‐protein signaling 1 is a marker for the architecture of immune tissues and lymphocyte migration and is associated with a poor prognosis in DLBCL. 21 , 41 , 42 The present results point out a relationship between RGS1 and the composition of the immune microenvironment. Regulator of G‐protein signaling 1 correlated with the expression of CD163 and PTX3 (P <.05), and with FOXP3 (P =.062).
A relationship has already been reported between the expression of CD163 and poor prognosis in DLBCL 43 , 44 ; however, we here revealed correlations with other macrophage markers representing different differentiation states. We examined several markers of pan‐macrophages, M1‐like, and M2‐like polarization, and Tregs. High infiltration of macrophages but low infiltration of Tregs were associated with poor prognosis. In the multivariate Cox regression analysis of OS between all macrophage markers and FOXP3, PTX3 and IL‐10 retained their prognostic value. Therefore, these markers are the most relevant in this prognostic model.
We determined the prognostic relevance of PTX3, which will be an important biological marker in lymphoma. Pentraxin 3 is a marker of immune regulatory M2c‐like polarization and NF‐κB activation. 5 , 6 , 7 , 8 The non‐GCB type can be stratified according to PTX3 expression levels: (a) GCB; (b) non‐GCB PTX3 low; and (c) non‐GCB PTX3 high. In comparisons with GCB, non‐GCB PTX3 high was associated with a poorer prognosis, with a hazard risk of 3.9 (P =.007). Although a previous study reported that PTX3 was associated with a poor prognosis in pancreatic carcinoma, 13 this is the first study to show this relationship in hematological malignancies.
We identified the IL‐10 marker. Interleukin‐10 is a major immune regulatory cytokine that acts on many cells in the immune system, in which it exerts marked antiinflammatory effects, thereby limiting excessive tissue disruption by inflammation. It downregulates the expression of Th1 cytokines, MHC class II antigens, and costimulatory molecules on macrophages. It also promotes B‐cell survival, proliferation, and Ab production. Interleukin‐10 contributes to peripherally derived Tregs. 45 It is also a marker of macrophage M2c polarization and many of the M2c‐specific genes identified to date have been implicated in angiogenesis, matrix remodeling, and phagocytosis, including CD163, MMP8, TIMP1, VCAN, SERPINA1, MARCO, PLOD2, PCOCLE2, and F5. 46 Interleukin‐10 also contributes to the pathogenesis and/or development of autoimmune diseases and cancer. 47 We recently reported the expression of IL‐10 in the DLBCL subtype of methotrexate‐associated lymphoproliferative disorder in patients with rheumatoid arthritis 22 ; however, its role in DLBCL NOS had not yet been investigated by our group. In this study, IHC revealed that IL‐10 marked cells with a macrophage morphology. In FL, the high expression of FOXP3 was associated with a good prognosis, 19 which is consistent with the present results for DLBCL.
We examined the differential gene expression between cases with high and low expression of PTX3, because PTX3 is a novel marker of prognosis and pathogenesis of DLBCL. We found that expression profiles differed between the two groups. Genes involved in innate and adaptive immune responses and macrophage and Toll‐like receptor pathways, including CYFIP2, NUBP1, CD53, IL2RG, IKBKE, BTK, SYK, MAP2K1, and MYD88, were upregulated in the high PTX3 group. Genes involved in regulation, interleukins, adhesion, and NK cell function pathways, including TFFB2, KIR2DS1, PPIA, IL22RA1, and ITGB4, were upregulated in the low PTX3 group.
The analysis of PTX3 in a validation series of cases confirmed the correlation with OS in DLBCL patients, including the stratification of the Hans classifier. Therefore, PTX3 is a relevant marker for the pathogenesis of DLBCL.
Epstein–Barr virus produces a specific cytotoxic T cell immunity response, 48 , 49 that plays an important role in controlling the proliferation of virus‐infected cells. Cytotoxic T cells detect EBV peptides bound to MHC molecules in the surface of affected cells, and eliminate these cells by direct lysis. Conversely, the presence of EBV within the lymphoma cell acts as a potential target, using mechanisms of immune evasion. Indeed, EBV‐positive DLBCL cases are associated with changes in the tumoral immune microenvironment. Histological analysis shows high infiltration of macrophages that express high levels of PD‐L1 and indoleamine 2,3‐dioxygenase 50 , 51 ; these factors contribute to the tumor immune escape, 52 much as iatrogenic immunosuppression does in EBV‐positive posttransplant lymphoproliferative disorders. 53 , 54 , 55 , 56 The recruitment of macrophages in the immune microenvironment by EBV is related to vascular endothelial growth factor (VEGF) and granulocyte/macrophage colony‐stimulating factor (GM‐CSF). Keane et al recently reported that EBV+ DLBCL was characterized by higher mRNA expression of CD163. 57 , 58 This result agrees with our IHC results. Nevertheless, in that research only the CD163 M2‐like macrophage marker was analyzed, so we do not know the RNA levels of PTX3 or IL‐10 in their series.
We have recently described the importance of PD‐L1, CSF1R, TNFAIP8, and CASP8 in the pathogenesis and prognosis of DLBCL, and shown how systems biology can help improve the understanding of disease mechanisms. 59 , 60 , 61 , 62 , 63 , 64 , 65 In this research, we showed how PD‐L1 correlated with CD16, RGS1, and IL‐10. The relationship with PTX3 will be analyzed in future publications.
Finally, we used the gene expression data of the GSE10846 dataset, which included 233 cases of DLBCL patients treated with a R‐CHOP‐like therapy. Using this independent series, we confirmed that high gene expression of PTX3 correlated with poor OS of patients.
In conclusion, we showed that the M2c immune regulatory pathway is associated with an unfavorable prognosis in de novo DLBCL. These results provide novel insights into the pathogenesis of DLBCL with applicability to current and future clinical trials.
CONFLICT OF INTEREST
None to declare.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by grants KAKENI 24590430, 15K19061, and 18K15100 by the Japan Society for the Promotion of Science (JSPS) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT). Joaquim Carreras had a Postdoctoral Research Fellowship at Tokai University, School of Medicine with Professor Naoya Nakamura, and he is funded by Tokai University School of Medicine, research incentive assistant plan 2021‐B04. Rifat Hamoudi was funded by Al‐Jalila Foundation (grant number AJF201741), the Sharjah Research Academy (grant number MED001), and University of Sharjah (grant number 1901090258). We want to thank Dr Juan Fernando Garcia (Department of Pathology, MD Anderson Cancer Center Madrid, Madrid, Spain) for the development and characterization of the CSF1R antibody.
Carreras J, Kikuti YY, Hiraiwa S, et al. High PTX3 expression is associated with a poor prognosis in diffuse large B‐cell lymphoma. Cancer Sci.2022;113:334–348. 10.1111/cas.15179
Funding information
The Ministry of Education, Culture, Sports, Science and Technology (MEXT) and The Japan society for the promotion of science (JSPS KAKENI), Grant/Award Number: 24590430, 15K19061, and 18K15100; Tokai University School of Medicine, Research incentive assistant plan Grant/Award Number: 2021‐B04; Al‐Jalila Foundation, Grant/Award Number: AJF201741, AJF2018090; Sharjah Research Academy, Grant/Award Number: MED001; University of Sharjah, Grant/Award Number: 1901090258
REFERENCES
- 1. Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375‐2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large‐B‐cell lymphoma. N Engl J Med. 2002;346:1937‐1947. [DOI] [PubMed] [Google Scholar]
- 3. Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large‐B‐cell lymphomas. N Engl J Med. 2008;359:2313‐2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell‐like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861‐1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor‐associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549‐555. [DOI] [PubMed] [Google Scholar]
- 6. Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231‐237. [DOI] [PubMed] [Google Scholar]
- 7. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677‐686. [DOI] [PubMed] [Google Scholar]
- 8. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889‐896. [DOI] [PubMed] [Google Scholar]
- 9. Kambayashi T, Laufer TM. Atypical MHC class II‐expressing antigen‐presenting cells: can anything replace a dendritic cell? Nat Rev Immunol. 2014;14:719‐730. [DOI] [PubMed] [Google Scholar]
- 10. Kim HS, Lee JH, Han HD, et al. Autocrine stimulation of IL‐10 is critical to the enrichment of IL‐10‐producing CD40(hi)CD5(+) regulatory B cells in vitro and in vivo. BMB Rep. 2015;48:54‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Germano G, Frapolli R, Simone M, et al. Antitumor and anti‐inflammatory effects of trabectedin on human myxoid liposarcoma cells. Cancer Res. 2010;70:2235‐2244. [DOI] [PubMed] [Google Scholar]
- 12. Diamandis EP, Goodglick L, Planque C, Thornquist MD. Pentraxin‐3 is a novel biomarker of lung carcinoma. Clin Cancer Res. 2011;17:2395‐2399. [DOI] [PubMed] [Google Scholar]
- 13. Kondo S, Ueno H, Hosoi H, et al. Clinical impact of pentraxin family expression on prognosis of pancreatic carcinoma. Br J Cancer. 2013;109:739‐746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Q, Ni H, Lan L, Wei X, Xiang R, Wang Y. Fra‐1 protooncogene regulates IL‐6 expression in macrophages and promotes the generation of M2d macrophages. Cell Res. 2010;20:701‐712. [DOI] [PubMed] [Google Scholar]
- 15. Komohara Y, Niino D, Ohnishi K, Ohshima K, Takeya M. Role of tumor‐associated macrophages in hematological malignancies. Pathol Int. 2015;65:170‐176. [DOI] [PubMed] [Google Scholar]
- 16. Takeya M, Komohara Y. Role of tumor‐associated macrophages in human malignancies: friend or foe? Pathol Int. 2016;66:491‐505. [DOI] [PubMed] [Google Scholar]
- 17. Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117:5019‐5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carreras J, Lopez‐Guillermo A, Kikuti YY, et al. High TNFRSF14 and low BTLA are associated with poor prognosis in follicular lymphoma and in diffuse large B‐cell lymphoma transformation. J Clin Exp Hematop. 2019;59:1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carreras J, Lopez‐Guillermo A, Fox BC, et al. High numbers of tumor‐infiltrating FOXP3‐positive regulatory T cells are associated with improved overall survival in follicular lymphoma. Blood. 2006;108:2957‐2964. [DOI] [PubMed] [Google Scholar]
- 20. Carreras J, Lopez‐Guillermo A, Roncador G, et al. High numbers of tumor‐infiltrating programmed cell death 1‐positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J Clin Oncol. 2009;27:1470‐1476. [DOI] [PubMed] [Google Scholar]
- 21. Carreras J, Kikuti YY, Bea S, et al. Clinicopathological characteristics and genomic profile of primary sinonasal tract diffuse large B cell lymphoma (DLBCL) reveals gain at 1q31 and RGS1 encoding protein; high RGS1 immunohistochemical expression associates with poor overall survival in DLBCL not otherwise specified (NOS). Histopathology. 2017;70:595‐621. [DOI] [PubMed] [Google Scholar]
- 22. Carreras J, Yukie Kikuti Y, Miyaoka M, et al. Genomic profile and pathologic features of diffuse large B‐cell lymphoma subtype of methotrexate‐associated lymphoproliferative disorder in rheumatoid arthritis patients. Am J Surg Pathol. 2018;42:936‐950. [DOI] [PubMed] [Google Scholar]
- 23. Ogura G, Kikuti YY, Kikuchi T, Carreras J, Sato T, Nakamura N. MYD88 (L265P) mutation in malignant lymphoma using formalin‐fixed paraffin‐embedded section. J Clin Exp Hematop. 2013;53:175‐177. [DOI] [PubMed] [Google Scholar]
- 24. Aguirre‐Gamboa R, Gomez‐Rueda H, Martínez‐Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsuda S, Carreras J, Kikuti Y, et al. Prediction of steroid demand in the treatment of patients with ulcerative colitis by immunohistochemical analysis of the mucosal microenvironment and immune checkpoint: role of macrophages and regulatory markers in disease severity. Pathol Int. 2019;69:260‐271. [DOI] [PubMed] [Google Scholar]
- 26. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267‐273. [DOI] [PubMed] [Google Scholar]
- 28. Carreras J, Hamoudi R, Nakamura N. Artificial intelligence analysis of gene expression data predicted the prognosis of patients with diffuse large B‐cell lymphoma. Tokai J Exp Clin Med. 2020;20(45):37‐48. [PubMed] [Google Scholar]
- 29. Carreras J, Kikuti YY, Miyaoka M, et al. A single gene expression set derived from artificial intelligence predicted the prognosis of several lymphoma subtypes; and high immunohistochemical expression of TNFAIP8 associated with poor prognosis in diffuse large B‐cell lymphoma. AI. 2020;1:342‐360. [Google Scholar]
- 30. Lopez‐Guillermo A, Colomo L, Jimenez M, et al. Diffuse large B‐cell lymphoma: clinical and biological characterization and outcome according to the nodal or extranodal primary origin. J Clin Oncol. 2005;23:2797‐2804. [DOI] [PubMed] [Google Scholar]
- 31. Ichiki A, Carreras J, Miyaoka M, et al. Clinicopathological analysis of 320 cases of diffuse large B‐cell lymphoma using the hans classifier. J Clin Exp Hematop. 2017;57:54‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Busuttil RA, George J, Tothill RW, et al. A signature predicting poor prognosis in gastric and ovarian cancer represents a coordinated macrophage and stromal response. Clin Cancer Res. 2014;20:2761‐2772. [DOI] [PubMed] [Google Scholar]
- 33. Tang X. Tumor‐associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013;332:3‐10. [DOI] [PubMed] [Google Scholar]
- 34. Zhang B, Yao G, Zhang Y, et al. M2‐polarized tumor‐associated macrophages are associated with poor prognoses resulting from accelerated lymphangiogenesis in lung adenocarcinoma. Clinics (Sao Paulo). 2011;66:1879‐1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cook J, Hagemann T. Tumour‐associated macrophages and cancer. Curr Opin Pharmacol. 2013;13:595‐601. [DOI] [PubMed] [Google Scholar]
- 36. Farinha P, Masoudi H, Skinnider BF, et al. Analysis of multiple biomarkers shows that lymphoma‐associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL). Blood. 2005;106:2169‐2174. [DOI] [PubMed] [Google Scholar]
- 37. Klein JL, Nguyen TT, Bien‐Willner GA, et al. CD163 immunohistochemistry is superior to CD68 in predicting outcome in classical Hodgkin lymphoma. Am J Clin Pathol. 2014;141:381‐387. [DOI] [PubMed] [Google Scholar]
- 38. Komohara Y, Niino D, Saito Y, et al. Clinical significance of CD163⁺ tumor‐associated macrophages in patients with adult T‐cell leukemia/lymphoma. Cancer Sci. 2013;104:945‐951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugaya M, Miyagaki T, Ohmatsu H, et al. Association of the numbers of CD163(+) cells in lesional skin and serum levels of soluble CD163 with disease progression of cutaneous T cell lymphoma. J Dermatol Sci. 2012;68:45‐51. [DOI] [PubMed] [Google Scholar]
- 40. Hartmann S, Tousseyn T, Döring C, et al. Macrophages in T cell/histiocyte rich large B cell lymphoma strongly express metal‐binding proteins and show a bi‐activated phenotype. Int J Cancer. 2013;133:2609‐2618. [DOI] [PubMed] [Google Scholar]
- 41. Moratz C, Hayman JR, Gu H, et al. Abnormal B‐cell responses to chemokines, disturbed plasma cell localization, and distorted immune tissue architecture in Rgs1‐/‐ mice. Mol Cell Biol. 2004;24:5767‐5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han SB, Moratz C, Huang NN, et al. Rgs1 and Gnai2 regulate the entrance of B lymphocytes into lymph nodes and B cell motility within lymph node follicles. Immunity. 2005;22:343‐354. [DOI] [PubMed] [Google Scholar]
- 43. Nam SJ, Go H, Paik JH, et al. An increase of M2 macrophages predicts poor prognosis in patients with diffuse large B‐cell lymphoma treated with rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone. Leuk Lymphoma. 2014;55:2466‐2476. [DOI] [PubMed] [Google Scholar]
- 44. Wada N, Zaki MA, Hori Y, et al. Osaka Lymphoma Study Group. Tumour‐associated macrophages in diffuse large B‐cell lymphoma: a study of the Osaka Lymphoma Study Group. Histopathology. 2012;60:313‐319. [DOI] [PubMed] [Google Scholar]
- 45. Schmetterer KG, Pickl WF. The IL‐10/STAT3 axis: contributions to immune tolerance by thymus and peripherally derived regulatory T‐cells. Eur J Immunol. 2017;47:1256‐1265. [DOI] [PubMed] [Google Scholar]
- 46. Lurier EB, Dalton D, Dampier W, et al. Transcriptome analysis of IL‐10‐stimulated (M2c) macrophages by next‐generation sequencing. Immunobiology. 2017;222:847‐856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mannino MH, Zhu Z, Xiao H, Bai Q, Wakefield MR, Fang Y. The paradoxical role of IL‐10 in immunity and cancer. Cancer Lett. 2015;367:103‐107. [DOI] [PubMed] [Google Scholar]
- 48. Bouwstra R, He Y, de Boer J, et al. CD47 expression defines efficacy of rituximab with CHOP in non‐germinal center B‐cell (Non‐GCB) diffuse large B‐cell lymphoma patients (DLBCL), but not in GCB DLBCL. Cancer Immunol Res. 2019;7:1663‐1671. [DOI] [PubMed] [Google Scholar]
- 49. Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein‐Barr virus. Annu Rev Immunol. 2007;25:587‐617. [DOI] [PubMed] [Google Scholar]
- 50. Nicolae A, Pittaluga S, Abdullah S, et al. EBV‐positive large B‐cell lymphomas in young patients: a nodal lymphoma with evidence for a tolerogenic immune environment. Blood. 2015;126(7):863‐872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen BJ, Chapuy B, Ouyang J, et al. PD‐L1 expression is characteristic of a subset of aggressive B‐cell lymphomas and virus‐associated malignancies. Clin Cancer Res. 2013;19(13):3462‐3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nicholas NS, Apollonio B, Ramsay AG. Tumor microenvironment (TME)‐driven immune suppression in B cell malignancy. Biochim Biophys Acta. 2016;1863(3):471‐482. [DOI] [PubMed] [Google Scholar]
- 53. Gandhi MK. Epstein‐Barr virus‐associated lymphomas. Expert Rev Anti Infect Ther. 2006;4(1):77‐89. [DOI] [PubMed] [Google Scholar]
- 54. Jones K, Nourse JP, Morrison L, et al. Expansion of EBNA1‐specific effector T cells in posttransplantation lymphoproliferative disorders. Blood. 2010;116(13):2245‐2252. [DOI] [PubMed] [Google Scholar]
- 55. Nourse JP, Jones K, Gandhi MK. Epstein‐Barr Virus‐related posttransplant lymphoproliferative disorders: pathogenetic insights for targeted therapy. Am J Transplant. 2011;11(5):888‐895. [DOI] [PubMed] [Google Scholar]
- 56. Shannon‐Lowe C, Rickinson AB, Bell AI. Epstein‐Barr virusassociated lymphomas. Philos Trans R Soc Lond B Biol Sci. 2017;372(1732):20160271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang DI, Song S‐J, Wu Z‐Z, et al. Epstein–Barr Virus‐Induced VEGF and GM‐CSF drive nasopharyngeal carcinoma metastasis via recruitment and activation of macrophages. Can Res. 2017;77(13):3591‐3604. [DOI] [PubMed] [Google Scholar]
- 58. Keane C, Tobin J, Gunawardana J, et al. The tumour microenvironment is immuno‐tolerogenic and a principal determinant of patient outcome in EBV‐positive diffuse large B‐cell lymphoma. Eur J Haematol. 2019;103(3):200‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Carreras J, Hamoudi R, Nakamura N. Artificial intelligence analysis of gene expression data predicted the prognosis of patients with diffuse large B‐cell lymphoma. Tokai J Exp Clin Med. 2020;45(1):37‐48. [PubMed] [Google Scholar]
- 60. Carreras J, Kikuti YY, Miyaoka M, et al. Single gene expression set derived from artificial intelligence predicted the prognosis of several lymphoma subtypes; and high immunohistochemical expression of TNFAIP8 associated with poor prognosis in diffuse large B‐cell lymphoma. AI. 2020;1:342‐360. 10.3390/ai1030023 [DOI] [Google Scholar]
- 61. Carreras J, Kikuti YY, Miyaoka M, et al. Artificial intelligence analysis of the gene expression of follicular lymphoma predicted the overall survival and correlated with the immune microenvironment response signatures. Mach Learn Knowl Extr. 2020;2:647‐671. 10.3390/make2040035 [DOI] [Google Scholar]
- 62. Carreras J, Kikuti YY, Miyaoka M, et al. A combination of multilayer perceptron, radial basis function artificial neural networks and machine learning image segmentation for the dimension reduction and the prognosis assessment of diffuse large B‐cell lymphoma. AI. 2021;2:106‐134. 10.3390/ai2010008 [DOI] [Google Scholar]
- 63. Carreras J, Kikuti YY, Miyaoka M, et al. Integrative statistics, machine learning and artificial intelligence neural network analysis correlated CSF1R with the prognosis of diffuse large B‐cell lymphoma. Hemato. 2021;2:182‐206. 10.3390/hemato2020011 [DOI] [Google Scholar]
- 64. Carreras J, Kikuti YY, Roncador G, et al. High expression of caspase‐8 associated with improved survival in diffuse large B‐cell lymphoma: machine learning and artificial neural networks analyses. BioMedInformatics. 2021;1:18‐46. 10.3390/biomedinformatics1010003 [DOI] [Google Scholar]
- 65. Carreras J, Hamoudi R. Artificial neural network analysis of gene expression data predicted non‐hodgkin lymphoma subtypes with high accuracy. Mach Learn Knowl Extr. 2021;3:720‐739. 10.3390/make3030036 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


