Abstract
Background:
Next generation sequencing is increasingly used in prenatal diagnosis. Targeted gene panels and exome sequencing are both available, but the comparative diagnostic yields of these approaches are not known.
Objective:
We compared the diagnostic yield of exome sequencing with simulated application of commercial targeted gene panels in a large cohort of fetuses with nonimmune hydrops fetalis (NIHF).
Study Design:
This is a secondary analysis of a cohort study of exome sequencing for NIHF, in which recruitment, exome sequencing, and phenotype-driven variant analysis was completed in 127 pregnancies with features of NIHF. An internet search was performed to identify commercial laboratories that offer targeted gene panels for prenatal evaluation of NIHF or for specific disorders associated with NIHF using the terms “non-immune hydrops fetalis”, “fetal non-immune hydrops”, “hydrops”, “cystic hygroma”, “lysosomal storage disease”, “metabolic disorder”, “inborn error of metabolism”, “RASopathy”, and “Noonan”. Our primary outcome was the proportion of all genetic variants identified through exome sequencing that would have been identified if a targeted gene panel had instead been used. Secondary outcomes were the proportion of genetic variants that would have been identified by type of targeted gene panel (general NIHF, RASopathy, or metabolic), and the percent of variants of uncertain significance that would have been identified on the panels, assuming 100% analytical sensitivity and specificity of panels for variants in the included genes.
Results:
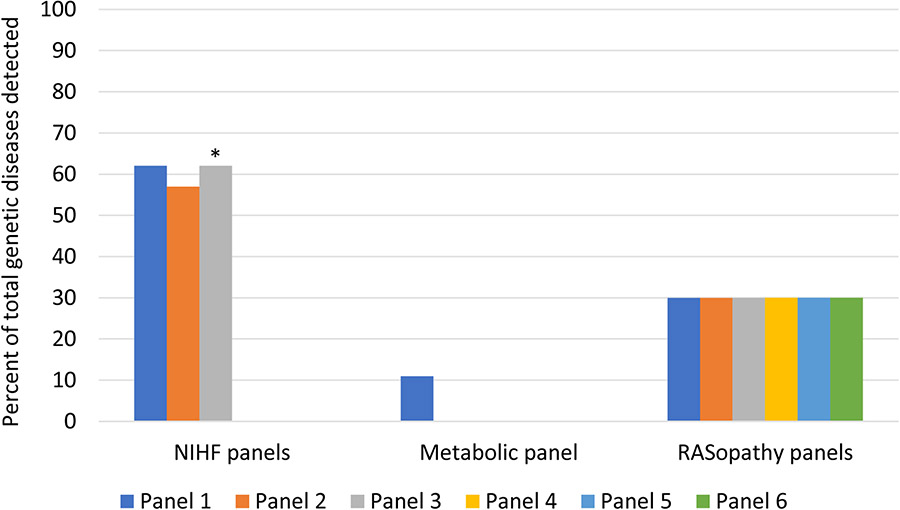
Exome sequencing identified a pathogenic or likely pathogenic variant in 37/127 (29%) cases in a total of 29 genes. A variant of uncertain significance, strongly suspected to be associated with the phenotype, was identified in another 12 (9%) cases. We identified 7 laboratories that offer 10 relevant targeted gene panels; 6 are described as RASopathy panels, 3 as NIHF panels, and one as a metabolic panel. The median number of genes included on each of these panels is 22, ranging from 11 to 148. Had an NIHF targeted gene panel been used instead of exome sequencing, 13 to 15 (45-52%) of the 29 genes identified in our NIHF cohort would have been sequenced, and 19 to 24 (51-62%) of the pathogenic variants would have been detected. The yield was predicted to be lowest with the metabolic panel (11%) and highest with the largest NIHF panel (62%). The largest NIHF targeted gene panel would have had a diagnostic yield of 18% as compared to 29% with exome sequencing. The exome sequencing platform used provides 30X or more coverage for all of the exons on the commercial targeted gene panels, supporting our assumption of 100% analytical sensitivity for exome sequencing.
Conclusion:
The broader coverage of exome sequencing for genetically heterogeneous disorders such as NIHF makes it a superior alternative to targeted gene panel testing.
Condensation:
Exome sequencing can detect a pathogenic or likely pathogenic genetic variant in a significantly greater number of prenatal cases of nonimmune hydrops fetalis when compared to targeted gene panels.
Introduction
Next generation sequencing is increasingly used for evaluation of fetal structural anomalies.1-4 Historically, molecular testing for Mendelian disorders involved analysis of a single gene, and molecular genetic diagnoses were rarely made prior to birth in the absence of a family history. With the advent of next generation sequencing and the capability to test simultaneously for a large number of genes either as part of a targeted gene panel or with exome sequencing, these methods are increasingly being applied to establish genetic diagnoses in the prenatal setting.
Clinical sequencing in the prenatal period often involves choosing between a targeted gene panel that targets a group of selected genes associated with a similar phenotype or exome sequencing to examine a broader array of genes. Focusing on a restricted set of genes enables greater depth of coverage and therefore can potentially provide greater analytic sensitivity and specificity, particularly for challenging variants such as small (exon-level) deletions/duplications.5 Exome sequencing, in contrast, involves sequencing of all of the known protein-coding regions of genes that make up 1-2% of the entire genome; this approach is often applied to clinical disorders with a broad differential diagnosis.6,7 In addition, providers often raise concerns that exome sequencing may be more likely to report variants of uncertain significance that may be difficult to interpret, particularly in a prenatal setting, although data supporting these concerns are limited.8,9. Furthermore, whether targeted gene panels or exome sequencing are used in a given case is often decided based on insurance coverage or cost considerations. There are limited data comparing the clinical benefits and diagnostic yield of targeted gene panels versus exome sequencing for pediatric populations, and a lack of data comparing these approaches for fetal anomalies.10,11 Importantly, there is also a lack of data comparing each of these approaches by phenotype, as some phenotypes are associated with a wider differential diagnosis than others.
Nonimmune hydrops fetalis (NIHF) is a complex disorder caused by a broad range of genetic disorders that can manifest with abnormal fetal fluid collection(s) early or late in gestation. This condition affects 1/1700-3000 pregnancies, and is associated with a high risk of stillbirth, preterm birth, and neonatal complications or death. 4 While cases due to aneuploidy can be diagnosed with karyotype or chromosomal microarray analysis, most cases remain of uncertain etiology after standard evaluation.12,13. As the range of single gene disorders associated with NIHF has been increasingly recognized, and these are not detected with karyotype or chromosomal microarray, genomic sequencing is more often employed for euploid cases. A number of laboratories now offer targeted gene panels for evaluation of NIHF; the included genes vary greatly across panels and may include those associated with RASopathies, inborn errors of metabolism, and other categories of disorders. Importantly, the diagnostic yield using these targeted gene panels for unexplained NIHF cases remains unclear. Exome sequencing has been used to assess a large cohort of pregnancies affected with NIHF and identified a causative gene variant in 29% of cases.4,14
Our goal was to compare the diagnostic yield of targeted gene panels and exome sequencing in unexplained NIHF. We performed a secondary analysis of a large cohort that underwent exome sequencing for NIHF to determine the predicted diagnostic yield had targeted gene panels been used. We hypothesized that exome sequencing would identify many additional single gene disorders beyond those detected through targeted gene panels. Given the importance of cost considerations when choosing a testing strategy, we also collected data on the costs of the included targeted gene panels and of prenatal exome sequencing.
Methods
Study design and participants
This was a secondary analysis of a cohort of prenatally diagnosed NIHF cases that underwent exome sequencing. The findings of the primary study have been published previously.4 The cohort included cases with abnormal fetal effusions including one or more of increased nuchal translucency (NT) ≥3.5 mm, cystic hygroma, pleural effusion, pericardial effusion, ascites, or skin edema. This range in phenotypes was included as literature supporting the traditional criteria of ≥2 abnormal fluid collections for a diagnosis of NIHF is lacking. Further, many genetic disorders associated with abnormal fetal effusions can present early in pregnancy with increased NT or cystic hygroma, or later in pregnancy with NIHF as traditionally defined.4,15-16 Eligible patients had a nondiagnostic karyotype or chromosomal microarray analysis.
Procedures
Details regarding the exome sequencing are provided in the prior report but briefly, trio exome sequencing using DNA from prenatal diagnosis samples was performed in most cases. The UCSF Genomic Medicine Laboratory performed exome sequencing with the Illumina HiSeq 2500 or the Illumina NovaSeq 6000 sequencing system. Mean sample exome coverage was 80X for the HiSeq and 148X for NovaSeq. Variant call format files were uploaded for variant filtering into Ingenuity Variant Analysis (Qiagen) or Moon (Diploid, Diagnosing Rare Diseases; Invitae), clinical informatics experts manually curated the variants, and a multidisciplinary review of curated variants in the context of phenotypic features was performed for each case. Genetic variants were classified according to recommendations of the American College of Medical Genetics and Genomics (ACMG) and Association for Medical Pathology.17 In situations where the gene-disease relationship was high but the ACMG criteria for pathogenicity were not met for the specific variant and there was evidence to support a strong potential for clinical significance, the laboratory reported as a variant of uncertain significance (VUS).
We identified commercial laboratories that provide targeted gene panel testing for prenatal evaluation of NIHF. These laboratories were identified through a general internet search and through query of the Concert Genetics search engine18, using terms including “non-immune hydrops fetalis”, “hydrops fetalis”, “fetal non-immune hydrops”, “hydrops”, “cystic hygroma”, “nuchal translucency”, “lysosomal storage disease”, “metabolic disorder”, “inborn error of metabolism”, “RASopathy”, and “Noonan”. We included panels that test for genes associated with NIHF, as well as genes causative of disorders known to be associated with NIHF, such as RASopathies, lymphedema disorders, and lysosomal storage diseases. The genes included on each targeted panel were identified on each laboratory’s website. Some laboratories offer more than one relevant panel, for example a general NIHF panel as well as a more specific RASopathy panel. In such cases, both targeted gene panels were analyzed and reported separately.
Outcomes
The primary outcome was the proportion of all pathogenic or likely pathogenic genetic variants identified through exome sequencing that would have been identified if a targeted gene panel had instead been used. Secondary outcomes were the hypothetical proportions of genetic variants that would have been identified by type of targeted gene panel (general NIHF, RASopathy, or metabolic), the percent of VUS detected by exome that would have been identified on the panels, and the proportion of variants that would have been identified through panels for isolated NIHF cases compared to those with additional structural anomalies. These calculations were done assuming 100% analytical sensitivity and specificity.5,7 Genetic variants identified by exome were classified as pathogenic or likely pathogenic by ACMG criteria, and as a VUS when ACMG criteria for pathogenicity were not met but the multidisciplinary review determined the variant to be suspicious and likely to be associated with the phenotype.
In order to assess the ability of exome sequencing to detect variants identified through targeted gene panels, we also determined the coverage of exome sequencing for all genes on the panels, including for genes not identified in any cases in our exome sequencing cohort. In order to compare costs, we contacted the laboratories that provide targeted gene panels or prenatal exome sequencing and collected data on the costs of each of these tests.
Statistical analysis
Primary and secondary outcomes were reported as proportions. Statistical analyses were performed in Excel. Approval was obtained through the UCSF institutional review board for the primary study; as this secondary analysis utilized publicly available information from commercial laboratories about targeted gene panels, additional IRB approval was not necessary.
Results
The cohort is described in Table 1. Of the 127 cases, the majority had fluid collections in 2 or more cavities (77, 61%) while 21 (17%) had a single fetal effusion such as isolated ascites and 29 (23%) presented with early enlarged nuchal translucency or cystic hygroma (of which 15 were isolated without other anomalies or additional abnormal fluid effusions). Overall, 64 (50%) had a concurrent structural anomaly.
Table 1.
Demographics of exome cohort
| Demographic | Value (N=127) |
|---|---|
| Median maternal age (IQR) | 32 years(29-35) |
| Nulliparous | 45% (57/127) |
| Median gestational age at diagnosis of NIHF (range) | 20.0 weeks (13.4-24.6) |
| Any concurrent anomaly | 50% (64/127) |
| Maternal race/ethnicity | |
| White | 58% (74/127) |
| Asian | 15% (19/127) |
| Multiracial | 14% (18/127) |
| Hispanic or Latina | 9% (12/127) |
| Black | 2% (3/127) |
| Unknown | 1% (1/127) |
| Type of abnormal fetal effusion | |
| Early onset (increased NT or cystic hygroma) | 23% (29/127) |
| Single abnormal fetal effusion | 17% (21/127) |
| Traditionally defined NIHF with ≥2 abnormal effusions | 61% (77/127) |
Exome sequencing identified a pathogenic or likely pathogenic variant in 37/127 (29%) cases. In all, 29 genes were represented including 6 RASopathy genes, 4 for musculoskeletal disorders, 3 for inborn errors of metabolism, 3 for lymphedema disorders, 3 for neurodevelopmental disorders, 3 for cardiovascular disorders, 2 for hematologic disorders, 2 for immunologic disorders, and one each for renal, ciliopathy, overgrowth, and CHARGE (Coloboma of the eye, Heart defects, Atresia of the choanae, Retardation of Growth and development, and Ear abnormalities and deafness) syndromes. Four genes were implicated multiple times, including 4 cases with variants in PTPN11, 3 with HRAS, 3 with PIEZO1, and 2 with GUSB. Among the 37 pathogenic or likely pathogenic variants, 16 (43%) were identified in cases with isolated NIHF and 21 (57%) in cases with concurrent structural anomalies. Overall, 9 (24%) presented early with cystic hygroma or increased NT, 2 (5%) with a later single abnormal fetal effusion, and 26 (70%) with fluid effusions in 2 or more cavities.4
We identified 7 laboratories that offer 10 relevant targeted gene panels; 6 were described as RASopathy panels, 3 as NIHF panels, and one as a metabolic panel. The median number of genes on the RASopathy panels was 19 (11-23). The largest NIHF panel (Laboratory 3) was updated to include additional genes after publication of our primary analysis;4 the median number of genes on NIHF panels was 87 (66-128) prior to this update and 87 (66-148) afterwards. The one metabolic panel included 51 genes. (Table 2). Overall, the targeted gene panels included 169 unique genes, none of the 169 genes were included on all panels, and 57 genes were represented only on a single gene panel.
Table 2.
Laboratory targeted gene panels and included genes
| Laboratory | Targeted gene panel description |
No. of genes included |
Disorders* | Total genes vs UCSF exome (n=29 genes) |
Total detection vs UCSF exome (n=37 variants) |
|---|---|---|---|---|---|
| Laboratory 1 | Non-immune hydrops | 87 | RASopathies, skeletal dysplasias, metabolic disorders, arthrogryposes, multiple congenital anomaly syndromes | 15 (52%) | 23 (62%) |
| RASopathy | 23 | RASopathies | 6 (21%) | 11 (30%) | |
| Laboratory 2 | Fetal hydrops | 66 | RASopathies | 13 (45%) | 21 (57%) |
| RASopathy | 19 | RASopathies | 6 (21%) | 11 (30%) | |
| Laboratory 3 | Non-immune hydrops | Pre-update**: 128 | RASopathies, skeletal dysplasias, metabolic disorders, congenital anemias, arthrogryposes, multiple congenital anomaly syndromes | 15 (52%) | 23 (62%) |
| Post-update**: 148 | 29 (100%) | 37 (100%) | |||
| RASopathy | 20 | RASopathies | 6 (21%) | 11 (30%) | |
| Laboratory 4 | Metabolic nonimmune fetal hydrops | 51 | Metabolic disorders only; cases not associated with malformations | 3 (10%) | 4 (11%) |
| Laboratory 5 | Prenatal Noonan syndrome | 19 | RASopathies | 6 (21%) | 11 (30%) |
| Laboratory 6 | Prenatal Noonan spectrum disorders | 11 | RASopathies | 6 (21%) | 11 (30%) |
| Laboratory 7 | Noonan spectrum disorders | 16 | RASopathies | 6 (21%) | 11 (30%) |
Disorders covered as described on laboratory website.
The largest NIHF panel was updated to include additional genes after publication of our primary analysis.4
Prior to the update for Laboratory 3, had an NIHF targeted gene panel been used instead of exome sequencing, 13 to 15 (45-52%) of the 29 genes identified in our NIHF cohort would have been sequenced depending on the specific panel, and 19 to 23 (51-62%) of the 37 pathogenic or likely pathogenic variants would have been detected. After the update for Laboratory 3, had an NIHF targeted gene panel been used instead of exome sequencing, 13 to 29 (45-100%) of the 29 genes identified in our NIHF cohort would have been sequenced, and 19 to 37 (51-100%) of the pathogenic or likely pathogenic variants would have been detected. In comparison, while RASopathies were the most common genetic disorder in our NIHF cohort, the 6 RASopathy panels include only 21% (6/29) of the total genes detected by exome sequencing and would have diagnosed only 30% (11/37) of the pathogenic or likely pathogenic variants identified in cases in our cohort. Similarly, the metabolic panel includes 3 of the genes detected in our cohort and would have diagnosed 11% (4/37) of NIHF cases with pathogenic or likely pathogenic variants.
Exome sequencing identified a VUS in 12 cases including 12 different genes, in which ACMG criteria for pathogenicity were not met but the clinical team was suspicious that the variant was likely to be causative of the phenotype. One targeted gene panel did not include any of these genes, while the remainder included 1 to 5 (8-42%) of these 12 genes.
Overall, there were 169 unique genes and 2823 exons on the targeted gene panels we evaluated. The average mean target coverage of these exons with exome sequencing through the UCSF Genomic Medicine Laboratory is 135.4X. All exons have 30X or more coverage (the minimum average exon coverage is 33.4X), and over 98% of the exons (2773/2823) have >50X coverage on the NovaSeq 6000 sequencing system. This high coverage supports our assumption that variants in any of the genes included on the targeted panels would have been detected through exome sequencing had they been present in our cohort.
Finally, the costs of targeted gene panels varied significantly, from $640 for the least expensive RASopathy panel to $3500 for the most expensive NIHF panel. The cost of prenatal exome sequencing ranged from $2458 to $7500.
Discussion:
Principal Findings
We found that exome sequencing has a substantially higher yield than targeted gene panels for NIHF, including those panels with a large number of genes covering a wide spectrum of single gene disorders. Exome sequencing had good coverage of genes examined by targeted gene panels, supporting our assumption that exome sequencing would likely have detected any relevant variants in genes included on the panels had they been present in cases in our cohort. This higher detection may have come at a higher cost, although this depends on the selected laboratory and testing approach, as there was some overlap in the comparative costs of targeted panels and exome sequencing.
Results in the Context of What is Known
Few studies have compared exome sequencing to targeted gene panels for the clinical evaluation of specific disorders, and there is a paucity of such comparisons for prenatal phenotypes specifically. A study by Dillon et al. compared the diagnostic yield of exome sequencing with simulated application of available targeted gene panels in children with genetically heterogeneous conditions. Exome sequencing identified causative genes that would not have been detected on available gene panels in 23% of children and, based on 20X exome coverage, the authors calculated that the likelihood of missing a clinically relevant variant using exome sequencing was maximally 8%. Furthermore, in 26% of cases, the least costly panel would have been more expensive than exome sequencing.11 Another study compared targeted panels to exome sequencing in patients with primary immunodeficiency. Targeted panels identified a disease-causing variant in 56% of the 878 probands, while exome sequencing detected single gene disorders in 18 additional cases resulting from novel genes. The authors also noted that performing exome alone had a simplified workflow and resultant cost savings when compared to targeted panels followed by exome for nondiagnostic cases.19
Clinical Implications
Some geneticists have recommended targeted gene panels as the first-tier test for some diseases based on diagnostic rate, coverage, depth, and costs.20,21 However, the genetic causes underlying many disorders, including NIHF, are not completely elucidated. Therefore, a targeted panel focused on genes that are already known to be associated with NIHF will not be as comprehensive as exome sequencing. At the very least, the targeted list of genes included on panels should be regularly edited based on findings reflecting newly established associations of NIHF with additional genes.
It has also been suggested that targeted gene panel sequencing is not indicated for individuals with less differentiated clinical phenotypes.21 NIHF falls in this category, given its significant genetic heterogeneity. There is also substantial phenotypic variability, as many genetic diseases underlying NIHF can present with early or later onset of one or more abnormal fetal fluid collections. Because the phenotype of NIHF is nonspecific in the absence of additional anomalies, sonographic information cannot identify the better option. Several laboratories offer gene panels targeted to disorders known to be associated with NIHF, such as RASopathies or metabolic disorders. However, it is not clear under what circumstances these more focused panels would be appropriate in a prenatal setting, given that phenotypic features of hydrops most often do not point to a specific category of disorders. The in utero phenotype of single gene disorders that can present with NIHF is incompletely understood, leading to the potential for missed diagnoses when a broad approach to the diagnostic evaluation is not pursued.
It has also been suggested that targeted gene panels may be more sensitive for a given variant due to superior coverage of the included genes and that clinicians must weigh higher coverage with targeted panels versus a greater number of genes included in exome sequencing when considering their testing approach. However, one study assessing coverage among 100 individual exome samples for each pathogenic variant (153,300 individual assessments), found that 99.7% (n = 152,798) would likely have been detected by exome sequencing.5 Likewise, in our cohort, we confirmed that the genes included on the targeted panels all had adequate coverage (>30X) on the exome platform such that all exceeded minimum coverage recommendations for next generation sequencing and variants in these genes would almost certainly have been identified.22,23 Finally, it has been suggested that exome sequencing might produce more VUS results, adding to patient anxiety. We were not able to directly compare this outcome as laboratories do not routinely provide data on VUS rates. However, rates of VUS as high as 58.1% have been published based on commercial hydrops panels,24 as compared to 9% in our exome cohort.
And finally, while targeted gene panels are typically less expensive than exome sequencing, this lower cost is offset by lower detection rates compared to the higher yield of exome sequencing. Importantly, these direct cost comparisons do not take into account downstream effects such as further pre- and postnatal testing required when panels or exome do not identify a diagnosis, medical costs resulting from early as compared to delayed diagnosis and treatment, and many other considerations. A formal cost effectiveness analysis is necessary to assess the tradeoff in these outcomes.
Research Implications
The ongoing process of gene and variant discovery will continue to increase our understanding of NIHF and improve our ability to diagnose the causes of this disorder. To this end, we identified a gene variant in 9% of cases that was classified as a VUS based on ACMG criteria, but that was strongly suspected by our multidisciplinary team to be associated with the phenotype. A small fraction of these VUS were captured by commercial targeted gene panels, but arguably these are important in expanding our understanding of NIHF. Furthermore, variant reclassification is often pursued after both exome sequencing and gene panels as additional information becomes available. Re-analysis of unsolved cases will likely have a higher yield following exome sequencing, as novel genes are discovered, and more causative variants are published. This is anticipated to be particularly important in NIHF pregnancies, given the significant genetic heterogeneity of this complex phenotype. It is also of note that one laboratory updated their largest hydrops panel to add the genes and variants that we reported in our exome sequencing cohort4. This further illustrates the utility of exome for gene discovery, and for expanding our understanding of the genetic disorders that are associated with hydrops. Targeted gene panels, by definition, rely on sequencing of previously reported genes, and will always have a lower detection rate.
Strengths and Limitations
This study adds to the limited data comparing these two approaches for prenatal diagnosis, and specifically for the evaluation of NIHF. A strength of our study is that it includes comprehensive genetic data on a large number of NIHF cases that underwent exome sequencing. The study, however, is not without limitations. While exome sequencing was performed in all reported cases, the targeted gene panel results were modeled based on the genes listed on each laboratory’s website with an assumption of 100% analytic sensitivity and specificity. It is not known with certainty that a targeted gene panel would detect all variants in a gene, even if that gene is included on a panel. Likewise, it is not certain that our exome would have detected all variants in the genes on each panel had a variant in one of these genes been present. Neither exome sequencing nor panels will detect all disease-causing variants, and copy number variants, indels, variants in non-coding regions, and other types of variants may not be identified with either test. Some commercial laboratories offering targeted gene panels will concurrently evaluate for copy number variants in the targeted list of genes, which is not routinely performed with exome sequencing, and was not considered in our calculations. Despite these limitations, these data provide a clinically useful comparison of the differences in the genes assessed through these different testing modalities.
Conclusions
Overall, use of targeted gene panels has been preferred because of lower costs, shorter turnaround time, and the perception of lower rates of nonspecific or incidental results. However, targeted gene panels are less likely to diagnose variants associated with NIHF when compared to exome sequencing. Panels have limited utility for discovering new genes or for expanding the phenotype for known genes not previously associated with NIHF. For disorders such as NIHF with significant genetic heterogeneity and less clear in utero phenotypes of underlying genetic diseases, the broader coverage of exome sequencing makes it a superior option to targeted gene panel testing.
Supplementary Material
Figure 1.
Percent of total genetic diseases detected by targeted gene panels compared to exome sequencing in cases of NIHF
* This NIHF panel was updated to include additional genes after publication of our primary analysis.4 After this update, the percent of total genetic diseases that would have detected by this panel was 100%, including the full list of genes we reported. (see text)
AJOG at a glance:
A. Why was this study conducted?
Nonimmune hydrops is a heterogeneous condition that can manifest in the setting of a broad array of genetic disorders. Targeted gene panels and exome sequencing are both options for evaluation of affected fetuses, and it is not known how the diagnostic yield differs.
B. What are the key findings?
In a cohort of 127 fetuses with nonimmune hydrops, we determined that the use of available targeted gene panels would have detected a pathogenic or likely pathogenic variant in 11- 62% of the 37 cases that received a genetic diagnosis with exome sequencing, or 3-18% of the total cases.
C. What does this study add to what is already known?
The diagnostic yield of exome sequencing in nonimmune hydrops has been reported to be 29%; use of targeted gene panels instead of exome sequencing will diagnose substantially fewer cases.
Disclosures:
Dr. Norton has received research funding from Natera and is a consultant to Invitae; the other authors report no conflicts of interest.
Funding support:
Supported by the University of California, San Francisco (UCSF) Center for Maternal–Fetal Precision Medicine, the Brianna Marie Foundation in collaboration with the Fetal Health Foundation, Ultragenyx (for studies conducted through the UCSF Center for Maternal–Fetal Precision Medicine), and grants (5K12HD001262-18, supporting Dr. Sparks, and U01HG009599, to Dr. Norton) from the National Institutes of Health. The funding sources had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Glossary
- Coverage
The number of times a portion of the genome is sequenced in a sequencing reaction. Often expressed as “depth of coverage” and numerically as 1X, 2X, 3X, etc
- Exome
The portion of the genome consisting of protein-coding sequences (as opposed to introns or noncoding DNA between genes)
- Exome sequencing
A technique for sequencing just the protein-coding regions of genes in a genome (known as the exome)
- Gene variant
A permanent change in the DNA sequence of a gene. Previously referred to as a gene mutation, but because changes in DNA do not always cause disease, gene variant is considered a more accurate term
- Genetic heterogeneity
A phenotype caused by more than one gene
- Next generation sequencing
DNA sequencing technology that permits rapid sequencing of large portions of the genome, greatly increasing the throughput over classic Sanger sequencing
- Pathogenic and likely pathogenic variant
Classifications of gene variants meeting specific American College of Medical Genetics and Genomics (ACMG) criteria. A pathogenic variant is thought to directly contribute to the development of disease while a likely pathogenic variant has a high likelihood (greater than 90%) to be disease-causing.
- Phenotype
The total observable characteristics of an individual, resulting from interaction of the genotype with the environment
- RASopathy
A group of developmental syndromes caused by variants in genes that alter the Ras subfamily and mitogen-activated protein kinases that control signal transduction. Examples of RASopathy disorders include Noonan syndrome and neurofibromatosis type 1
- Targeted gene panel
Sequencing approach that analyzes a select set of genes or gene regions that have known or suspected associations with the disease or phenotype under study
- Trio exome sequencing
An approach to exome sequencing in which the affected individual and their unaffected parents are all studied. Trio study design (father, mother, and child) can identify inherited/non-inherited or de novo variants and aid in classification of putative causal variants
- Variant call format files
The Variant Call Format (VCF) specifies the format of a text file used in bioinformatics for storing gene sequence variations
- Variant filtering
A secondary genomic sequencing analysis step that consists of identifying highly confident variants and removing the ones that are falsely called
- Variant curation
A process of using information from publicly available resources and internal laboratory data to assess a variant-disease relationship. A classification for each variant is assigned based on ACMG evidence codes and strength
- Variant of unknown significance (VUS)
Genetic variant that cannot be definitively determined to be associated with a specific phenotype
Footnotes
The findings were presented in poster format at the 30th World Congress of the International Society of Ultrasound in Obstetrics and Gynecology held virtually October 16-18, 2020.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.International Society for Prenatal Diagnosis; Society for Maternal and Fetal Medicine; Perinatal Quality Foundation. Joint Position Statement from the International Society for Prenatal Diagnosis (ISPD), the Society for Maternal Fetal Medicine (SMFM), and the Perinatal Quality Foundation (PQF) on the use of genome-wide sequencing for fetal diagnosis. Prenat Diagn. 2018. Jan;38(1):6–9. [DOI] [PubMed] [Google Scholar]
- 2.Petrovski S, Aggarwal V, Giordano JL, et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet 2019; 393: 758–67. [DOI] [PubMed] [Google Scholar]
- 3.Lord J, McMullan DJ, Eberhardt RY, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ul-trasonography (PAGE): a cohort study. Lancet 2019; 393: 747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sparks TN, Lianoglou BR, Adami RR, et al. Exome Sequencing for Prenatal Diagnosis in Nonimmune Hydrops Fetalis. N Engl J Med. 2020. Oct 29;383(18):1746–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaDuca H, Farwell KD, Vuong H, et al. Exome sequencing covers >98% of mutations identified on targeted next generation sequencing panels. PLoS One 2017. Feb 2;12(2):e0170843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams DR, Eng CM. Next-Generation Sequencing to Diagnose Suspected Genetic Disorders. N Engl J Med. 2018. Oct 4;379(14):1353–1362. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Ruivenkamp CAL, Hoffer MJV, et al. Next-generation diagnostics: gene panel, exome, or whole genome? Hum Mutat 2015;36(6):648–655. [DOI] [PubMed] [Google Scholar]
- 8.Lionel AC, Costain G, Monfared N, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med 2018. Apr;20(4):435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirino AL, Lakdawala NK, McDonough B, et al. A Comparison of Whole Genome Sequencing to Multigene Panel Testing in Hypertrophic Cardiomyopathy Patients. Circ Cardiovasc Genet 2017. Oct;10(5):e001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han JY, Lee IG. Genetic tests by next-generation sequencing in children with developmental delay and/or intellectual disability. Clin Exp Pediatr 2020. Jun;63(6):195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillon JO, Lunke S, Stark Z, et al. Exome sequencing has higher diagnostic yield compared to simulated disease-specific panels in children with suspected monogenic disorders. Eur J Hum Genet 2018. May;26(5):644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sparks TN, Thao K, Lianoglou BR, et al. Nonimmune hydrops fetalis: identifying the underlying genetic etiology. Genet Med 2019;21:1339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mardy AH, Rangwala N, Hernandez-Cruz Y, et al. Utility of chromosomal microarray for diagnosis in cases of nonimmune hydrops fetalis. Prenat Diagn 2020; 40:492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mone F, Eberhardt RY, Hurles ME, et al. Fetal hydrops and the Incremental yield of Next generation sequencing over standard prenatal Diagnostic testing (FIND) study: prospective cohort study and meta-analysis. Ultrasound Obstet Gynecol 2021;April 13. doi: 10.1002/uog.23652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mardy AH, Chetty SP, Norton ME, Sparks TN. A system-based approach to the genetic etiologies of nonimmune hydrops fetalis. Prenat Diagn 2019;39:732–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croonen EA, Nillesen WM, Stuurman KE, et al. Prenatal diagnostic testing of the Noonan syndrome genes in fetuses with abnormal ultrasound findings. Eur J Hum Genet 2013;21:936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.https://www.concertgenetics.com (last accessed 3/21/2021)
- 19.Platt CD, Zaman F, Bainter W, et al. Efficacy and economics of targeted panel versus whole-exome sequencing in 878 patients with suspected primary immunodeficiency. J Allergy Clin Immunol. 2020. Sep 2:S0091-6749(20)31226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saudi Mendeliome Group. Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol 2015. Jun 26;16(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consugar MB, Navarro-Gomez D, Place EM, Bujakowska KM, Sousa ME, Fonseca-Kelly ZD, et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet Med. 2015;17:253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meynert AM, Ansari M, FitzPatrick DR, et al. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics 2014;15(1):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehm HL, Bale SJ, Bayrak-Toydemir P, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013;15(9):733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leach NT, Wilson Mathews DR, Rosenblum LS, Zhou Z, Zhu H, Heim RA. Comparative assessment of gene-specific variant distribution in prenatal and postnatal cohorts tested for Noonan syndrome and related conditions. Genet Med 2019. Feb;21(2):417–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.