Abstract
CD28, one of the costimulatory molecules, has a pivotal role in T‐cell activation, and its expression is strictly regulated in normal T cells. Gain‐of‐function genetic alterations involving CD28 have been frequently observed in adult T‐cell leukemia/lymphoma (ATLL). These abnormalities, such as CD28 fusions and copy number variations, may not only confer continuous, prolonged, and enhanced CD28 signaling to downstream pathways but also induce overexpression of the CD28 protein. In this study, 120 ATLL cases were examined by immunohistochemistry for CD28 and its ligands CD80 and CD86, and their expression on tumor cells was semiquantitatively evaluated. CD28 was overexpressed in 55 (46%) cases, and CD80 or CD86 (CD80/CD86) was infrequently overexpressed in 12 (11%). Compared with non‐overexpressers, CD28 overexpressers showed a higher frequency of CD28 genetic alterations and had an increased number of CD80/CD86‐positive non‐neoplastic cells infiltrating tumor microenvironment. In the entire ATLL patient cohort, CD28 overexpressers showed a significantly poorer overall survival (OS) compared with non‐overexpressers (P = .001). The same was true for a subgroup who were treated with multidrug regimens with or without mogamulizumab. CD28 overexpression had no prognostic impact in the group who received allogeneic hematopoietic stem cell transplantation. In the multivariate analysis for OS, CD28 overexpression was selected as an independent risk factor. These results suggest ATLL patients with CD28 overexpression have more aggressive clinical course and are more refractory to treatment with multidrug chemotherapy. CD28 overexpression appears to be a novel unfavorable prognostic marker in ATLL patients, and further prospective studies are warranted to establish its prognostic significance.
Keywords: adult T‐cell leukemia/lymphoma, CD28, CD80 (B7‐1), CD86 (B7‐2), costimulatory molecule, genetic alterations, immunohistochemistry, overexpression, prognosis
The aim of the present study was to determine the clinicopathological significance of CD28 overexpression in adult T‐cell leukemia/lymphoma (ATLL). CD28 overexpression is associated with unfavorable prognosis of ATLL patients who were treated with multidrug regimens including mogamulizumab; thus, CD28 overexpression appears to be a novel unfavorable prognostic marker in ATLL.
Abbreviations
- ATLL
adult T‐cell leukemia/lymphoma
- CNV
copy number variation
- CTLA4
cytotoxic T‐lymphocyte–associated antigen 4
- HSCT
hematopoietic stem cell transplantation
- HTLV‐1
human T‐cell leukemia virus type‐1
- ICOS
inducible T‐cell costimulator
- OS
overall survival
- SNV
single‐nucleotide variant
1. INTRODUCTION
CD28 is a costimulatory molecule expressed on T cells which has a pivotal role in T‐cell biology. In normal T cells, CD28 is expressed on the cell surface, providing an essential costimulatory signal for T‐cell activation upon ligation by CD80 (B7‐1) and CD86 (B7‐2) on antigen‐presenting cells (APCs). After ligation, the accessory molecule cytotoxic T‐lymphocyte–associated antigen 4 (CTLA4) is upregulated. Normally, CTLA4 transmits negative signals to T cells. Because CTLA4 binds to CD80 and CD86 with significantly higher affinity than to CD28, when CTLA4 is upregulated, CD28 is immediately downregulated through a mechanism of competitive inhibition of the ligands to CD28 by CTLA4. 1 , 2 , 3 , 4 The importance and influence of CD28 signaling in T‐cell activation was dramatically demonstrated in a clinical trial. After receiving injections of the humanized monoclonal CD28 agonist antibody TGN1412, the six volunteers became desperately ill, had multiple‐organ failure, and were transferred to an intensive care unit with what has been described as a form of cytokine release syndrome. 5 , 6
In this context, T‐cell–activating alterations to the CD28 gene have been reported in some types of peripheral T‐cell malignancies such as adult T‐cell leukemia/lymphoma (ATLL), angioimmunoblastic T‐cell lymphoma, peripheral T‐cell lymphoma (PTCL), not otherwise specified, and cutaneous T‐cell lymphoma. 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 ATLL is a peripheral T‐cell neoplasm caused by human T‐cell lymphotropic virus type‐1 (HTLV‐1) and has a poor prognosis. 15 , 16 , 17 , 18 We previously reported that CD28 gene–related alterations were more frequent in this tumor, compared with other types of peripheral T‐cell neoplasms, and that these alterations were associated with a worse prognosis in ATLL patients. 19 Genetic alterations involving CD28 include gene fusions such as CTLA4–CD28 and inducible T‐cell costimulator (ICOS)–CD28, activating single‐nucleotide variants (SNVs) such as F51I/V, D124V/E, or T195I/L/P, and copy number variations (CNVs) such as gain and amplification. Of these abnormalities, CD28 gene fusions and CNVs may not only confer continuous, prolonged, and enhanced CD28 signaling to the downstream pathways 7 , 8 , 9 , 10 , 11 , 20 but also are most likely to induce overexpression of the CD28 molecule. In fact, in other PTCL tumors, it has been reported that gene fusions related to CD28 were associated with CD28 overexpression, 8 , 21 probably because the expression of CD28 transcripts may be driven by ICOS or CTLA4 promoters.
In addition, there have been recent reports of ectopic expression of CD80 and CD86 on tumor cells in some ATLL cases, although these molecules were originally expressed on APCs and not on T cells. 22 , 23 These observations suggest that intracellular and intercellular interactions via the CD80/CD86‐CD28 pathway may exist and contribute to ATLL pathogenesis.
The aims of this study were to clarify the clinicopathological and prognostic significance of overexpression of CD28, CD80, and CD86 in ATLL and to examine the association between the expression of these molecules and the genetic status of CD28.
2. MATERIALS AND METHODS
2.1. ATLL patients
From the 144 ATLL patients enrolled in our previous study, 19 we selected 120 patients whose affected tissues were available for immunohistochemical analysis. The organ tissues consisted of lymph node (n = 63), skin (n = 34), and bone marrow (n = 6), and other tissues included stomach (n = 6), pharynx (n = 5), and other tissue types (n = 6). Details are available in the supporting information file. 16 , 18 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32
2.2. Immunohistochemistry for CD28, CD80, and CD86
Immunohistochemistry for CD28 was performed on formalin‐fixed paraffin‐embedded (FFPE) sections of the affected tissues of ATLL patients, using a BOND‐III fully automated IHC and ISH staining system (Leica Biosystems) and a BOND polymer refine detection kit (Leica Biosystems). The rabbit anti‐human CD28 monoclonal antibody (clone, EPR22076; Abcam) used was diluted to 1:50 in BOND primary antibody diluent (Leica Biosystems), and BOND epitope retrieval solution 2 (Leica Biosystems) was employed for heat‐induced antigen retrieval. Areas showing representative immunohistochemical CD28 expression on the cytomembrane in each section were chosen, and the expression was evaluated semiquantitatively by the signal intensity on the tumor cells. Using a scoring scale of 0, 1+, 2+, and 3+, as described in a previous study, 33 we conducted CD28 immunostaining scoring as follows: 0, no staining; 1+, complete or incomplete membrane staining that is weak or only faintly perceptible; 2+, complete membrane staining of moderate intensity; and 3+, complete membrane staining of strong intensity. Scoring of CD28 expression was performed by experienced hematopathologists (A.M. and H.I.), and when the scoring was discordant, a consensus score was reached. The cutoff point was set between 0/1+ and 2+/3+ for CD28 expression, as this cutoff point allowed for the best segregation into prognostic groups. Based on these observations, we defined ATLL cases that were scored as 0 or 1+ as lower CD28 expressers and those that were scored as 2+ or 3+ as CD28 overexpressers.
We also performed immunohistochemistry for CD80 and CD86 and evaluated their expression on tumor cells in the same manner. In addition, non‐neoplastic cells expressing CD80 or CD86 (CD80/CD86) in the tumor microenvironment were counted in three representative high‐powered fields, and the average number for each sample was calculated, respectively. Microenvironmental expression was defined as positive if 10 or more non‐neoplastic cells expressing CD80/CD86 were observed per high‐powered field. 23 CD80/CD86 expression on microenvironmental cells was distinguished from that on tumor cells by cytomorphology using such parameters as a low nuclear/cytoplasmic ratio and the presence of nuclei without atypia. All types of microenvironmental cells, including macrophages, dendritic cells, and others, were counted. These analyses were carried out using a rabbit anti‐human CD80 monoclonal antibody (clone, EPR1157(2); Abcam) and a rabbit anti‐human CD86 monoclonal antibody (clone, E2G8P; Cell Signaling Technology INC.), respectively.
2.3. RNA extraction and quantitative RT‐PCR for CD28
A real‐time quantitative reverse‐transcription polymerase chain reaction (RT‐PCR) assay for CD28 mRNA was carried out with a QuantStudio™ 12K Flex real‐time PCR system (Thermo Fisher Scientific) using GeneAce SYBR® qPCR Mix α (NIPPON GENE Co., Ltd.) according to the manufacturer’s instructions. Details are available in the supporting information file. 34 , 35
2.4. Detection of CD28 gene–related activating alterations
Details are available in the supporting information file. 19
2.5. Detection of CCR4 protein expression and CCR4 gene mutations
Details are available in the supporting information file. 33 , 36
2.6. Statistical analysis
Details are available in the supporting information file.
3. RESULTS
3.1. Clinical and genetic features of ATLL patients
The cohort of the present study consisted of 120 ATLL patients, whose clinical characteristics are summarized in Table S1. There were 53 female patients (44%) and 67 male patients (56%), and their median age at the time of tumor sampling was 65 (range, 41‐90 years). Patient subtypes were classified as acute (n = 65), lymphoma (n = 40), unfavorable chronic (n = 3), and smoldering (n = 12). No patient was assigned to a favorable chronic subtype.
CD28‐related fusions were detected in eight cases (7%), of which all eight had ICOS (ex1)‐CD28 (ex2) and one had the dual fusions ICOS (ex1)‐CD28 (ex2) and CTLA4 (ex3)‐CD28 (ex4). CD28‐activating SNVs were present in two cases (2%), one with p.F51I and one with p.D124V. CD28 CNV was observed in 28 cases (23%), including 18 gains and 10 amplifications. Among these cases, one also had genetic alterations of an ICOS (ex1)‐CD28 (ex2) fusion and a CNV gain. Collectively, alterations of a type involving the CD28 gene were found in 37 cases (31%) (data not shown).
CCR4 mutations were found in 39 cases (33%), comprising p.R323fs in two, p.F326fs in two, p.C329fs in one, p.C329* in nine, p.Q330fs in one, p.Q330* in three, p.Y331fs in two, p.Y331* in 10, p.Q336* in five, p.I337fs in one, and p.S345fs in three (data not shown).
3.2. Characteristics of ATLL patients stratified by CD28 or CD80/CD86 protein expression
On immunohistochemistry for CD28, 20 cases were scored as having no or faint expression, 45 were scored as 1+, 31 as 2+, and 24 were scored as 3+ (Figure S1). Eventually, 55 (46%) cases with scores of 2+ or 3+ were considered to be CD28 overexpressers. Representative immunohistochemical CD28 expression is shown in Figure 1A‐D. We examined the association between CD28 overexpression and the following clinical factors: sex, age, clinical variant, Eastern Cooperative Oncology Group (ECOG) performance status (PS), Ann Arbor stage, serum soluble interleukin‐2 receptor (sIL‐2R) level, serum‐adjusted calcium (Ca), serum albumin (Alb), white blood cell count, hemoglobin, and platelet count and found no clinical feature showing a significant association with CD28 overexpression (Table 1).
FIGURE 1.
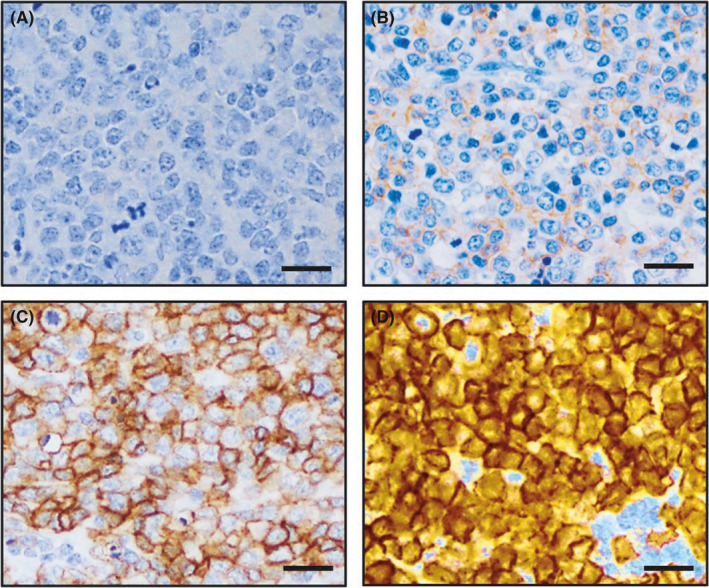
CD28 expression scores of adult T‐cell leukemia/lymphoma (ATLL) cases. Representative immunohistochemical CD28 expression of four individual ATLL cases with intensities scored as 0 (negative, A), 1+ (faint or weak, B), 2+ (moderate, C), and 3+ (strong, D), respectively. The scale bars represent 25 µm
TABLE 1.
Clinical characteristics of ATLL patients according to CD28 expression
| Characteristics | CD28 overexpression | P‐value | |
|---|---|---|---|
| Absence | Presence | ||
| Number (%) | 65 (54) | 55 (46) | |
| Sex | |||
| Females | 35 (54) | 32 (58) | .713 |
| Males | 30 (46) | 23 (42) | |
| Clinical variant | |||
| Indolent | 8 (12) | 4 (7) | .543 |
| Aggressive | 57 (88) | 51 (93) | |
| ECOG PS a | |||
| 0, 1 | 51 (78) | 39 (72) | .521 |
| 2, 3, 4 | 14 (22) | 15 (28) | |
| Ann Arbor stage | |||
| I, II | 11 (17) | 8 (15) | .805 |
| III, IV | 54 (83) | 47 (85) | |
| Age (years) | |||
| ≤70 | 44 (68) | 42 (76) | .317 |
| >70 | 21 (32) | 13 (24) | |
| Serum sIL‐2R (U/mL) b | |||
| ≤20 000 | 41 (68) | 31 (57) | .249 |
| >20 000 | 19 (32) | 23 (43) | |
| Serum Ca (mg/dL) c , e | |||
| ≤11 | 54 (90) | 47 (85) | .572 |
| >11 | 6 (10) | 8 (15) | |
| Serum albumin (g/dL) d | |||
| ≥3.5 | 46 (75) | 35 (64) | .224 |
| <3.5 | 15 (35) | 20 (36) | |
| WBC (/μL) d | |||
| Mean | 12,129 | 16,928 | .355 |
| Median | 7,240 | 8,400 | |
| Range | 3,430‐68,400 | 2,500‐232,100 | |
| Hb (g/dL) d | |||
| Mean | 12.7 | 13 | .349 |
| Median | 12.8 | 13.2 | |
| Range | 8.9‐16 | 7.9‐17.1 | |
| Plt (×103/μL) d | |||
| Mean | 231 | 218 | .853 |
| Median | 214 | 212 | |
| Range | 15‐622 | 29‐444 | |
Abbreviations: Alb, albumin; ATLL, adult T‐cell leukemia/lymphoma; Ca, calcium; ECOG, Eastern Cooperative Oncology Group; Hb, hemoglobin; Plt, platelet count.; PS, performance status; sIL‐2R, soluble interleukin‐2 receptor; WBC, white blood cell count.
One patient's data were missing.
Six patients' data were missing.
Five patients' data were missing.
Four patients' data were missing.
When the serum Alb level was less than 4.0 g/dL, serum Ca was adjusted by the concentration of serum Alb as follows: adjusted Ca level (mg/dL) = measured Ca level (mg/dL) + [4‐ albumin level (g/dL)].
Figure 2 depicts the expression of CD80/CD86 on tumor cells or microenvironmental cells, CD28 gene–related alterations, CCR4 expression, and CCR4 mutations in ATLL cases according to the CD28 protein expression status. CD28 overexpressers had CD28 genetic abnormalities of any type more frequently than non‐overexpressers (30/55 [55%] vs 7/65 [11%], P < .0001). On the other hand, neither CCR4 mutations (CD28 overexpression vs its absence, 22/55 [40%] vs 17/65 [26%], P = .121) nor CCR4 protein expression (CD28 overexpression vs its absence, 50/55 [91%] vs 61/65 [94%], P = .731) was significantly different on stratification according to CD28 overexpression (Figure 2).
FIGURE 2.
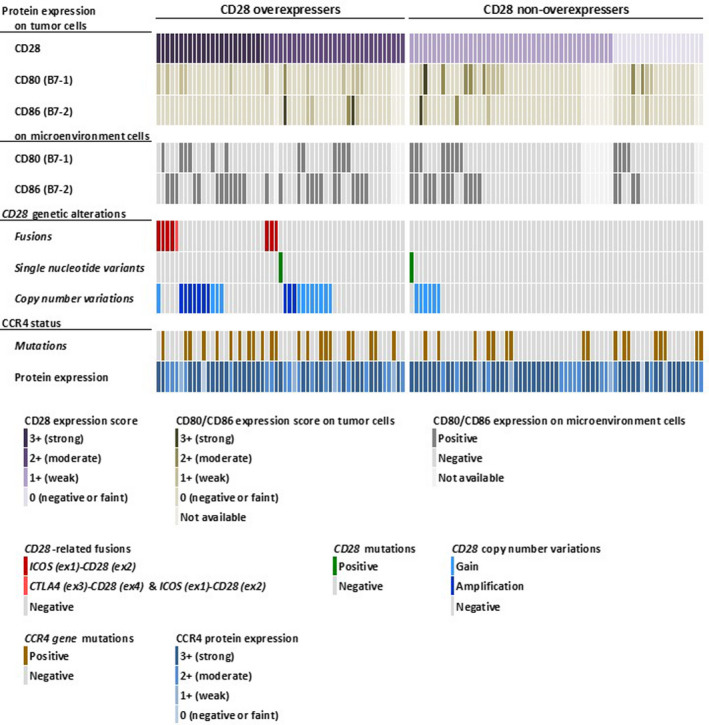
Landscape of protein expression of CD28, CD80 (B7‐1), CD86 (B7‐2), and CCR4 and genetic alterations of CD28 and CCR4 in adult T‐cell leukemia/lymphoma (ATLL) cases. Each column in the panel represents an individual ATLL case. Each row shows, from above, scores of CD28, CD80, and CD86 protein expression on tumor cells, CD80 and CD86 expression on microenvironmental cells, genetic alterations of CD28 (fusions, single‐nucleotide variants, and copy number variations), CCR4 mutations, and CCR4 protein expression
Overexpression of CD80/CD86 protein on tumor cells was not frequent in the ATLL cases (12/107, 11%; Figure 2 and Figure S2A‐D). CD28 overexpression and CD28 genetic alterations were not significantly different on stratification according to the CD80/CD86 expression status of the tumor cells. No significant association was found between CD80/CD86 overexpression and the clinical features analyzed in this study (Table S2).
On the other hand, CD80 and CD86 expression on microenvironmental cells was observed in 24 (22%) and 41 (38%) of 107 ATLL cases, respectively (Figure S2E‐H). Collectively, 54 (50%) cases were positive for microenvironmental CD80/CD86 expression. CD80/CD86 expression on microenvironmental cells was significantly associated with the presence of CD28 overexpression (33/54 [61%] vs 18/53 [34%], P = .007) and CD28 gene–related alterations (26/54 [48%] vs 10/53 [19%], P = .002) (Figure 2).
3.3. CD28 mRNA levels in ATLL cases
To assess CD28 mRNA levels, real‐time quantitative RT‐PCR analysis was performed in quantifiable cases (n = 112). As shown in Figure 3A, relative CD28 mRNA levels were significantly higher in CD28 protein overexpressers than in non‐overexpressers (median, 149 vs 40, respectively, P < .0001). Furthermore, relative CD28 mRNA levels were positively associated with the degree of the CD28 protein expression score (Figure S3). Relative CD28 mRNA levels were significantly higher in ATLL cases with CD28 gene–related alterations than in those without (median, 144 vs 47, respectively, P = .007; Figure 3B).
FIGURE 3.
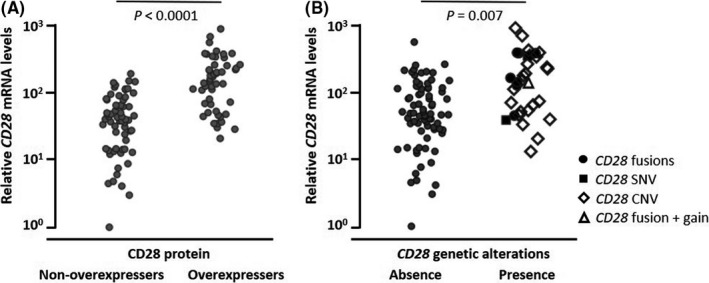
Correlations of CD28 messenger RNA (mRNA) levels, CD28 protein expression, and CD28 genetic alterations. Relative CD28 mRNA levels stratified according to CD28 protein expression (A) or CD28 genetic alterations (B). Relative CD28 mRNA levels are plotted on the y‐axis. Each dot plot in the panels represents an individual adult T‐cell leukemia/lymphoma (ATLL) case. The significance of the differences in CD28 mRNA levels was assessed by the Mann‐Whitney U test, and P‐values are indicated in each panel. CNV, copy number variation; SNV, single‐nucleotide variant
3.4. OS of ATLL patients stratified by CD28 or CD80/CD86 protein expression
The 5‐year OS of the ATLL patients was 42.3% (95% confidence interval [CI], 32.3%‐53.1%, Figure 4A). The 5‐year OS of the CD28 overexpressers was 24.2% (n = 55, 95% CI, 13.0%‐40.6%), which was significantly poorer than that (57.8%) of the non‐overexpressers (n = 65, 95% CI, 43.9%‐70.6%, P = .001; Figure 4B). As shown in Figure 4C, the 5‐year OS (37.2%) was significantly worse in patients with an aggressive variant (n = 108, 95% CI, 27.1%‐48.6%), compared with those (90.9%) with an indolent variant (n = 12, 95% CI, 56.1‐98.7%, P = .027). Therefore, we performed survival analysis according to the clinical subtype. Among the ATLL patients with an aggressive variant, the 5‐year OS for CD28 overexpressers was 22.0% (n = 51, 95% CI, 11.1%‐38.8%), which was significantly less than that (51.4%) for non‐overexpressers (n = 57, 95% CI, 36.8%‐65.8%, P = .008; Figure 4D). The same was true in the analysis of patients with an indolent variant, whose 5‐year OS for CD28 overexpressers and non‐overexpressers was 66.7% (n = 4, 95% CI, 15.4%‐95.7%) and 100% (n=8, 95% CI, not reached), respectively (P = .032; Figure 4E). As for CD80/CD86 expression on tumor cells, the 5‐year OS for overexpressers was 71.4% (n = 12, 95% CI, 32.7%‐92.8%), not significantly different from that (36.8%) for non‐overexpressers (n = 95, 95% CI, 26.3%‐48.7%, P = .077; Figure S4A).
FIGURE 4.
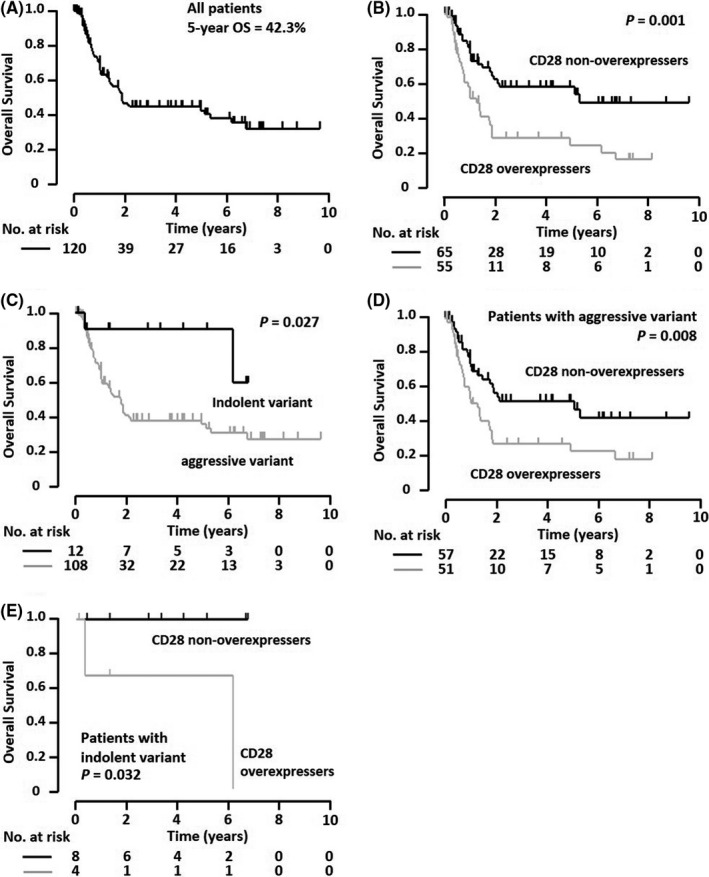
Overall survival (OS) of the entire adult T‐cell leukemia/lymphoma (ATLL) patient cohort stratified by CD28 protein expression. A, OS of all ATLL patients enrolled in this study (n = 120). B, OS of the 55 patients with CD28 overexpression and the 65 with non‐overexpression. C, OS of the 108 ATLL patients with an aggressive variant and the 12 with an indolent variant. D, Among the ATLL patients with an aggressive variant, OS of the 51 patients with CD28 overexpression and the 57 with non‐overexpression. E, Among the ATLL patients with an indolent variant, OS of the four patients with CD28 overexpression and the eight with non‐overexpression. OS curves were compared using the log‐rank test, and the P‐value is indicated in each panel. No., number
3.5. OS of ATLL patients not receiving allogeneic HSCT, stratified by CD28 or CD80/CD86 protein expression
Because it is generally accepted that allogeneic hematopoietic stem cell transplantation (HSCT) is still the only curative treatment for ATLL patients, 18 , 26 , 27 , 28 we evaluated the impact of CD28 overexpression on patients not receiving allogeneic HSCT.
The 5‐year OS for the 92 patients not receiving allogeneic HSCT was 41.5% (95% CI, 29.9%‐54.2%; Figure 5A). In this cohort, the 5‐year OS (21.7%) for CD28 overexpressers (n = 42, 95% CI, 9.6%‐42.1%) was significantly less than that (57.2%) for non‐overexpressers (n = 50, 95% CI, 41.3%‐71.7%, P = .004; Figure 5B). In this non‐HSCT cohort, in ATLL patients with an aggressive variant, the 5‐year OS (18.3%) for CD28 overexpressers (n = 38, 95% CI, 7.0%‐39.9%) was significantly worse than that (51.3%) for non‐overexpressers (n = 44, 95% CI, 34.9%‐67.4%, P = .017; Figure 5C). Among the non‐HSCT patients with an indolent variant, the 5‐year OS for CD28 overexpressers and non‐overexpressers was 66.7% (n = 6, 95% CI, 15.4%‐95.7%) and 100% (n = 4, 95% CI, not reached), respectively. Despite the small numbers analyzed, this difference was statistically significant (P = .046; Figure 5D). As for CD80/CD86 expression on tumor cells, the 5‐year OS for overexpressers (n = 10) and non‐overexpressers (n = 75) was 80.0% (95% CI, 30.9%‐97.3%) and 36.3% (95% CI, 24.5%‐50.1%), respectively (not significantly different, P = .087; Figure S4B).
FIGURE 5.
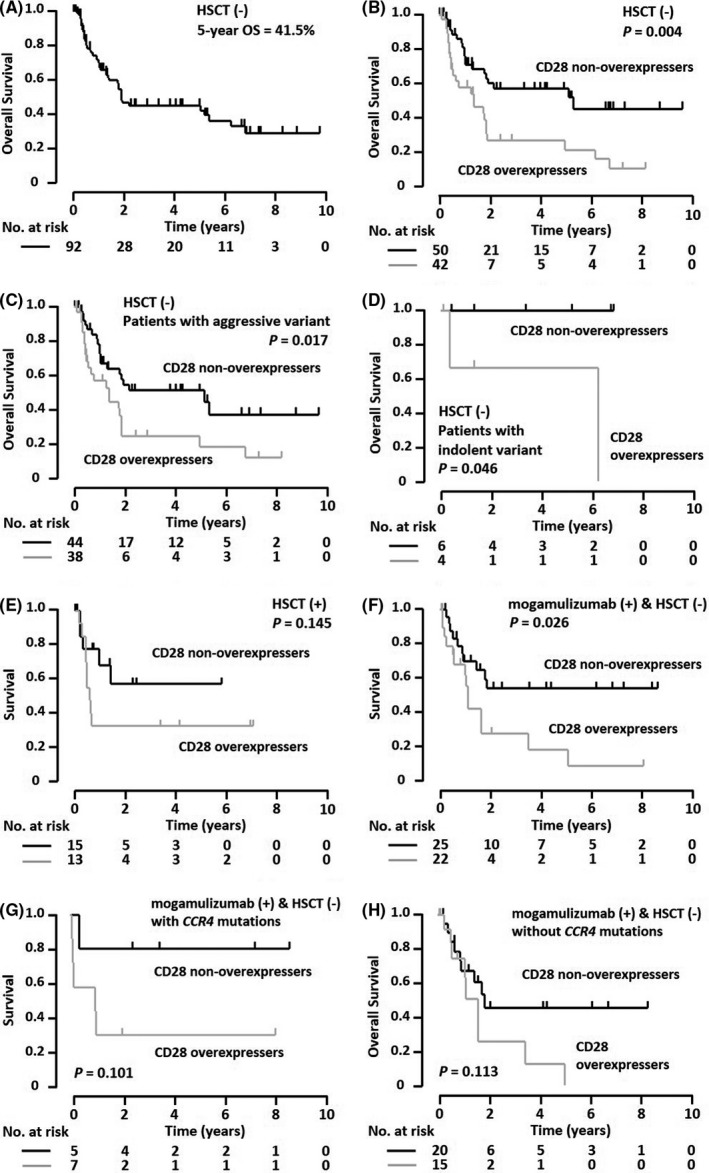
Survival of adult T‐cell leukemia/lymphoma (ATLL) patients according to treatment modalities, stratified by CD28 protein expression. A, Overall survival (OS) of the ATLL patients who did not receive allogeneic hematopoietic stem cell transplantation (HSCT) (n = 92). B, OS of the 42 patients with CD28 overexpression and the 50 with non‐overexpression. C, Among the nontransplanted ATLL patients with an aggressive variant, OS of the 38 patients with CD28 overexpression and the 44 with non‐overexpression. D, Among the nontransplanted ATLL patients with an indolent variant, OS of the four patients with CD28 overexpression and the six with non‐overexpression. E, Among the ATLL patients who received allogeneic HSCT, survival from the day of HSCT in the 13 patients with CD28 overexpression and the 15 with non‐overexpression. F, Among the ATLL patients who received mogamulizumab‐containing treatment, but not allogeneic HSCT, survival from the day of the first dose of mogamulizumab in the 22 patients with CD28 overexpression and the 25 with non‐overexpression. G, For patients who received mogamulizumab‐containing treatment but not allogeneic HSCT and who had CCR4 mutations, survival from the day of the first dose of mogamulizumab in the seven patients with CD28 overexpression and the five with non‐overexpression is shown. H, For patients who received mogamulizumab‐containing treatment but not allogeneic HSCT and who lacked CCR4 mutations, survival from the day of the first dose of mogamulizumab in the 15 patients with CD28 overexpression and the 20 with non‐overexpression is shown. Survival curves were compared using the log‐rank test, and the P‐value is indicated in each panel. No., number
3.6. Survival of ATLL patients who received allogeneic HSCT, stratified by CD28 or CD80/CD86 protein expression
The 5‐year survival of the 28 patients from the day of allogeneic HSCT was 44.4% (95% CI, 26.4%‐64.0%; data not shown), and that of CD28 overexpressers and non‐overexpressers was 30.8% (n = 13, 95% CI, 12.0%‐59.1%) and 56.1% (n = 15, 95% CI, 27.5%‐81.2%), respectively. This difference was not statistically significant (Figure 5E). As for tumor cell CD80/CD86 expression, the 5‐year survival from the day of allogeneic HSCT was not attained by any of the overexpressers, also not significantly different from that (40.6%) of non‐overexpressers (n = 20, 95% CI, 21.0%‐63.8%, P = .549; Figure S4C).
3.7. Survival of ATLL patients who received mogamulizumab‐containing treatment but not allogeneic HSCT, stratified by CD28 or CD80/CD86 protein expression
We next investigated the survival of patients who received mogamulizumab‐containing regimens but not allogeneic HSCT. The 5‐year survival of the 47 patients from the first dose of mogamulizumab was 39.3% (95% CI, 24.5%‐56.3%; data not shown). In this cohort, the 5‐year survival from the first dose of antibody for the subgroup of the CD28 overexpressers and non‐overexpressers was 18.9% (n = 22, 95% CI, 5.5%‐48.4%) and 54.3% (n = 25, 95% CI, 33.4%‐73.8%), respectively, and the difference between the two groups was significant (P = .026; Figure 5F). In this non‐HSCT and mogamulizumab cohort, ATLL cases with an aggressive variant revealed a 5‐year survival from the day of the first antibody dose of 24.2% (n = 19, 95% CI, 8.3%‐52.9%) and 54.3% (n = 25, 95% CI, 33.4%‐73.8%) for CD28 overexpressers and non‐overexpressers, respectively, and the difference was statistically significant (P = .036; data not shown). The 5‐year survival from the day of antibody administration to the three patients with an indolent variant could not be estimated. All three patients were assigned to the CD28 overexpresser group, and two died within 3.5 years after the initiation of mogamulizumab treatment. As a close association between CCR4 gene mutations and a superior responsiveness of mogamulizumab was reported in ATLL patients, 36 we also performed survival analyses for the patients receiving mogamulizumab independent of their CCR4 gene status. As a result, we found that among patients with CCR4 mutations who received mogamulizumab‐containing regimens but not allogeneic HSCT (n = 12), the 5‐year survival from the day of the first dose of antibody for CD28 overexpressers and non‐overexpressers was 28.6% (95% CI, 30.9%‐97.3%) and 80.0% (95% CI, 24.4%‐75.6%), respectively (not significantly different, P = .101; Figure 5G). Among patients without CCR4 mutations who received a mogamulizumab‐containing regimen but not allogeneic HSCT (n = 35), the 5‐year survival from the day of the first dose of antibody for CD28 overexpressers and non‐overexpressers was 0.00% (95% CI, not reached) and 45.5% (95% CI, 23.3%‐69.7%), respectively (also not significantly different, P = .113; Figure 5H). As for CD80/CD86 expression on tumor cells, the 5‐year survival from the first dose of antibody in the overexpressers was 75.0% (n = 5, 95% CI, 23.8%‐96.6%), again not significantly different from that (33.0%) of the non‐overexpressers (n = 39, 95% CI, 18.4%‐51.8%, P = .186; Figure S4D).
3.8. Prognostic significance of CD28 overexpression in ATLL patients
Multivariate analysis of the OS of the entire ATLL patient cohort (n = 113, seven patients were excluded because of missing data) was performed and included the following variables: ECOG PS (0‐1 or 2‐4), ATLL clinical variant (indolent or aggressive), age (≤ 70 or > 70 years), serum sIL‐2R (≤ 20 000 or >20 000 U/mL), serum Alb (≥ 3.5 or <3.5 g/dL), and CD28 expression (overexpression or non‐overexpression). Of these six variables, a poor PS (HR, 3.369, 95% CI, 1.492‐7.612) and CD28 overexpression (HR, 2.149, 95% CI, 1.207‐3.827) were selected as independent risk factors for a worse OS (Table 2).
TABLE 2.
Prognostic factors affecting OS in the entire ATLL patient cohort
| Variables | Number a | Univariate | Multivariate | ||
|---|---|---|---|---|---|
| HR (95% CI) | P‐value | HR (95% CI) | P‐value | ||
| ECOG PS | |||||
| 0, 1 | 87 | 1.000 | Reference | 1.000 | Reference |
| 2, 3, 4 | 26 | 3.272 (1.789‐5.985) | .0001 | 3.369 (1.492‐7.612) | .004 |
| Clinical variant | |||||
| Indolent | 10 | 1.000 | Reference | 1.000 | Reference |
| Aggressive | 103 | 2.997 (0.729‐12.319) | .128 | 2.207 (0.523‐9.314) | .281 |
| Age (years) | |||||
| ≤70 | 80 | 1.000 | Reference | 1.000 | Reference |
| >70 | 33 | 1.138 (0.627‐2.068) | .671 | 1.363 (0.717‐2.593) | .345 |
| Serum sIL‐2R (U/mL) | |||||
| ≤20 000 | 71 | 1.000 | Reference | 1.000 | Reference |
| >20 000 | 42 | 1.908 (1.099‐3.309) | .022 | 1.194 (0.632‐2.255) | .585 |
| Serum Alb (g/dL) | |||||
| ≥3.5 | 80 | 1.000 | Reference | 1.000 | Reference |
| <3.5 | 33 | 1.807 (1.033‐3.162) | .038 | 1.502 (0.669‐3.373) | .324 |
| CD28 protein overexpression | |||||
| Absence | 60 | 1.000 | Reference | 1.000 | Reference |
| Presence | 53 | 2.371 (1.368‐4.109) | .002 | 2.149 (1.207‐3.827) | .009 |
Abbreviations: Alb, albumin; ATLL, adult T‐cell leukemia/lymphoma; CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio; OS, overall survival; PS, performance status; sIL‐2R, soluble interleukin‐2 receptor.
Of the 120 patients, seven were excluded because of missing data.
Multivariate analysis of OS in the nontransplanted patient cohort (n = 92, six patients were excluded because of missing data) was also performed using the same variables. Of these, a poor PS (HR, 4.540, 95% CI, 1.495‐13.791) and CD28 overexpression (HR, 2.137, 95% CI, 1.118‐4.084) were selected as independent risk factors for a worse OS (Table 3).
TABLE 3.
Prognostic factors affecting OS in the nontransplanted ATLL patient cohort
| Variables | Number a | Univariate | Multivariate | ||
|---|---|---|---|---|---|
| HR (95% CI) | P‐value | HR (95% CI) | P‐value | ||
| ECOG PS | |||||
| 0, 1 | 63 | 1.000 | Reference | 1.000 | Reference |
| 2, 3, 4 | 23 | 3.981 (2.004‐7.906) | <.0001 | 4.540 (1.495‐13.791) | .008 |
| Clinical variant | |||||
| Indolent | 8 | 1.000 | Reference | 1.000 | Reference |
| Aggressive | 78 | 2.235 (0.539‐9.273) | .268 | 1.601 (0.371‐6.911) | .528 |
| Age (years) | |||||
| ≤70 | 53 | 1.000 | Reference | 1.000 | Reference |
| >70 | 33 | 1.046 (0.552‐1.979) | .891 | 1.333 (0.668‐2.662) | .415 |
| Serum sIL‐2R (U/mL) | |||||
| ≤20 000 | 53 | 1.000 | Reference | 1.000 | Reference |
| >20 000 | 33 | 2.142 (1.139‐4.031) | .018 | 1.241 (0.606‐2.540) | .555 |
| Serum Alb (g/dL) | |||||
| ≥3.5 | 63 | 1.000 | Reference | 1.000 | Reference |
| <3.5 | 23 | 2.322 (1.184‐4.557) | .014 | 1.402 (0.478‐4.115) | .538 |
| CD28 protein overexpression | |||||
| Absence | 46 | 1.000 | Reference | 1.000 | Reference |
| Presence | 40 | 2.323 (1.184‐4.557) | .014 | 2.137 (1.118‐4.084) | .022 |
Abbreviations: Alb, albumin; ATLL, adult T‐cell leukemia/lymphoma; CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio; OS, overall survival; PS, performance status; sIL‐2R, soluble interleukin‐2 receptor.
Of the 92 patients, six were excluded because of missing data.
Finally, multivariate analyses of the OS in the cohort with an aggressive variant (n = 108; five patients were excluded because of missing data) and in the nontransplanted cohort with an aggressive variant (n = 82; four patients were excluded because of missing data) were performed using the five variables: ECOG PS (0‐1 or 2‐4), age (≤ 70 or > 70 years), serum sIL‐2R (≤ 20 000 or > 20 000 U/mL), serum Alb (≥3.5 or <3.5 g/dL), and CD28 expression (overexpression or non‐overexpression). In both cohorts, the prognostic significance of CD28 overexpression was again retained (HR, 1.973, 95% CI, 1.099‐3.539; Table S3, and HR, 1.964, 95% CI, 1.014‐3.801; Table S4, respectively).
4. DISCUSSION
In the current study, we examined CD28 and CD80/CD86 expression in 120 ATLL cases and correlated the expression with the clinical and genetic features and prognosis of these patients. The cases were divided into overexpressers and non‐overexpressers based on immunohistochemistry for CD28, and the protein expression was quite consistent with CD28 mRNA expression obtained by real‐time quantitative RT‐PCR analysis. Overexpression of CD80/CD86 on tumor cells was not frequent in our ATLL cohort. On the other hand, CD80/CD86‐positive microenvironmental cells were frequent, and were significantly associated with the presence of CD28 overexpression and CD28 gene–related alterations. We found that, in the entire ATLL cohort, CD28 overexpressers showed a worse OS relative to non‐overexpressers. The same was true of the patient cohort not receiving allogeneic HSCT and of those receiving mogamulizumab but not HSCT. These results suggest that ATLL with CD28 overexpression has a more aggressive clinical course and is more refractory to treatment with multidrug chemotherapy and regimens that include mogamulizumab. On the other hand, among the ATLL cohort receiving allogeneic HSCT, the survival impact of CD28 overexpression was not significant. This observation is not very surprising because HSCT is a drastic strategy by which hematopoietic and immune systems are completely replaced by healthy donor–derived cells.
We demonstrated that CD28 overexpression is an independent prognostic factor for a worse prognosis in ATLL patients. The importance of aberrant CD28 overexpression has been well recognized in multiple myeloma (MM). In this tumor, CD28 overexpression is one of the major prognostic predictors for a poor clinical outcome following high‐dose chemotherapy. 37 , 38 , 39 Furthermore, CD28 overexpression highly correlates with disease progression and relapse 40 and has been shown to deliver a prosurvival signal to MM cells, in conjunction with CD86 overexpression on tumor cells, not only through enhanced activation of multiple downstream effectors, such as nuclear factor‐kB (NF‐kB) and phosphatidyl inositol 3‐kinase (PI3K)/Akt, but also via suppression of transcription factors including FoxO3a. 40 , 41 , 42 , 43 In ATLL, multistep oncogenic events are involved in the development of the full malignant phenotype in HTLV‐1–infected cells after a long latency. The cumulative incidence of ATLL development is estimated to be 2%‐7% among HTLV‐1 carriers in Japan, 17 , 18 and half of patients with an indolent variant go on to develop a more aggressive form. 44 Interestingly, CD28 overexpression was not significantly associated with any of the clinical characteristics analyzed in this study, suggesting that no obvious clinical features would help distinguish CD28 overexpressers from non‐overexpressers. In addition, the incidence of CD28 overexpression in aggressive and indolent variants was not significantly different. These results suggest that CD28 overexpression may represent biological behavior rather than evident clinical progression.
Although the underlying mechanisms responsible for CD28 expression in ATLL remain to be elucidated, we demonstrated that CD28 overexpression showed a significant correlation with genetic alterations involving CD28. Of these alterations, all of the cases harboring CD28‐related fusions were categorized immunohistochemically as CD28 overexpressers. With regard to CD28 CNV, all of the cases showing CD28 amplification were assigned to the CD28 overexpressers, but some cases with a CD28 gain were not. Another factor contributing to CD28 overexpression may be microenvironmental cells expressing CD80/CD86, as these cells were significantly associated with CD28 overexpression on tumor cells in this study. The reason why CD28 overexpression contributed to a poorer prognosis in ATLL remains unclear at present. Its overexpression likely reflects aberrant activation of CD28 signaling, resulting in more aggressive behavior and an extremely poor prognosis. Unlike in the case of MM, however, we failed to show an association between CD86 overexpression on ATLL tumor cells and patient survival. In addition, microenvironmental cells expressing CD80/CD86 had no impact on the prognosis (data not shown). Further studies are warranted to clarify molecular mechanisms that could account for the association between CD28 overexpression in ATLL tumor cells and an unfavorable prognosis in the ATLL patients.
The present study offers significant observations regarding how CD28 expression affects the outcome and clinicopathological features of ATLL patients, but some limitations should be recognized other than the matter mentioned above. First, the significance of CD28 overexpression, especially in some cohorts stratified by treatment strategies and clinical variants, was not fully examined due to an insufficient number of patients. Second, it was difficult to estimate which of the CD28 genes, the altered type or wild type, contributed more to the CD28 overexpression. Third, consistent with earlier reports, 22 , 23 we definitively detected the presence of tumor cells ectopically overexpressing CD80/CD86 in some ATLL cases. Unfortunately, we failed to clarify the significance, as the number of such cases was small.
In conclusion, we showed that CD28 protein overexpression is a novel unfavorable prognostic marker in ATLL patients. In addition, we assessed the correlation between CD28 overexpression, CD80/CD86 expression on tumor cells and on microenvironment cells, as well as CD28 gene status, and CCR4 status and found that CD28 overexpressers harbored more frequent CD28 genetic abnormalities and had an increased number of CD80/CD86‐positive non‐neoplastic cells infiltrating the tumor microenvironment. Further studies are needed to clarify the overall mechanisms responsible for CD28 overexpression and its association with an unfavorable ATLL prognosis.
CONFLICT OF INTEREST
H. Iwasaki received research funding from Kyowa Kirin Co., Ltd. K. Yonekura received honoraria from AbbVie, Celgene, Daiichi Sankyo Co., Ltd., Eisai, Eli Lilly Japan, Janssen Pharmaceuticals, Kaken Pharmaceutical, Kyowa Kirin Co., Ltd., Maruho, Minophagen Pharmaceutical, Novartis, Sanofi, Taiho Pharmaceutical, Torii Pharmaceutical, and UCB Japan. S. Kusumoto received research funding from Chugai Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., and honoraria from Chugai Pharmaceutical Co., Ltd. and Kyowa Kirin Co., Ltd. S. Iida received research funding from Sanofi, Chugai Pharmaceutical Co., Ltd., Ono, Takeda, Kyowa Kirin Co., Ltd., Celgene, Janssen, Bristol‐Myers Squibb, Abbvie, and Glaxo‐Smithklein, and honoraria from Janssen, Celgene, Ono, Takeda, Sanofi, and Daiichi Sankyo Co., Ltd. A. Utsunomiya received honoraria from Kyowa Kirin Co., Ltd., Daiichi Sankyo Co., Ltd., Bristol‐Myers, and Celgene, and consulting fees from HUYA Japan, JIMRO, Meiji Seika Pharma Co., Ltd., and Otsuka Medical Devices Co., Ltd. R. Ueda received research funding from Kyowa Kirin Co., Ltd., Chugai Pharmaceutical Co., Ltd., and Ono Pharmaceutical Co., Ltd. No other authors have any conflict of interest.
AUTHOR CONTRIBUTIONS
Y. Sakamoto, T. Ishida, and H. Inagaki designed the research. Y. Sakamoto, T. Ishida, A. Masaki, M. Takeshita, H. Iwasaki, K. Yonekura, Y. Tashiro, A. Ito, S. Kusumoto, A. Utsunomiya, and S. Iida performed the experiments. T. Ishida, R. Ueda, and H. Inagaki analyzed and interpreted data. All authors wrote and approved the manuscript.
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
The present study was approved by the Institutional Ethics Committees of Nagoya City University Graduate School of Medical Sciences (Nagoya, Japan), Imamura General Hospital (Kagoshima, Japan), Fukuoka University (Fukuoka, Japan), and National Hospital Organization Kyushu Medical Center (Fukuoka, Japan), and Nagoya University Graduate School of Medicine (Nagoya, Japan).
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants‐in‐aid for Early‐Career Scientists (20K16177 to Y. Sakamoto), for scientific research (from Aichi Cancer Research Foundation and the Nitto Foundation to Y. Sakamoto), grants‐in‐aid from the Japan Agency for Medical Research and Development (Nos. 19cm0106301h0004, and 20cm0106301h0005 to T. Ishida), and grants‐in‐aid from the National Cancer Research and Development Fund (29‐A‐3 and 2020‐J‐3 to S. Iida).
Sakamoto Y, Ishida T, Masaki A, et al. Clinicopathological significance of CD28 overexpression in adult T‐cell leukemia/lymphoma. Cancer Sci.2022;113:349–361. doi: 10.1111/cas.15191
DATA AVAILABILITY STATEMENT
All data generated or analyzed in the present study are included in this article and its supporting information files.
REFERENCES
- 1. Rudd CE, Taylor A, Schneider H. CD28 and CTLA‐4 coreceptor expression and signal transduction. Immunol Rev. 2009;229:12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen L, Flies DB. Molecular mechanisms of T cell co‐stimulation and co‐inhibition. Nat Rev Immunol. 2013;13:227–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. [DOI] [PubMed] [Google Scholar]
- 4. Esensten JH, Helou YA, Chopra G, Weiss A, Bluestone JA. CD28 costimulation: From mechanism to therapy. Immunity. 2016;44:973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wood AJJ, Darbyshire J. Injury to research volunteers––the clinical‐research nightmare. N Engl J Med. 2006;354:1869–1871. [DOI] [PubMed] [Google Scholar]
- 6. Suntharalingam G, Perry MR, Ward S, et al. Cytokine storm in a phase 1 trial of the anti‐CD28 monoclonal antibody TGN1412. N Engl J Med. 2006;355:1018–1028. [DOI] [PubMed] [Google Scholar]
- 7. Kataoka K, Nagata Y, Kitanaka A, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47:1304–1315. [DOI] [PubMed] [Google Scholar]
- 8. Rohr J, Guo S, Huo J, et al. Recurrent activating mutations of CD28 in peripheral T‐cell lymphomas. Leukemia. 2016;30:1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vallois D, Dupuy A, Lemonnier F, et al. RNA fusions involving CD28 are rare in peripheral T‐cell lymphomas and concentrate mainly in those derived from follicular helper T cells. Haematologica. 2018;103:e360–e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ungewickell A, Bhaduri A, Rios E, et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47:1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi J, Goh G, Walradt T, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47:1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watatani Y, Sato Y, Miyoshi H, et al. Molecular heterogeneity in peripheral T‐cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33:2867–2883. [DOI] [PubMed] [Google Scholar]
- 13. Vallois D, Dobay MPD, Morin RD, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T‐cell‐derived lymphomas. Blood. 2016;128:1490–1502. [DOI] [PubMed] [Google Scholar]
- 14. Wang L, Ni X, Covington KR, et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47:1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T‐cell leukemia: clinical and hematologic features of 16 cases. Blood. 1977;50(3):481–492. [PubMed] [Google Scholar]
- 16. Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T‐cell leukaemia‐lymphoma. A report from the Lymphoma Study Group (1984‐87). Br J Haematol. 1991;79:428‐437. [DOI] [PubMed] [Google Scholar]
- 17. Matsuoka M, Jeang KT. Human T‐cell leukaemia virus type 1 (HTLV‐1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–280. [DOI] [PubMed] [Google Scholar]
- 18. Ishitsuka K, Tamura K. Human T‐cell leukaemia virus type I and adult T‐cell leukaemia‐lymphoma. Lancet Oncol. 2014;15:e517–e526. [DOI] [PubMed] [Google Scholar]
- 19. Sakamoto Y, Ishida T, Masaki A, et al. Clinical significance of CD28 gene‐related activating alterations in adult T‐cell leukemia/lymphoma. Br J Haematol. 2021;192:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gmyrek GB, Pingel J, Choi J, Green JM. Functional analysis of acquired CD28 mutations identified in cutaneous T cell lymphoma. Cell Immunol. 2017;319:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoo HY, Kim P, Kim WS, et al. Frequent CTLA4‐CD28 gene fusion in diverse types of T‐cell lymphoma. Haematologica. 2016;101:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshida N, Shigemori K, Donaldson N, et al. Genomic landscape of young ATLL patients identifies frequent targetable CD28 fusions. Blood. 2020;135:1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takeuchi M, Miyoshi H, Nakashima K, et al. Comprehensive immunohistochemical analysis of immune checkpoint molecules in adult T cell leukemia/lymphoma. Ann Hematol. 2020;99:1093–1098. [DOI] [PubMed] [Google Scholar]
- 24. Yamada Y, Tomonaga M, Fukuda H, et al. A new G‐CSF‐supported combination chemotherapy, LSG15, for adult T‐cell leukaemia‐lymphoma: Japan Clinical Oncology Group Study 9303. Br J Haematol. 2001;113:375–382. [DOI] [PubMed] [Google Scholar]
- 25. Tsukasaki K, Utsunomiya A, Fukuda H, et al. VCAP‐AMP‐VECP compared with biweekly CHOP for adult T‐cell leukemia‐lymphoma: Japan Clinical Oncology Group Study JCOG9801. J Clin Oncol. 2007;25:5458–5464. [DOI] [PubMed] [Google Scholar]
- 26. Ishida T, Hishizawa M, Kato K, et al. Allogeneic hematopoietic stem cell transplantation for adult T‐cell leukemia‐lymphoma with special emphasis on preconditioning regimen: a nationwide retrospective study. Blood. 2012;120:1734–1741. [DOI] [PubMed] [Google Scholar]
- 27. Ishida T, Hishizawa M, Kato K, et al. Impact of graft‐versus‐host disease on allogeneic hematopoietic cell transplantation for adult T cell leukemia‐lymphoma focusing on preconditioning regimens: nationwide retrospective study. Biol Blood Marrow Transplant. 2013;19:1731–1739. [DOI] [PubMed] [Google Scholar]
- 28. Utsunomiya A. Progress in allogeneic hematopoietic cell transplantation in adult T‐Cell leukemia‐lymphoma. Front Microbiol. 2019;10:2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ishii T, Ishida T, Utsunomiya A, et al. Defucosylated humanized anti‐CCR4 monoclonal antibody KW‐0761 as a novel immunotherapeutic agent for adult T‐cell leukemia/lymphoma. Clin Cancer Res. 2010;16:1520–1531. [DOI] [PubMed] [Google Scholar]
- 30. Ishida T, Joh T, Uike N, et al. Defucosylated anti‐CCR4 monoclonal antibody (KW‐0761) for relapsed adult T‐cell leukemia‐lymphoma: a multicenter phase II study. J Clin Oncol. 2012;30:837–842. [DOI] [PubMed] [Google Scholar]
- 31. Ishida T, Jo T, Takemoto S, et al. Dose‐intensified chemotherapy alone or in combination with mogamulizumab in newly diagnosed aggressive adult T‐cell leukaemia‐lymphoma: a randomized phase II study. Br J Haematol. 2015;169:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishida T, Fujiwara H, Nosaka K, et al. Multicenter phase II study of lenalidomide in relapsed or recurrent adult T‐Cell leukemia/lymphoma: ATLL‐002. J Clin Oncol. 2016;34:4086–4093. [DOI] [PubMed] [Google Scholar]
- 33. Fujii K, Sakamoto Y, Masaki A, et al. Immunohistochemistry for CCR4 C terminus predicts CCR4 mutations and mogamulizumab efficacy in adult T cell leukemia/lymphoma. J Pathol Clin Res. 2021;7:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Inagaki H, Okabe M, Seto M, Nakamura S, Ueda R, Eimoto T. API2‐MALT1 fusion transcripts involved in mucosa‐associated lymphoid tissue lymphoma: multiplex RT‐PCR detection using formalin‐fixed paraffin‐embedded specimens. Am J Pathol. 2001;158:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li C, Takino H, Eimoto T, et al. Prognostic significance of NPM‐ALK fusion transcript overexpression in ALK‐positive anaplastic large‐cell lymphoma. Mod Pathol. 2007;20:648–655. [DOI] [PubMed] [Google Scholar]
- 36. Sakamoto Y, Ishida T, Masaki A, et al. CCR4 mutations associated with superior outcome of adult T‐cell leukemia/lymphoma. Blood. 2018;132:758–761. [DOI] [PubMed] [Google Scholar]
- 37. Almeida J, Orfao A, Ocqueteau M, et al. High‐sensitive immunophenotyping and DNA ploidy studies for the investigation of minimal residual disease in multiple myeloma. Br J Haematol. 1999;107:121–131. [DOI] [PubMed] [Google Scholar]
- 38. Mateo G, Castellanos M, Rasillo A, et al. Genetic abnormalities and patterns of antigenic expression in multiple myeloma. Clin Cancer Res. 2005;11:3661–3667. [DOI] [PubMed] [Google Scholar]
- 39. Paiva B, Vidriales MB, Cerveró J, et al. Multiparameter flow cytometric remission is the most relevant prognostic factor for multiple myeloma patients who undergo autologous stem cell transplantation. Blood. 2008;112:4017–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robillard N, Jego G, Pellat‐Deceunynck C, et al. CD28, a marker associated with tumoral expansion in multiple myeloma. Clin Cancer Res. 1998;4:1521–1526. [PubMed] [Google Scholar]
- 41. Bahlis NJ, King AM, Kolonias D, et al. CD28‐mediated regulation of multiple myeloma cell proliferation and survival. Blood. 2007;109:5002–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nair JR, Carlson LM, Koorella C, et al. CD28 expressed on malignant plasma cells induces a prosurvival and immunosuppressive microenvironment. J Immunol. 2011;187:1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murray ME, Gavile CM, Nair JR, et al. CD28‐mediated pro‐survival signaling induces chemotherapeutic resistance in multiple myeloma. Blood. 2014;123:3770–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takasaki Y, Iwanaga M, Imaizumi Y, et al. Long‐term study of indolent adult T‐cell leukemia‐lymphoma. Blood. 2010;115:4337–4343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data generated or analyzed in the present study are included in this article and its supporting information files.


