Abstract
Soybean cyst nematode is the most economically damaging pathogen of soybean and host resistance is a core management strategy. The SCN resistance QTL cqSCN-006, introgressed from the wild relative Glycine soja, provides intermediate resistance against nematode populations including those with increased virulence on the heavily used rhg1-b resistance locus. cqSCN-006 was previously fine-mapped to a genome interval on chromosome 15. The present study determined that Glyma.15G191200 at cqSCN-006, encoding a ɣ-SNAP (gamma-SNAP), contributes to SCN resistance. CRISPR/Cas9-mediated disruption of the cqSCN-006 allele reduced SCN resistance in transgenic roots. There are no encoded amino acid polymorphisms between resistant and susceptible alleles. However, other cqSCN-006-specific DNA polymorphisms in the Glyma.15G191200 promoter and gene body were identified, and we observed differing induction of ɣ-SNAP protein abundance at SCN infection sites between resistant and susceptible roots. We identified alternative RNA splice forms transcribed from the Glyma.15G191200 ɣ-SNAP gene and observed differential expression of the splice forms two days after SCN infection. Heterologous overexpression of ɣ-SNAPs in plant leaves caused moderate necrosis, suggesting that careful regulation of this protein is required for cellular homeostasis. Apparently, certain G. soja evolved quantitative SCN resistance through altered regulation of ɣ-SNAP. Previous work has demonstrated SCN resistance impacts of the soybean α-SNAP proteins encoded by Glyma.18G022500 (Rhg1) and Glyma.11G234500. The present study shows that a different type of SNAP protein can also impact SCN resistance. Little is known about ɣ-SNAPs in any system, but the present work suggests a role for ɣ-SNAPs during susceptible responses to cyst nematodes.
INTRODUCTION
Soybean cyst nematode (Heterodera glycines; SCN) is consistently the most economically damaging pathogen of soybean (Glycine max), one of the world’s most important crops (Allen et al., 2017). This soilborne pathogen infects soybean roots and establishes complex feeding sites known as syncytia. Nematode-secreted effectors dramatically reprogram host cells to develop into these syncytia from which nematodes feed throughout development (reviewed in Gheysen & Mitchum, 2011). Adult females will continue to feed at the syncytium for three to four weeks, during which time they are fertilized by males that have left the root (Niblack et al., 2002). The body cavity of fertilized females fills with eggs and eventually their bodies harden to form cysts. Cysts with viable eggs can endure in the soil for more than a decade, providing a continual source of inoculum that is difficult to eradicate.
Due to the persistent nature of this pathogen and the expense, non-target toxicity and/or low efficacy of available nematicides, SCN management has relied primarily on crop rotation and host resistance. For decades, resistance in commercial varieties in the United States has been based heavily on a single source, Rhg1 (Resistance to Heterodera glycines 1) from G. max accession PI 88788 (Rincker et al., 2017). As a result, nematode populations with an increasing capacity to develop on soybean lines that carry Rhg1 have emerged across the nation (Acharya et al., 2016; Chen et al., 2010; Faghihi et al., 2010; Hershman et al., 2008; McCarville et al., 2017; Mitchum et al., 2007; Niblack et a,l, 2008). The identification of new sources of resistance is needed to sustain soybean production.
Wild relatives of domesticated species are commonly exploited for novel traits (Dempewolf et al., 2017). Resistance to SCN has previously been identified in Glycine soja accession PI 468916 (Kabelka et al., 2005). This trait can be bred into commercially relevant G. max varieties without any apparent negative agronomic consequences (Kabelka et al., 2006). Two independent SCN resistance quantitative trait loci (QTL), cqSCN-006 and cqSCN-007, have been fine mapped in this accession to chromosome 15 and 18, respectively, and each QTL can confer resistance on its own (Kim & Diers, 2013; Yu & Diers, 2017). When originally mapped, QTL cqSCN-006 on chr 15 explained 23% of the variation for SCN resistance and cqSCN-007 on chr 18 explained 27% of the variation (Wang et al., 2001). When these loci are stacked with other resistance loci, including or not including Rhg1, resistance to highly virulent nematode populations can be achieved (Brzostowski & Diers, 2017), suggesting these QTL could be important for cultivar improvement to thwart evolving SCN populations.
Fine mapping has narrowed the cqSCN-006 locus to a genetic interval corresponding in the SCN-susceptible Williams 82 reference genome to a 212 kilobase (kb) segment on chromosome 15 that contains three functionally annotated genes and three additional genes with unknown function (Supplemental Table S1; Yu & Diers, 2017). Included in this interval is Glyma.15G191200 that encodes a ɣ-SNAP (gamma-soluble NSF (n-ethylmaleimide sensitive fusion protein) attachment protein). This protein bears some resemblance to the product of one of the three genes involved in resistance at Rhg1, Glyma.18G022500, which encodes an α-SNAP protein (Cook et al., 2012). SNAP proteins are involved in vesicle trafficking as part of the 20S complex that consists of SNAPs, the ATPase NSF (N-ethylmaleimide sensitive fusion protein), and membrane bound SNARE (SNAP-receptor) proteins of various types (Clary et al., 1990). The 20S complex forms after vesicle fusion has occurred and unwinds the membrane bound SNARE bundles that drew the two membranes together, releasing the SNARE proteins for subsequent rounds of vesicle fusion. While there has been extensive research exploring the function of α-SNAPs in this and other processes, there has been strikingly little research in any system about the role of ɣ-SNAPs. ɣ-SNAPs share about 23-25% amino acid sequence similarity to the other SNAPs, α-SNAP and β-SNAP, but the known tertiary structure is highly similar to that of α-SNAPs (Bitto et al., 2007). Their participation in 20S complexes has also been demonstrated (Whiteheart et al., 1993). It has been proposed that in mammalian cells ɣ-SNAPs are involved in only a subset of vesicle fusion events, namely endocytosis-related trafficking, based on the limited set of SNAREs with which ɣ-SNAP was observed to interact (Inoue et al., 2015).
The α-SNAPs encoded at SCN resistance conferring Rhg1 alleles contain amino acid polymorphisms that interfere with vesicle trafficking events and 20S complex formation (Cook et al., 2014). The toxic Rhg1 α-SNAP accumulates to elevated levels in syncytia during SCN infection (Bayless et al., 2016), presumably disrupting vesicle trafficking events required for syncytium vitality. Findings about Rhg1 α-SNAP indicate that vesicle trafficking is a key process that can be targeted to disrupt the host biotrophic interface that some pathogens, like SCN, require to complete their lifecycle. It is possible, therefore, that the ɣ-SNAP from cqSCN-006 has also evolved to target this process in a unique way.
In this study, we report a role for the cqSCN-006 ɣ-SNAP in the SCN resistance conferred by the locus, potentially due to altered regulation and response to nematode infection. CRISPR/Cas9-mediated gene editing of the resistance allele increased SCN susceptibility. From there, we identify genetic and regulatory differences of ɣ-SNAP in cqSCN-006 resistant plants compared to susceptible varieties. This includes promoter and intronic SNPs that appear to impact protein accumulation and gene expression of previously unidentified splice forms. Both protein levels and splice form expression differ between resistant and susceptible roots and both respond to SCN infection. This work identifies a unique role for ɣ-SNAPs, an understudied protein, in plant-nematode interactions.
RESULTS
Glyma.15G191200 contributes to SCN resistance at cqSCN-006
The SCN resistance allele from cqSCN-006 was previously bred into G. max from G. soja accession PI 468916. Prior fine-structure genetic mapping had identified a genetic interval spanning six predicted genes, Glyma.15G191000 to Glyma.15G191500, as the source of cqSCN-006 resistance (Kabelka et al., 2005; Kim & Diers, 2013; Yu & Diers, 2017). Three of the genes in this interval had annotated functions, including Glyma.15G191200 that encodes a ɣ-SNAP (Supplemental Table S1). A whole genome sequence (WGS) of PI 468916 was generated using paired-read Illumina technology and assembled de novo to identify genomic sequence and structural changes between PI 468916 and the susceptible reference Williams 82 (Schmutz et al., 2010). Additionally, cloned fosmids from a fosmid library generated from PI 468916 were used along with the WGS to identify any major structural variation at cqSCN-006. This provided substantial but incomplete coverage of the candidate genetic interval, and identified numerous single nucleotide and indel variants, but the analysis did not direct strong attention toward a particular candidate gene. Work was initiated to silence the expression of a few individual candidate genes in transgenic soybean roots and then test those roots for altered SCN resistance. None of the tested genes gave preliminary results confirming altered SCN resistance, but none were tested with sufficient replication to draw strong conclusions in those initial efforts. In light of previous results identifying an encoded α-SNAP variant as a contributor to resistance at the Rhg1 locus of soybean (Bayless et al., 2016; Cook et al., 2012), Glyma.15G191200 was then investigated in greater detail as the strongest candidate gene at the cqSCN-006 locus.
A CRISPR/Cas9 gene-editing system was employed to test the hypothesis that Glyma.15G191200 contributes to SCN resistance in soybean lines such as LD10-30110 that carry the resistant genotype at cqSCN-006. Transgenic roots were generated expressing Cas9 and two guide-RNAs (gRNAs) specific to Glyma.15G191200 (Jacobs et al., 2015; Supplemental figure S1A). As controls, empty vector (EV) roots expressing Cas9 but not gRNAs in resistant (LD10-30110) and LD10-30092, a closely related line with the susceptible genotype at cqSCN-006, were also generated. Heteroduplex analysis was used to identify roots from the resistant line LD10-30110 that carried successful editing (mutated alleles of Glyma.15G191200; Supplemental Figure S1B)
Roots were then tested in SCN infection assays in four biological replicate experiments. Roots with mutations in Glyma.15G191200 were more susceptible to SCN, observed as a greater proportion of nematodes maturing past the J2 stage 12-14 days post inoculation relative to the resistant EV control (Figure 1). Resistant LD10-30110 roots with detectable gene editing were also more susceptible to SCN than resistant LD10-30110 roots that had been transformed with the Glyma.15G191200-targeting gRNAs but for which no editing was detected (Figure 1).
Figure 1. SCN resistance of cqSCN-006 soybean roots with edited Glyma.15G191200 allele.
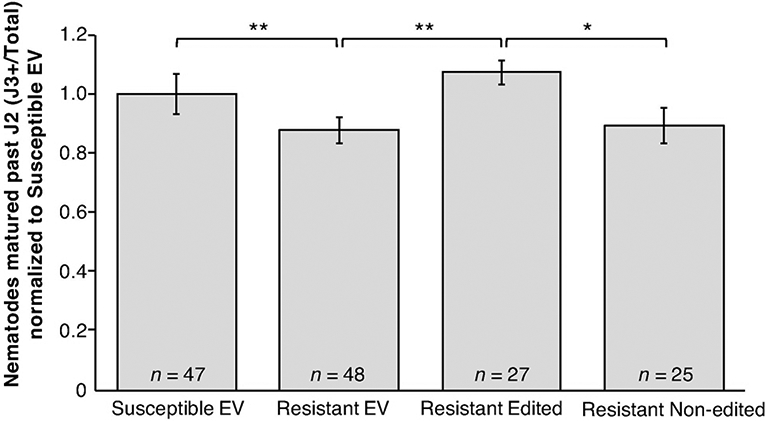
SCN development on transgenic roots of cqSCN-006 line LD10-30110 (SCN resistant) carrying CRISPR/Cas9-induced mutations in one or both alleles of Glyma.15G191200 (Resistant Edited), compared to control LD10-30092 (SCN susceptible) roots transformed with empty vector (Susceptible EV), LD10-30110 with empty vector (Resistant EV), or LD10-30110 transformed with the gene editing construct but not carrying mutations in the Glyma.15G191200 target region (Resistant Non-edited). Combined data from four independent experiments are shown; total n for each treatment is shown along with mean and standard error of the mean. Asterisks identify all significant differences (* = P < 0.1, ** = P < 0.05, *** = P < 0.01, One-tailed Welch’s t-test).
Promoter, intronic, and exonic SNPs are associated with cqSCN-006
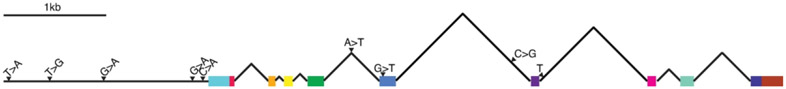
Genomic DNA sequence data from the cloned and isolated PI 468916 fosmids and PI 468916 WGS scaffolds covered the entire ɣ-SNAP candidate gene Glyma.15G191200 together with flanking intergenic regions. Comparison between the PI 468916 and Williams 82 sequences of Glyma.15G191200 revealed just one single nucleotide variant (SNV) within an exon; this change is synonymous. However, we identified multiple SNVs between PI 468916 and Williams 82 which were located in the inferred regulatory region upstream of Glyma.15G191200 and within the introns of Glyma.15G191200. These SNVs were verified by PCR and Sanger sequencing directly from the susceptible and resistant soybean experimental lines LD10-30080 and LD10-30110 (Figure 2). When compared to 18 other soybean lines, all but one of the SNVs associated with Glyma.15G191200006 were found to be unique to PI 468916 and the resistant line derived therefrom, LD10-30110. (Supplemental Table S2). The PI 468916 specific SNVs were also unique when compared to the recently sequenced G. soja accession PI 483463 (The Glycine max cv Lee and Glycine soja PI 483463 sequencing consortium, pre-publication). The two G. soja lines only share one SNV, located 180bp upstream of the predicted transcriptional start site.
Figure 2. Genetic polymorphisms unique to cqSCN-006.
Gene model depicting exons (colored bars), and introns (black lines) of Glyma.15G191200. Arrows indicate nucleotide polymorphisms unique to cqSCN-006 compared to susceptible Williams 82 with difference (the sequence of the PI 468916 allele) noted after the corresponding Williams 82 nucleotide. One letter indicates insertion of that nucleotide.
In addition to the 296 amino acid ɣ-SNAP protein encoded on chromosome 15, the Williams 82 reference genome also encodes an ɣ-SNAP on chromosome 9 at Glyma.09G083100. The two homeologous genes share 90% nucleotide sequence identity. The chromosome 15 ɣ-SNAP and the Williams 82 soybean α-SNAP within the Rhg1 locus on chromosome 18 share 25% predicted protein identity. When the derived amino acid sequence of Glyma.15G191200 is submitted to the protein structure prediction program Phyre2 (Kelley et al, 2015), 92% of the amino acid sequence maps with 100% confidence to the solved crystal structure 2IFU of a ɣ-SNAP from zebrafish (Danio rerio; Bitto et al., 2007).
Further bioinformatic analysis using PlantPAN2.0 (Chow et al., 2016) revealed the presence of a unique predicted GATA transcription factor binding site in the PI 468916 allele of Glyma.15G191200, resulting from a G>A SNV 153 bp upstream of the transcription start site (Figure 2). There is no comparable GATA binding site upstream of Glyma.15G191200 in the susceptible Williams 82 reference genome, nor in the susceptible breeding line LD10-30080.
There are multiple alternative splice forms of Glyma.15G191200
Publicly available soybean RNAseq data on the Phytozome genome browser (Goodstein et al., 2012) indicated the presence of RNA reads mapping to the fifth intron of Glyma.15G191200, although no alternative splice form was annotated. These reads were unique to the chromosome 15 ɣ-SNAP and did not map to the homeolog on chromosome 9. To detect transcription of the predicted intron 5 retention splice form, RT-PCR was performed using primers within the retained intron (intron 5) and the exon immediately 5’ to that intron (exon 5). Expression of the alternative transcript above background was detected in cDNA libraries prepared from resistant (LD10-30110) and susceptible (LD10-30092) roots, with or without nematode infection (Figure 3A).
Figure 3. RT-PCR to detect alternative splice forms of Glyma.15G191200.
(A) Predicted intron 5 retention transcript model based on published RNAseq data (Phytozome). (B) RT-PCR using primers specific to alternative splice form (arrows in A) detected the intron 5 retention splice form in all tested contexts: resistant (LD10-30110), susceptible (LD10-30092), with or without SCN infection. No reverse transcriptase (No RT) controls test that amplification is from cDNA and not background genomic DNA. GmSKP16 serves as internal control for cDNA quality and abundance (C) Transcript models for primary fully spliced transcript and additional splice forms revealed upon cloning of the intron retention splice form shown in (A) carrying an intron retention and/or exon exclusion. SNPs unique to cqSCN-006 indicated on gene models. The locations of the premature stop codons are indicated with a red asterisk.
The alternative splice form was cloned from multiple contexts, including resistant and susceptible genotypes, with or without nematode infection. Overlapping primers within the retained intron were used along with primers in the 5’ and 3’ UTR to specifically amplify the alternative transcript in two pieces that were subsequently recombined using Gibson assembly (Gibson et al., 2009). Cloning and subsequent sequencing revealed an additional splice form that, along with the retained fifth intron, also had a partial exclusion of the third exon, where only eight nucleotides from this exon were included (Figure 3B). These two splice forms have been named Glyma.15G191200.2, the intron 5 retention, and Glyma.15G191200.3, the intron 5 retention and exon 3 exclusion. Both transcripts contain premature stop codons (Figure 3C). If translated, these transcripts would generate a 166 amino acid and a 46 amino acid peptide, respectively.
The available RNAseq data also indicated some expression of the sixth intron (Goodstein et al., 2012). RT-PCR primers designed to amplify this splice form could detect minimal expression of this transcript above background. (Supplemental Figure S2A). However, both the RNAseq data and the RT-PCR result suggested that expression of this transcript is relatively low. This transcript has been named Glyma.15G191200.4. The retained intron introduces a premature stop codon and if translated, the transcript would encode a 283 amino acid peptide (Supplemental Figure S2B).
Expression of the alternative splice forms responds to nematode infection at two and five days after inoculation
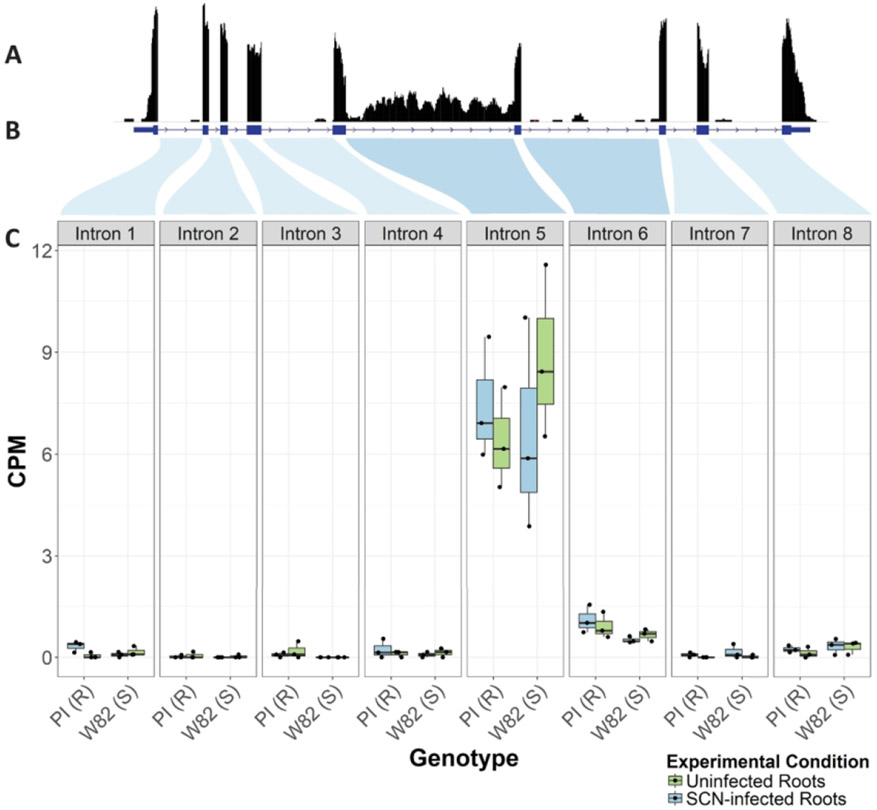
Because we detected DNA SNVs in the introns and exons surrounding the Glyma.15G191200 splice locations, we hypothesized that there may be differences in splice form expression between resistant and susceptible varieties. Reads from transcripts showing retention of intron 5 (Glyma.15G191200.2/Glyma.15G191200.3), and to a smaller extent intron 6 (Glyma.15G191200.4) were then further investigated using Illumina RNASeq data. RNA was extracted from replicated samples of entire roots from PI 468916 and Williams 82 grown under controlled conditions either without nematodes, or 8h post inoculation with J2 SCN. Uniquely mapping reads were found in significant amounts to each of the introns in Glyma.15G191200 (Figure 4). These data suggest that there is retention of intron 5 (Glyma.15G191200.2) and to a lesser extent intron 6 (Glyma.15G191200.4) both in infected and uninfected tissue 8 hours post infection)in resistant and susceptible genotypes (PI 468916 and Williams 82). However, the data at 8 hours post inoculation did not indicate a statistically significant change in expression between the infected and uninfected plants of either genotype.
Figure 4. Reads mapped to introns of Glyma.15G19200.
Reads from RNASeq analysis of RNA from uninfected or 8h post SCN inoculation root tissue of susceptible (Williams 82) or resistant (PI 468916) soybean plants were uniquely mapped the genomic region of the gamma-SNAP gene Glyma.15G19200. (A) Uniquely mapped reads to this region are shown as a coverage graph of the genic region. (B) The annotated gene exon/intron borders of Glyma.15G19200 are shown as blue bars and lines. Shaded regions indicate where introns map to box plots in (C). (C) Normalized CPM (counts per million) of reads as normalized mapping to each intron of the gamma-SNAP gene (box plots shown means, 25th, and 75th percentiles and 1.5 IQR (Interquartile Range).
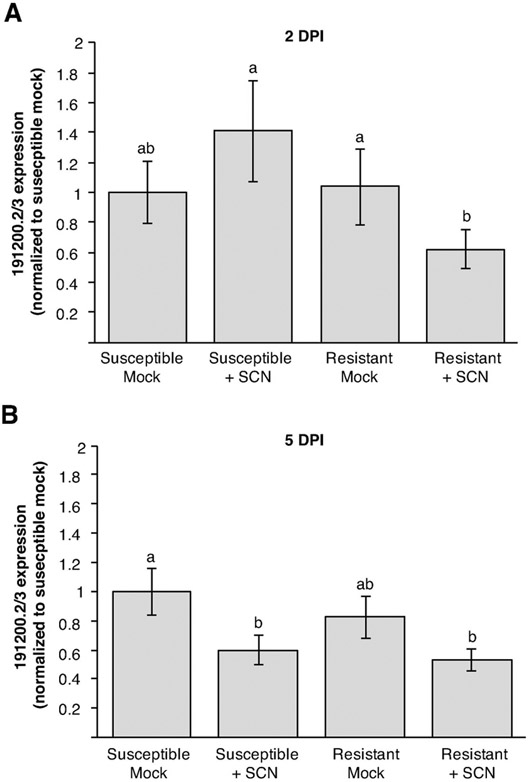
We then hypothesized that the alternative splice forms may change in response to infection, but within cells only adjacent to the infection sites and/or later on in the infection process. Samples of infected tissues were taken at two and five days post inoculation (dpi). At two dpi, successful members of the primary wave of infecting soybean cyst nematodes are expected to have selected a feeding cell and started the reprogramming of host cells to form the syncytium nematode feeding site. By five dpi on susceptible plants many nematodes will have established a syncytium and will have enlarged, with some nematodes progressing through the second molt (Ithal et al., 2007). Thus, the expression of Glyma.15G191200.2 and Glyma.15G191200.3 in plants at these stages of infection was analyzed using qRT-PCR with a primer set that would detect both alternative transcripts. For this experiment our samples were enriched for syncytia by collecting tissues near the area of inoculation. Tissues from similar areas on mock inoculated roots for each time point were sampled as controls.
At two dpi, abundance of the alternative transcripts in the infected cqSCN-006 resistant line (LD10-30110) was significantly down-regulated compared to the resistant mock-inoculated line (Figure 5A; P < 0.05, ANOVA Tukey). The susceptible infected roots did not show this reduction of alternative transcript abundance. By five days post inoculation, abundance of the alternative transcripts was significantly decreased in both susceptible and resistant lines compared to the susceptible mock control variety, albeit under slightly relaxed criteria (P = 0.06 for susceptible and P = 0.06 for resistant inoculated treatments in ANOVA Tukey; Figure 5B). No significant difference in overall expression of all Glyma.15G191200 transcripts was observed at either time point (Supplemental Figure S3). Thus, differential expression of specific splice forms was observed, but not differential overall levels of transcript expression.
Figure 5. Gene expression of alternative splice forms Glyma.15G191200.2/3 during SCN infection.
qPCR data for Glyma.15G191200.2/3 during SCN infection (+ SCN) or mock inoculation (mock) susceptible (LD10-30080) and resistant (LD10-30110) roots at two (A) and five (B) days post infection. Expression of the alternative transcript was measured relative to constitutively expressed GmSKP16. Data show mean and standard error of the mean of expression values normalized to susceptible mock within each experiment. Statistical analysis was performed on raw data. Treatments with same letter are not significantly different (P > 0.05 ANOVA Tukey (A); P > 0.1 ANOVA Tukey (B)). Data are for four independent experiments from different dates that each contained all treatments. Total n for each treatment was: (A) susceptible mock, 14; susceptible +SCN, 13; resistant mock, 11; resistant +SCN, 15. (B) susceptible mock, 14; susceptible +SCN, 16; resistant mock, 13; resistant +SCN, 15.
cqSCN-006 resistant lines exhibit delayed nematode-induced accumulation of ɣ-SNAP protein in infection sites
To more directly observe changes in ɣ-SNAP protein levels during infection, ɣ-SNAP protein levels over the course of infection were measured by immunoblots with a custom ɣ-SNAP antibody. Samples were taken and two, five and ten dpi and total protein was extracted and used for immunoblots (Figure 6 B,D). The antibody was determined, using recombinant proteins, to be specific for ɣ-SNAP rather than α-SNAP (Supplemental Figure S4). The antibody was further determined to be approximately five-fold more sensitive in detecting the ɣ-SNAP encoded at the cqSCN-006 locus on chromosome 15 relative to the ɣ-SNAP encoded on chromosome 09 (Supplemental Figure S4).
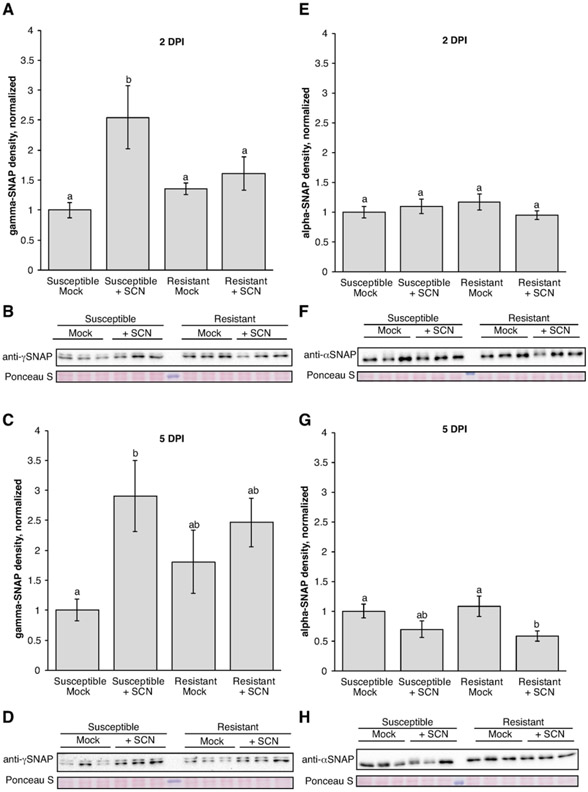
Figure 6. ɣ-SNAP and α-SNAP protein levels measured during SCN infection.
(A, C, E, G) Densitometry analysis of SNAP protein levels from the indicated 2 dpi or 5 dpi time point. n = 12 samples for each treatment (3 each from four independent biological replicate experiments). Mean and standard error of mean are for all data points normalized to mean for susceptible mock within each experiment. Means of treatments with the same letter are not significantly different (P > 0.01 for all except P > 0.05 for G) ANOVA Tukey, analysis performed on non-normalized data). (B, D, F, H) Representative SNAP immunoblots for each time point. Ponceau S stain tests for equal loading across samples. For ɣ-SNAP blots, the lower band that aligns with the predicted size of ɣ-SNAP was measured.
Densitometry analyses of ɣ-SNAP immunoblots from four independent SCN infection experiments, consisting of three samples for each treatment in each experiment, revealed that resistant and susceptible roots exhibited differences in ɣ-SNAP protein accumulation behavior. At two days after nematode inoculation a statistically significant induction of ɣ-SNAP protein in susceptible infection sites compared to the susceptible mock control was detected (Figure 6 A,B; P < 0.01 ANOVA Tukey). No comparable increase in ɣ-SNAP protein was observed in resistant roots at this time point (Figure 6 A,B). At five dpi, ɣ-SNAP protein levels remained elevated in susceptible infection sites relative to susceptible mock-inoculated control (Figure 6 C,D; P < 0.01 ANOVA Tukey). However, we still observed no differences in ɣ-SNAP accumulation in SCN-infected resistant roots compared to the resistant mock-inoculated roots (Figure 6 C,D). By ten dpi ɣ-SNAP levels were diminished and difficult to detect (Supplemental Figure S5).
To determine if the induction of ɣ-SNAP is unique to this SNAP family member, changes in α-SNAP levels were also investigated with immunoblots and subsequent densitometry analyses. Both lines tested have the Williams 82 (susceptible-type) α-SNAP allele. Blots previously probed for ɣ-SNAP were stripped of antibody under mild conditions. Stripping was verified and blots were subsequently probed using a custom antibody that recognize wild-type (Williams 82-type) α-SNAP proteins (Figure 6 F,H; Bayless et al., 2016). No significant changes in α-SNAP protein levels at 2 dpi in any treatment were detected (Figure 6 E, P > 0.1 ANOVA Tukey). Meanwhile, at 5 dpi α-SNAP protein levels are significantly lower in resistant infected treatments compared to the resistant and susceptible mock treatments, although under more relaxed criteria than those used previously (Figure 6 G; P < 0.05 ANOVA Tukey). However, further analysis revealed that this antibody also detects ɣ-SNAP (Supplemental Figure S4) and therefore this signal may be better interpreted as overall SNAP protein levels. Any decrease in α-SNAP levels may be partially masked by small or large increases in ɣ-SNAP levels, so the actual changes in α-SNAP level may be even greater.
A balance between ɣ-SNAP and α-SNAP levels required for homeostasis
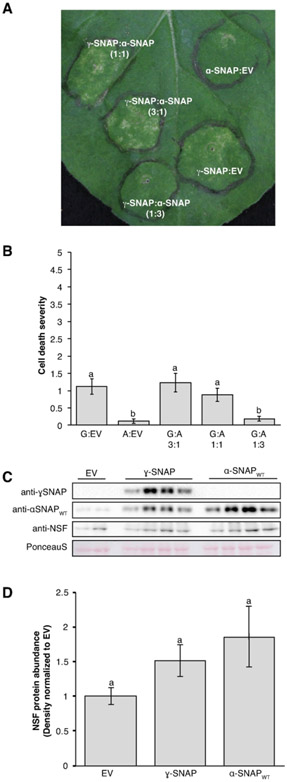
It had been reported previously that in mammalian systems, either overexpression of ɣ-SNAP or its opposite, silencing of ɣ-SNAP, induce the same defects in vesicle trafficking, observed as enlarged early endosomes (Inoue et al., 2015). This suggests that modifying ɣ-SNAP levels can interfere with normal vesicle trafficking. Transient expression of 20S complex components in Nicotiana benthamiana leaves has been used previously to study the function and impact of particular variants of the 20S complex (Bayless et al., 2016, 2018). For example, the unusual Rhg1 α-SNAP causes dose-dependent necrotic flecking in the N. benthamiana assay that is not observed when expressing wild-type soybean α-SNAPs (Bayless et al., 2016, 2018). Therefore, this system was used to investigate a potential impact of overexpressing ɣ-SNAP in plant cells.
Overexpression of ɣ-SNAP induced a moderate cell death phenotype in N. benthamiana leaves that consisted of chlorosis and necrotic flecking (Figure 7 A). When cell death of infiltrated areas was rated on a 0-5 scale, the moderate phenotype was found to be statistically significant across four biological replicates (Figure 7 B, P < 0.05 Pairwise Wilcoxon rank sum test). This cell death was relieved by co-expression of wild-type soybean α-SNAP in a three-fold excess relative to ɣ-SNAP expression, but not when ɣ-SNAP transient expression abundance exceeded that of α-SNAP or if both were expressed at a 1:1 ratio of Agrobacterium strains (Figure 7 A, B). The relief by wild-type α-SNAP of the toxic ɣ-SNAP-induced phenotypes suggests that α-SNAP and ɣ-SNAP compete for similar sites of action (see also Bayless et al., 2016, 2018).
Figure 7. Transient expression of ɣ-SNAP in N. benthamiana leaves.
(A) Representative N. benthamiana leaf expressing ɣ-SNAP and wildtype soybean α-SNAP 8 days after infiltration with Agrobacterium strains at the indicated ratios. (B) Cell death ratings on a 0-5 scale, for multiple independent leaves. Means for treatments with the same letter are not statistically different (P > 0.05, pairwise Wilcoxon rank sum test). Data shown are mean and standard error of the mean; n = 26 for all but G:A 1:3 for which n = 21. G: ɣ-SNAP; A: α-SNAP. (C) Representative immunoblots verifying expression of the indicated proteins and used to measure NSF levels in leaf areas expressing ɣ-SNAP or wildtype soybean α-SNAP. NSF antibody detects N. benthamiana NSF. Blots were first probed for ɣ-SNAP and then stripped and simultaneously reprobed for α-SNAP and NSF. Ponceau S stain tests for equal loading across samples. (D). Densitometry analysis of NSF protein levels across three independent biological replicates consisting of three or four samples each (n = 10). Data were normalized to the empty vector (EV) control for each replicate; mean and standard error of the mean are shown. Means for treatments with the same letter are not statistically different (P > 0.1, ANOVA Tukey).
Previous work also revealed that ectopic expression of the unusual and toxic α-SNAPs from Rhg1 SCN resistance loci strongly induces expression of N. benthamiana NSF (Bayless et al., 2016). To analyze if expression of ɣ-SNAP produced a similar response, NSF protein levels during ɣ-SNAP expression were measured using immunoblots (Figure 6C). Across three biological replicates, no significant increase in NSF was observed (P > 0.1 ANOVA Tukey; Figure 7 C, D). While there appeared to be a slight increase in NSF protein abundance upon expression of either ɣ-SNAP or wild-type soybean α-SNAP, this was not statistically significant, and did not approach the obvious induction of NSF by expression of α-SNAPRhg1 (Supplemental Figure S6). In addition, transient expression in N. benthamiana of either of the two above-discussed alternative transcripts in N. benthamiana did not result in the moderate cell death phenotype observed from expression of full length ɣ-SNAP (Supplemental Figure S7). To summarize, differences in the modulation of ɣ-SNAP protein levels at SCN infection sites were observed between susceptible roots and those resistant due to cqSCN-006, and altered ɣ-SNAP levels did perturb cell health in N. benthamiana leaves, but the severity of the response was not as strong as that previously observed for the Rhg1-encoded α-SNAP proteins.
DISCUSSION
Identification of novel SCN-resistance genetics is crucial for sustaining soybean production across the United States. Through this work we have identified a gene that contributes to the resistance mechanisms conferred by the SCN resistance QTL cqSCN-006, originally derived from a wild Glycine soja accession. CRISPR/Cas9 mediated gene editing revealed a role of the ɣ-SNAP encoded by Glyma.15G191200 of cqSCN-006 in the SCN-resistance conferred by this locus. Successful CRISPR editing of the resistant allele increased nematode susceptibility compared to resistant empty vector roots as well as roots that were expressing gRNAs but for which no editing was detected.
ɣ-SNAP proteins are part of the 20S complex that recycles membrane-bound SNAREs for subsequent rounds of vesicle trafficking (Clary et al., 1990; Whiteheart et al., 1993). A variant form of an α-SNAP protein, part of the same complex, has been found to confer resistance from the major SCN resistance locus Rhg1 (Bayless et al., 2016; Cook et al., 2012). The identification of an SCN resistance phenotype conferred by an alternative allele of another SNAP protein from an unrelated locus points to the likely importance of the 20S complex in cyst nematode pathogenesis and host resistance. Apparently, the Glycine genus has independently evolved multiple mechanisms of SCN resistance that impact the vesicle trafficking system during SCN infection
Little is known about the function of ɣ-SNAPs in any eukaryotic system. ɣ-SNAPs have greater amino acid sequence divergence from the other SNAP family proteins, α-SNAP and β-SNAP, than they have between themselves, but all three have similar structures (Bitto et al., 2007). ɣ-SNAPs are found in plants and animals, but no ɣ-SNAPs are encoded in unicellular yeasts, (Clary et al., 1990). In mammalian cells, ɣ-SNAPs were only observed to interact with a subset of SNAREs, including those involved in endocytosis (Inoue et al., 2015). While ɣ-SNAPs have been marginally associated with salt stress in Arabidopsis through transcriptomics (Sottosanto et al., 2004) and seed germination in rice through proteomics (Han et al., 2014), no functional studies of ɣ-SNAPs in plants have yet been published. In this work we learned that expression of a ɣ-SNAP protein and its associated transcripts responds to SCN infection during a compatible interaction, and, alteration of those responses occurs during the SCN resistance response mediated by cqSCN-006.
Previously characterized resistance to SCN has been conferred, at least partially, by amino acid polymorphisms in cellular proteins (Cook et al., 2014; Liu et al., 2012). For example, the α-SNAP at Rhg1 has amino acid polymorphisms at key residues in the functional C-terminus that renders it toxic to cells (Bayless et al., 2016; Cook et al., 2014). Whole genome and fosmid sequencing found no nonsynonymous single nucleotide variants in exons of Glyma.15G191200 relative to the susceptible reference. However, multiple SNVs throughout the Glyma.15G191200 promoter and gene body were found to be unique to PI 468916 when compared to 18 other soybean sequences and to a separate G. soja accession. One of these distinctive SNVs, a G>A 153bp upstream of the predicted transcriptional start site, introduces a strong consensus sequence for a GATA transcription factor binding site (Figure 2). In soybean, GATA transcription factors have been associated with nitrogen metabolism (Zhang et al., 2015), salt stress, and perhaps auxin and gibberellic acid signaling (Liu et al., 2018). Moreover, nematode-induced transcriptional reprogramming has been observed during syncytial development (Gheysen & Mitchum, 2011). For example, SCN effector GLAND4 was recently demonstrated to bind to promoter elements of defense related genes to repress transcription (Barnes et al., 2018). Therefore, changes in transcription factor binding sites, be they the inferred new GATA transcription factor binding site or other unidentified sites, may influence the ability of the nematode to induce expression changes in Glyma.15G191200 expression required for syncytia establishment.
We observed differential regulation of ɣ-SNAP protein levels in resistant cqSCN-006 lines compared to SCN-susceptible lines. In a susceptible soybean root, ɣ-SNAP protein levels were found to increase during SCN infection relative to mock inoculated controls at two and five days post inoculation. This significant induction was not observed in cqSCN-006 resistant roots at either time point (Figure 6 A,C). It appears that cqSCN-006 resistant plants avoid the nematode-induced elevation of ɣ-SNAP levels within or near feeding sites, suggesting that induction of ɣ-SNAP helps to promote SCN infection.
No changes in α-SNAP levels were observed at 2dpi, but there does appear to be a significant decrease in α-SNAP protein at 5dpi in resistant lines. (Figure 6 E,G). These results are confounded by the fact that the α-SNAP antibody also detects ɣ-SNAP (Supplemental Figure S4). Therefore, the decrease in α-SNAP could be even greater in both susceptible and resistant lines. This indicates that SCN may specifically induce ɣ-SNAP and not α-SNAP during a compatible interaction, suggesting a particular role for ɣ-SNAP in SCN pathogenesis. The differential protein accumulation may be the result of the SNVs found in the promoter region of Glyma.15G191200 in cqSCN-006, or at presently unidentified sites that also reside outside of the gene coding sequence. Promoter mutations have been associated with disease resistance, such as in the xa13 rice bacterial bight resistance (Chu, 2006). These promoter changes allow the resistant plant to escape pathogen induced expression of the downstream gene (Yuan et al., 2009).
Our observation of significant SCN-induced accumulation of ɣ-SNAP only in susceptible roots might seem to run counter to the results of the CRISPR/Cas9 virulence assay. In CRISPR edited roots that mutate the ɣ-SNAP gene, reduction of ɣ-SNAP protein levels might be hypothesized to increase resistance if ɣ-SNAP is a straightforward susceptibility factor. However, there is precedent for the same functional outcome or organismal phenotype being caused by increases or decreases in a particular protein, particularly those in macromolecular complexes such as SNAPs, including a specific precedent with ɣ-SNAP protein (Inoue et al., 2015; Veitia et al., 2008; Wang et al., 2014). It has been observed previously in mammalian cells that both overexpression and RNAi-mediated silencing of ɣ-SNAP in various cell lines caused enlarged endosomes (Inoue et al., 2015). This also suggests that a certain level of ɣ-SNAP is required for cellular homeostasis. In mammalian cells, when this balance was upset, cells responded with modified trafficking through enlarged endosomes (Inoue et al., 2015). In soybean roots or developing syncytia, CRISPR mediated reduction in ɣ-SNAP may have resulted in similar vesicle trafficking changes as those naturally induced by elevated ɣ-SNAP protein levels during SCN infection of susceptible roots. If this change promotes infection, both instances result in SCN susceptibility. Meanwhile, the cqSCN-006 resistant roots maintained steady ɣ-SNAP protein levels during early stages of infection, which apparently is associated with SCN resistance.
We did observe that overexpression of ɣ-SNAP in N. benthamiana leaves caused stress that was relieved by an abundance of α-SNAP (Figure 7 A, B). This supports a required balance between these two SNAPs in plants, as was observed in mammalian cells (Inoue et al., 2015). The overexpression of ɣ-SNAP does not appear to cause the same stress to the plant cell that was caused by SCN resistance-associated α-SNAPs, as observed through an increase in NSF protein expression (Figure 7 C, D; Supplemental Figure S4). It appears that the effect of ɣ-SNAP overexpression is not as overtly lethal, but may be inducing changes that manifest as moderate cell death in the N. benthamiana assay. These changes may favor a developing syncytium (e.g. Siddique et al., 2014). Further research is required to determine what these vesicle trafficking changes or other changes may be and if they are occurring in susceptible, and not resistant, syncytia.
We identified the expression of at least three previously unannotated splice forms of Glyma.15G191200 (Figure 3, Figure 4, Supplemental Figure S2). These splice forms appear to be unique to the ɣ-SNAP encoded on chromosome 15 as no RNAseq reads mapping to the intronic regions were observed in the publicly available data for the homolog on chromosome 9. This strengthens the potential that the splice forms might participate in cqSCN-006 mediated resistance. Gene expression analyses revealed down regulation of the alternative splice form in resistant infected roots compared to resistant mock control at two days after inoculation, while this down-regulation was not observed for infected vs. mock-inoculated roots of the susceptible genotype (Figures 4, 5A). However, by five days post-inoculation both susceptible and resistant lines apparently exhibited reduced expression of the alternative transcript, to approximately the same level (Figure 5B). These root segment samples from SCN-infected regions contain many root cells at or within a few cells of infected areas but are diluted with a majority of cells from adjacent non-infected areas. The moderate significance (P = 0.06) that was observed in Figure 5B does indicate reproducibly measurable changes in expression, but finer-scale tissue sampling might reveal even greater changes in expression. Hence it may be that an early decrease in alternative transcript expression by 2 DPI in resistant infection sites limits SCN development. While no variation in protein-coding exon sequence was present between resistant and susceptible varieties, and no overall differences in gene expression were observed in bulk roots, we found evidence for altered gene expression via an alternative splicing mechanism, and altered protein abundance in syncytia-enriched tissues, and evidence that this altered protein expression contributes to SCN resistance. To reiterate, this was despite the absence of changes in the protein-coding sequence of the gene and despite evidence that overall transcript expression in the roots does not change significantly in the resistant genotype. Thus, an important trait is in this case associated with polymorphisms in introns and upstream of transcription start sites that, in many studies, would not be considered as “potentially important” sequence variants. These variants have important effects on the splicing and tissue-specific pathogen-responsive expression of the gene.
Although beyond the scope of this first report about ɣ-SNAP as a contributing factor in cqSCN-006-mediated SCN resistance, future studies might productively be focused on discovering how and if alternative splicing helps regulate the protein levels at this early infection stage. This may be an especially interesting point because a decrease in alternative splice forms 2/3 at 2 DPI in the resistant genotype might be expected to raise ɣ-SNAP protein levels, but instead we observed ɣ-SNAP protein abundance increase in the susceptible genotype. We hypothesize that avoiding the SCN-induced ɣ-SNAP protein accumulation that occurs during a compatible interaction is a mechanism of SCN resistance. Sequence analysis revealed cqSCN-006 specific SNVs in the introns and exons involved in the alternative splicing events, and nowhere else in the gene body, presenting potential genetic basis for the observed differential regulation (Figure 2). Overall transcription from the locus was not significantly different between resistant and susceptible roots, likely due to the confounding effects of the alternative splice forms (Supplemental Figure S3). Therefore, the protein levels that were measured are likely to provide a better picture of the ɣ-SNAP expression during infection. Additionally, the absence of notable overall changes in transcript abundance may suggest that protein levels are impacted by more than just transcript levels from the locus.
Recently, it has been shown that a cyst nematode effector, 30D08, interacts with Arabidopsis auxiliary spliceosomal protein SMU2 (Verma et al., 2018). Interaction of this effector impacts splicing and expression of certain genes. Therefore, splicing changes are a nematode-manipulated aspect of syncytia establishment (Verma et al., 2018). It is possible that cqSCN-006 escapes specific splicing changes stimulated by or even directly mediated by nematode infection, as observed through the more rapid decline of Glyma.15G191200.2/3 expression. The presence of the alternative transcript early during infection may promote syncytia development. Interestingly, it was also observed that heterologous overexpression of 30D08, which would be expected to promote parasitism, actually reduced nematode development (Verma et al., 2018). This further supports a model where careful regulation of specific proteins, splicing, and gene expression is required for nematode infection. Again, CRISPR/Cas9 editing of Glyma.15G191200 may disrupt this balance too dramatically and therefore result in susceptibility even though compatibility is associated with elevated alternative transcript and protein levels. Overall, it appears that careful regulation of ɣ-SNAP may be required for SCN infection, and that the ɣ-SNAP at cqSCN-006 has evolved subtle ways to disrupt this regulation.
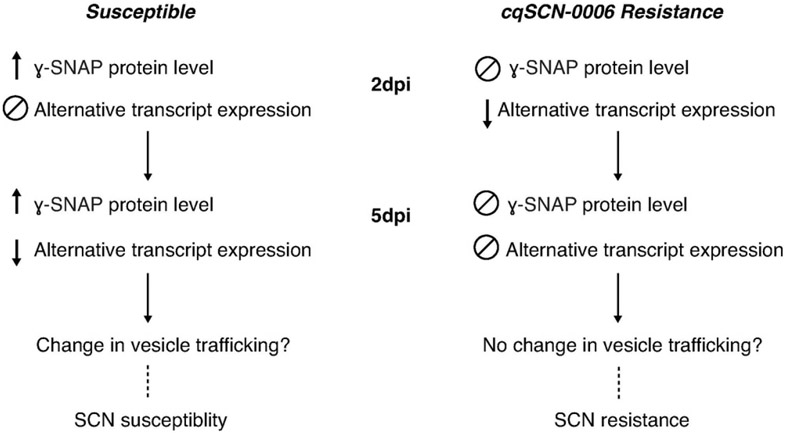
To summarize, we have identified another SNAP protein, a ɣ-SNAP, involved in SCN resistance. This trait derives from the resistance locus cqSCN-006 that originated from Glycine soja accession PI 468916. Interestingly, it appears soybean has evolved multiple mechanisms of SCN resistance that impact the 20S complex machinery and vesicle trafficking events. Resistance at cqSCN-006 seems to be, at least partially, conferred by altered regulation of the abundance of the ɣ-SNAP encoded by Glyma.15G191200, apparently as a result of promoter and intronic SNVs (Figure 8). This is observed through limited ɣ-SNAP protein accumulation during infection in resistant plants as well as differential expression of previously unidentified splice forms, particularly at early stages of infection. We hypothesize that changes in ɣ-SNAPs protein levels may elicit changes in vesicle trafficking elements that promote syncytia establishment and nematode development, which are avoided in a resistant plant. Incidentally, this work also reiterates the importance of considering the biological effects of polymorphisms outside the protein-encoding regions of genes, which for example can alter protein abundance even when neither the protein-encoding sequence nor the overall transcript levels are affected. Lastly, little is known about ɣ-SNAPs in any system, and the present study reveals a role for this protein and its associated transcripts in plant interactions with cyst nematodes.
Figure 8. Summary and proposed model for SCN resistance encoded at cqSCN-006.
Differential regulation of protein and alternative splice expression are observed between susceptible and cqSCN-006 resistant plants, particularly at early stages of infection (2 dpi). These changes may favor syncytia establishment and nematode development in susceptible lines, while SCN resistance may result from avoiding or delaying these changes.
METHODS
Plant material
Soybean lines in a commercially relevant G. max background with or without the cqSCN-006 resistance allele were generated in the breeding program of Dr. Brian Diers at the University of Illinois. Briefly, these lines were developed by first backcrossing the SCN resistance alleles cqSCN-006 and cqSCN-007 from G. soja PI 468916 into the G. max susceptible background A81-356022 (Diers et al., 2005). The allele was subsequently backcrossed into the background of LD00-3309 (Diers et al., 2006) and lines segregating for resistance alleles at Rhg1, cqSCN-006 and cqSCN-007 were identified with markers linked to each gene. The resistant line used throughout these experiments was LD10-30110, which has the resistance allele for cqSCN-006 and the susceptible allele for cqSCN-007 and Rhg1. The susceptible lines used were LD10-30080 and LD10-30092, sibling lines of LD10-30110 that have the susceptible allele for all three genes. For de novo and fosmid based sequence analysis, the G. soja accession PI 468916 was used directly
Nematode inoculum
SCN incoula from verified Hg 2.5.7 populations were obtained as eggs from Dr. Alison Colgrove at the University of Illinois Plant Clinic. Eggs were hatched in 3mM ZnCl2 for 4-6 days to obtain infectious second stage juveniles (J2). Four to six days after hatching, infectious J2 SCN were sterilized with 0.1g/L HgCl2 and 0.01% sodium azide for three minutes and washed twice in sterile water before resuspension in 0.05% sterile agarose water.
Vector construction
For editing of Glyma.15G191200, two gRNA targets were identified using CRISPR-P (Liu et al., 2017). The 19 base pair (bp) gRNAs were selected within the third exon (TCCGCTGCTGCTCTCGCAA) and the fourth exon (GTATTCTGTTGCAGCTAAT) 316 bp from each other and both on the positive strand. Exons towards the 5’ of the gene were selected as the targets to increase likelihood that editing would result in a deletion, insertion, or nonsynonymous mutation that would inhibit protein function. Additionally, the simultaneous editing at both target sites would result in a 350bp deletion, knocking out the protein completely. The p201G binary vector (a gift from Wayne Parrot, Addgene plasmid 59178) was used for expression of Cas9 and gRNAs in transgenic soybean roots with a GFP screenable marker (Jacobs et al., 2015). Assembly of gRNAs into p201G was performed using Gibson Assembly as previously reported (Gibson et al., 2009; Jacobs & Martin, 2016). The resulting plasmid, p201G_Cas9:191200_gRNAx2, was verified with restriction digest and sequencing and transformed into Agrobacterium rhizogenes strain ARqua 1 using a freeze-thaw method.
For transient expression of ɣ-SNAP in N. benthamiana leaves, Glyma.15G191200 CDS was amplified from resistant LD10-30110 cDNA with overhangs for Gibson assembly. Amplified ORFs were then placed between the soybean ubiquitin promoter (Hernandez-Garcia et al., 2010) and nos terminator in the vector pBluescript using Gibson assembly (Gibson et al., 2009) and sequence-verified. The overexpression cassette was digested from pBluescript using XbaI/PstI, gel extracted using the QIAquick Gel Extraction kit (Qiagen, Hilden, Germany), and ligated into the binary vector pSM101 using T4 DNA ligase (Promega, Madison, WI). Final constructs were transformed into Agobacterium tumefaciens strain GV3101 (pMP90) using the freeze-thaw method.
Generation of transgenic roots
Transgenic roots were generated with Agrobacterium rhizogenes strain ARqua1 as in Melito et al. (2010) Cotyledons from cqSCN-006 resistant (LD10-30110) and susceptible (LD10-30092) soybeans were transformed with either p201G_Cas9 empty vector or p201G_Cas9:191200_gRNAx2 to generate roots as previously described (Melito et al., 2010). Transformed roots were screened for GFP expression and subcultured on root media (RM) (4.3g/L MS salts, 2% sucrose, 1x Gamborg’s B5 vitamins, 6.7% agar, 0.13g/L cefotaxime, 0.13g/L carbenicillin pH 5.6) before use in subsequent experiments.
Heteroduplex analysis for identification of edited transgenic roots
Roots with successful editing (mutation) of Glyma.15G191200 were identified by heteroduplex analysis (Zhu et al., 2015). Genomic DNA was extracted from root samples with the DNeasy PowerPlant Pro kit (QIAGEN Hilden, Germany) according to manufacturer instructions. The targeted genomic site and 126 or 87bp respectively flanking the outside of the two target sites was amplified with KAPA HiFi polymerase (KAPA Biosystems Wilmington, MA) using specific primers. Amplification of the predicted 569bp band was verified on a 1.5% agarose gel stained with EtBr. After verification, amplicons were incubated at 95°C for five minutes to melt DNA and then allowed to reanneal by removal of tubes to room temperature for five minutes. Heteroduplexes were resolved on a 5% polyacrylamide gel and visualized with ethidium bromide and UV light. Mutations were detected by the presence of multiple bands indicative of the formation of heteroduplexes resulting from base substitutions or INDELs in the targeted region (Supplemental Figure 1). In some instances, smaller bands representing homoduplexes and/or larger deletions resulting from simultaneous editing at both gRNA target regions were observed. A subset of purified amplicons were sequenced to relate heteroduplex presence to editing and many complicated variants were seen, including a variety of deletions at either or both gRNA target sites (Supplemental Figure 1C).
SCN resistance assay
Resistance of transgenic roots expressing p201G_Cas9 or p201G_Cas9:191200_gRNAx2 was measured using a SCN demographics assay as in Melito et al. (2010). Transgenic roots with robust GFP expression were placed on individual tissue culture plates with RM medium (one root per plate) 24 hours before inoculation. Tissue samples for genotyping were collected from the basal end of each root at this stage, flash frozen in liquid N2, and stored at −80°C until processing. Roots were inoculated with 150-200 motile J2 at the root tip and incubated in the dark at 28°C. Nematodes on and within roots were stained with acid fuchsin 12-14 days post inoculation (Byrd et al., 1983). All nematodes on a single root were classified and counted by life stage as resembling J2, or as resembling J3 or more mature stages beyond J2. The number of nematodes resembling J3 or more mature stages as a percent of total number of nematodes on each root was determined as the index of SCN susceptibility (Cook et al., 2012; Melito et al., 2010). Means were compared using a one-way Welch’s t-test due to the unbalanced design that resulted from differential availabilities of transgenic roots within experiments and from separating the data for edited and non-edited CRISPR/Cas9 roots. To control for between-experiment variation, the result for each root was normalized to the mean for the susceptible controls within the same experiment prior to statistical analysis.
De novo sequencing and assembly of PI 468916
Whole genome sequencing of PI 468916 was performed using the HiSeq DNA sequencer (Illumina, San Diego, CA) at the Keck Biotechnology Center, University of Illinois Urbana-Champaign. DNA was extracted using a CTAB DNA extraction method (Cook et al., 2012). The raw reads were assembled using ABySS (Assembly by Short Sequences) (Simpson et al., 2009) to generate scaffolds. A total of 182,255,190 paired end reads were assembled with an N50 in the final assembly of 19,821 bp.
Fosmid library production, isolation and analysis
Genomic DNA was isolated from PI 468916 and a fosmid library was constructed as described in Cook et al., 2012. Fosmids were isolated from the fosmid library by a PCR-based isolation screening method. Primers were designed to the susceptible reference in the region where the QTL had been identified. These primers were then used for PCR on pools of the genomic library of PI 468916 each containing approximately 5,000 different fosmid clones. Subsequent rounds of PCR were performed on fosmid pools after dilution and regrowth in bacterial culture allowing for an eventual single fosmid clone to be identified that contained the amplicon. Isolated fosmids were then end-sequenced with vector-specific primers using Sanger sequencing at the Keck Biotechnology Center at the University of Illinois-Urbana Champaign to verify their physical location in the region of interest before further sequencing efforts. Isolated fosmids confirmed to be from the region of interested were then sequenced using either Sanger sequencing and primer walking or through the Illumina MiSeq at the Keck Biotechnology Center. Sequenced fosmids were assembled using ABySS.
Confirmation of SNVs in PI 468916 breeding lines
Genomic DNA was extracted from roots of resistant LD10-30110 grown in Metro Mix at 27°C/14hr daylight with the DNeasy PowerPlant Pro kit (QIAGEN Hilden, Germany) according to manufacturer’s instructions. Glyma.15G191200 promoter regions, including the 5’ UTR and the upstream 2kb, from susceptible (LD10-30080) and resistant (LD10-30110) lines were amplified with KAPA HiFi polymerase using specific primers. Purified PCR products were directly sequenced with multiple internal primers and compared to PI468916 and Williams82. Promoter sequences were obtained from fifteen additional genome sequences and compared to the PI 468916 and LD10-30110 gene sequences (Supplementary Table 1). The promoter sequence from G. soja accession PI 483463 was obtained from Soybase (Soybase.org: The Glycine max cv Lee and Glycine soja PI 483463 sequencing consortium, pre-publication)
Promoter sequences from resistant (LD10-30110) and susceptible (LD10-30080) soybean lines were analyzed for transcription factor binding sites using Plant Pan 2.0 (Chow et al., 2016). Predicted transcription factor binding site output was analyzed for differences between the two lines at areas where SNVs had been identified.
Root infections for protein and qPCR analyses
Resistant (LD10-30110) and susceptible (LD10-30080) soybean seeds were sterilized overnight with chlorine gas by adding 3.5 mL of HCl to beaker 100 mL bleach (8.5% sodium hypochlorite) in a desiccant jar. Seeds were plated on germination media (4.41 g/L MS salts with B5 vitamins, 2% sucrose, 7% agar, pH 5.8) and allowed to germinate at 24°C/16 hour daylight for 7 days. Roots from germinated seedlings were transferred to RM for four days in the dark at room temperature to allow for growth, or transferred and inoculated the next day. If grown for four days, roots were transferred to fresh RM the day prior to inoculation. All roots were cultured in 50 x 100 x 100 mm square petri dishes containing RM, four per plate during SCN infection. Roots were inoculated with 175-200 motile, surface disinfested J2 Hg 2.5.7 as above. Mock treated roots were inoculated with an equal volume of 0.05% agarose water. The point of inoculation was marked on the back of the plate to aid with sample collection. Tissue samples were taken at indicated time points. SCN-infested root regions, previously demonstrated to correspond to the area with subtle browning ~1 cm above the point of inoculation, were excised with a scalpel. Similar areas from mock inoculated roots were collected at each time point. Three to four inoculated zones or mock inoculated zones from independent roots growing on the same plate were pooled for each sample. Tissue samples were flash frozen in liquid N2 and stored at −80°C until sample preparation. Four independent biological replicate experiments were performed (each biological replicate utilized plant/nematode materials from a separate experiment performed on a separate date). Within each experiment, treatments generated three or four root samples and homogenized aliquots from the same sample were then analyzed in both immunoblot and qPCR studies.
RT-PCR and qRT-PCR
Total RNA from infected and mock treatments was extracted from root tissues using the Zymo RNA extraction kit (Zymo Research Irvine, CA) according to manufacturer’s instructions with on column DNase treatment. RNA quality was validated using a 1.5% agarose gel and concentration was measured on a Nandrop-1000 Spectrophotometer. cDNA was synthesized with the iScript cDNA synthesis kit (BioRad Hercules, CA) according to manufacturer’s instructions. No reverse transcriptase (RT) controls were run in the same conditions but without the RT enzyme.
For detection of the Glyma.15G191200 alternative splice form, RT-PCR using GoTaq master mix (Promega Madison, WI) using specific primers and GmSKP16 as internal control was performed. For cloning of Glyma.15G191200 alternative splice forms, KAPA HiFi polymerase (KAPA Biosystems Wilmington, MA) was used to amplify the predicted transcript in two segments with 158bp of overhang within the retained intron. The two purified products were recombined with Gibson assembly (Gibson et al., 2009). The assembled product was amplified with KAPA HiFi polymerase (KAPA Biosystems Wilmington, MA) using primers for Gibson assembly into a linearized pBluescript vector. Positive clones were sequenced with Sanger sequencing. Each alternative splice form was cloned from multiple cDNAs at least three independent times.
qRT-PCR was performed using Solis Biodyne Evagreen HOT FIREPol EvaGreen qPCR mix (Tatu, Estonia) in a BioRad CFX96 Real Time system per manufacturer’s instructions. No RT controls were run with the alternative splice specific primers. Samples with Cq values for the alternative transcript less than 1 cycle greater than the no RT control were eliminated from further data analysis. Data was analyzed with the −ΔCq method and these values were used in statistical analyses with ANOVA with Tukey test in JMP.
RNASeq analysis for intron retention.
Surface sterilized soybean seeds were germinated on petri dishes with autoclaved paper towels. After three days germinated seedlings were moved to new sterilized paper towel dishes and inoculated with second stage juvenile (J2) Hg type 0 nematodes. Roots from three biological replicates of both uninoculated and inoculated plants from the two genotypes were harvested and immediately frozen for RNA analysis 8 h after the inoculation of the inoculated plants. RNA was isolated from roots using the E-Z® 96 Plant RNA Kit (Omega Bio-Tek) and sequenced on an Illumina HiSeq 4000 to a depth of 14 million reads per sample. Reads were then uniquely mapped to the annotated intron sequences of Glyma.15G191200 using tophat2 (Kim et al., 2013). Reads were then counted using featureCount in R (Liao et al., 2014). Counts were normalized to create counts per million (CPM) with edgeR (Robinson et al., 2010).
Transient expression in N. benthamiana leaves
Agrobacterium-mediated transient expression was performed as in (Bayless et al., 2016). Within each experiment, two leaves on multiple plants were used for infiltration and with randomized locations between leaves. Images were collected 8-9 days after infiltration, using a Canon Rebel T3i, de-identified (masked to rater), and then each infiltrated area was rated on a standardized 0-5 scale with 0 being no visible signs of chlorosis or necrosis and 5 being complete death of the infiltrated tissues. Ratings from at least four independent experiments for Glyma.15G191200 were analyzed using the pairwise Wilcoxon rank sum test in R (R Core Team, 2013). For alternative splice forms, at least two independent experiments were performed.
Samples from infiltrated leaves for RT-PCR and immunoblot analyses were taken three days post infiltration. Entire spots were either taken individually or pooled with its match on the other infiltrated leaf of the same plant. Samples were immediately frozen and stored at −80°C until processing. Samples were processed as described above for protein and total RNA extraction. For immunoblots, total protein lysate was extracted and immunoblot was performed as above except the α-SNAP and NSF incubation was performed simultaneously overnight. The NSF antibody has been previously demonstrated to cross react with N. benthamiana NSF (Bayless et al., 2016). Expression of the alternative splice form was validated with RT-PCR on cDNA from total RNA extraction using primers specific for the alternative splice form and the EF_1a internal control for N. benthamiana.
Antibody production
Affinity-purified polyclonal antibodies were raised against a synthetic peptide “GDVSSLKANKAEDE” specific to soybean chromosome 15 ɣ-SNAP. The antibodies were cross purified with a peptide specific to chromosome 9 ɣ-SNAP “GDCSALKAEDEEA”. The ɣ-SNAP antibody was produced by GenScript. Specificity of the antibody for ɣ-SNAP and not α-SNAP was verified with recombinant proteins (Supplemental Figure S4). The α-SNAP and NSF antibodies were described in Bayless et al. (2016).
Purification of ɣ-SNAP protein
ɣ-SNAP protein encoded by Glyma.15G191200 was expressed from the pRham N-His-Sumo Kan expression vector (Lucigen) in E. coli strain ‘E. cloni 10G’. Recombinant protein was produced and purified as in Bayless et al. (2016). α-SNAP protein was previously purified (Bayless et al., 2016). A five-step, five-fold dilution series of both ɣ-SNAP and α-SNAP recombinant protein was used for immunoblot to determine specificity of the SNAP antibodies. Both antibodies were used at the experimentally relevant concentrations: 1:1000 for α-SNAP and 1:250 for ɣ-SNAP.
Immunoblots
Tissues were homogenized in 2mL screw cap tubes with porcelain beads at 4000 rpm with a PowerLyzer 24 (Mo Bio, St. Louis, MO) for 30 seconds and flash frozen. A volume of lysis buffer (50 mM Tris-Hcl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.2% Triton X-100, 10% (v/v) glycerol and 1x protease inhibitor) equal to 1.5x the mass of the tissue was added to the ground tissue and kept on ice. Samples were homogenized again at 4000 rpm for 30 seconds. The samples were centrifuged twice to remove residual debris. Bradford assays were performed with the final lysate and samples were normalized to contain the equal amounts of total protein for SDS/PAGE. Loading and transfer was verified with Ponceau S stain.
Immunoblots for ɣ-SNAP were blocked in 5%(w/v) nonfat dry milk TBS-T (50mM Tris, 150 mM NaCl, 0.05% Tween 20) and then incubated with primary antibody overnight at 4°C in 5% (w/v) nonfat dry milk TBS-T at 1:250 for most experiments. Membranes were washed several times with TBS-T and then incubated with secondary horseradish peroxidase-conjugated goat anti-rabbit IgG in 5% (w/v) at 1:5000 for one hour at room temperature. Membranes were again washed several times with TBS-T. Chemiluminescent detection was performed with SuperSignal Dura chemiluminescent substrate (Thermo Scientific Waltham, MA). Immunoblot signal was then recorded with a ChemiDoc MP chemiluminescent imager (Bio Rad Hercules, CA). For detection with α-SNAP antibody, membranes were stripped under mild conditions with stripping buffer (15g/L glycine, Ig/L SDS, 0.1% Tween20, pH 2.2) and verified with redevelopment. Stripped membranes were incubated with primary α-SNAP antibody (Bayless et al., 2016) at 1:1000 in 5% nonfat dry milk TBS-T for one hour at room temperature. Following several washes with TBS-T, membranes were incubated with secondary horseradish peroxidase-conjugated goat anti-rabbit IgG as above and developed under the same conditions.
For densitometry analysis, the blot image just prior to overexposure (saturation of any pixels within the band) of any single band of interest was used. Densitometry analysis was performed in ImageLab (Bio Rad). When doublets occur for ɣ-SNAP, only signal from the bottom band that corresponds with the expected size as observed with purified protein, was used. Raw densitometry data were analyzed by ANOVA with Tukey test in R. Supplemental Figures S8 and S9 show original (uncropped) whole-blot images for the immunoblots in Figures 6 and 7.
Oligonucleotides
All oligonucleotides used in this study are listed in supplemental table S2
Supplementary Material
AKNOWLEDGEMENTS
We thank Dr. Adam Bayless for generating the ɣ-SNAP E. Cloni recombinant protein expression vector, Dr. Tong Geon Lee for the production of the fosmid library of PI 468916, and Usawadee Chaiprom for assistance with data analysis.
Funding:
This project was funded primarily by grants to A.B., M.H. and B.D. from the United Soybean Board.
Footnotes
The authors declare no conflict of interest
LITERATURE CITED
- Acharya K, Tande C, & Byamukama E (2016). Determination of Heterodera glycines Virulence Phenotypes Occurring in South Dakota. Plant Disease, 100(11), 2281–2286. 10.1094/PDIS-04-16-0572-RE [DOI] [PubMed] [Google Scholar]
- Allen TW, Bradley CA, Sisson AJ, Byamukama E, Chilvers MI, Coker CM, … Wrather J. Allen. (2017). Soybean Yield Loss Estimates Due to Diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Progress. 18, 19–27. 10.1094/PHP-RS-16-0066 [DOI] [Google Scholar]
- Barnes SN, Wram CL, Mitchum MG, & Baum TJ (2018). The plant-parasitic cyst nematode effector GLAND4 is a DNA-binding protein: GLAND4 nematode effector: a DNA-binding protein. Molecular Plant Pathology, 10.1111/mpp.12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless AM, Smith JM, Song J, McMinn PH, Teillet A, August BK, & Bent AF (2016a). Disease resistance through impairment of α-SNAP-NSF interaction and vesicular trafficking by soybean Rhg1. Proceedings of the National Academy of Sciences of the United States of America, 113(47), E7375–E7382. 10.1073/pnas.1610150113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless AM, Smith JM, Song J, McMinn PH, Teillet A, August BK, & Bent AF (2016b). Disease resistance through impairment of α-SNAP-NSF interaction and vesicular trafficking by soybean Rhg1. Proceedings of the National Academy of Sciences of the United States of America, 113(41), E7375–E7382. 10.1073/pnas.1610150113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless AM, Zapotocny RW, Grunwald DJ, Amundson KK, Diers BW, & Bent AF (2018). An atypical N-ethylmaleimide sensitive factor enables the viability of nematode-resistant Rhg1 soybeans. Proceedings of the National Academy of Sciences, 201717070. 10.1073/pnas.1717070115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto E, Bingman CA, Kondrashov DA, McCoy JG, Bannen RM, Wesenberg GE, & Phillips GN (2007). Structure and dynamics of γ-SNAP: Insight into flexibility of proteins from the SNAP family. Proteins: Structure, Function, and Bioinformatics, 70(1), 93–104. 10.1002/prot.21468 [DOI] [PubMed] [Google Scholar]
- Brzostowski LF, & Diers BW (2017). Pyramiding of Alleles from Multiple Sources Increases the Resistance of Soybean to Highly Virulent Soybean Cyst Nematode Isolates. Crop Science, 57(6), 2932. 10.2135/cropsci2016.12.1007 [DOI] [Google Scholar]
- Byrd DWJ, Kirkpatrick T, & Barker KR (1983). An Improved Technique for Clearing and Staining Plant Tissues for Detection of Nematodes. Journal of Nematology, 15(1), 142–143. [PMC free article] [PubMed] [Google Scholar]
- Chen S, Potter B, & Orf J (2010). Virulence of the soybean cyst nematode has increased over years in Minnesota. Journal of Nematology, 42, 238. [Google Scholar]
- Chow C-N, Zheng H-Q, Wu N-Y, Chien C-H, Huang H-D, Lee T-Y, … Chang W-C (2016). PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Research, 44(D1), D1154–1160. 10.1093/nar/gkv1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes & Development, 20(10), 1250–1255. 10.1101/gad.1416306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary DO, Griff IC, & Rothman JE (1990). SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell, 61(4), 709–721. [DOI] [PubMed] [Google Scholar]
- Cook DE, Bayless AM, Wang K, Guo X, Song Q, Jiang J, & Bent AF (2014). Distinct Copy Number, Coding Sequence, and Locus Methylation Patterns Underlie Rhg1-Mediated Soybean Resistance to Soybean Cyst Nematode. PLANT PHYSIOLOGY, 165(2), 630–647. 10.1104/pp.114.235952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, … Bent AF (2012). Copy Number Variation of Multiple Genes at Rhg1 Mediates Nematode Resistance in Soybean. Science, 335(6111), 1206–1209. 10.1126/science.1228746 [DOI] [PubMed] [Google Scholar]
- Dempewolf H, Baute G, Anderson J, Kilian B, Smith C, & Guarino L (2017). Past and Future Use of Wild Relatives in Crop Breeding. Crop Science, 57(3), 1070. 10.2135/cropsci2016.10.0885 [DOI] [Google Scholar]
- Diers BW, Arelli PR, Carlson SR, Fehr WR, Kabelka EA, Shoemaker RC, & Wang D (2005) Registration of “LDX01-1-65’ soybean. Crop Science, 45:1671. [Google Scholar]
- Diers BW, Cary TR, Thomas DJ, & Nickell CD (2006) Registration of ‘LD00-3309’ soybean. Crop Science. 46:1384. [Google Scholar]
- Faghihi J, Donald PA, Welacky TW, & Ferris VR (2010). Soybean Resistance to Field Populations of Heterodera glycines in Selected Geographic Areas. Plant Health Progress. 10.1094/PHP-2010-0426-01-RS [DOI] [Google Scholar]
- Gheysen G, & Jones JT (2006). Molecular aspects of Plant-Nematode Interactions. In Perry R & Moens M (Eds.), Plant Nematology (pp. 234–254). [Google Scholar]
- Gheysen G, & Mitchum MG (2011). How nematodes manipulate plant development pathways for infection. Current Opinion in Plant Biology, 14(4), 415–421. 10.1016/j.pbi.2011.03.012 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, & Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6(5), 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, … Rokhsar DS (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research, 40(D1), D1178–D1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Yang P, Sakata K, & Komatsu S (2014). Quantitative Proteomics Reveals the Role of Protein Phosphorylation in Rice Embryos during Early Stages of Germination. Journal of Proteome Research, 13(3), 1766–1782. 10.1021/pr401295c [DOI] [PubMed] [Google Scholar]
- Hernandez-Garcia CM, Bouchard RA, Rushton PJ, Jones ML, Chen X, Timko MP, & Finer JJ (2010). High level transgenic expression of soybean (Glycine max) GmERF and Gmubi gene promoters isolated by a novel promoter analysis pipeline. BMC Plant Biology, 10(1), 237. 10.1186/1471-2229-10-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershman DE, Heinz RD, & Kennedy BS (2008). Soybean Cyst Nematode, Heterodera glycines, Populations Adapting to Resistant Soybean Cultivars in Kentucky. Plant Disease, 92(10), 1475–1475. 10.1094/PDIS-92-10-1475B [DOI] [PubMed] [Google Scholar]
- Inoue H, Matsuzaki Y, Tanaka A, Hosoi K, Ichimura K, Arasaki K, … Tagaya M (2015). γ-SNAP stimulates disassembly of endosomal SNARE complexes and regulates endocytic trafficking pathways. Journal of Cell Science, 128(15), 2781–2794. 10.1242/jcs.158634 [DOI] [PubMed] [Google Scholar]
- Ithal N, Recknor J, Nettleton D, Hearne L, Maier T, Baum TJ, & Mitchum MG (2007). Parallel genome-wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Molecular Plant-Microbe Interactions: MPMI, 20(3), 293–305. 10.1094/MPMI-20-3-0293 [DOI] [PubMed] [Google Scholar]
- Jacobs TB, LaFayette PR, Schmitz RJ, & Parrott WA (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnology, 15(1), 16. 10.1186/s12896-015-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs TB, & Martin GB (2016). High-throughput CRISPR Vector Construction and Characterization of DNA Modifications by Generation of Tomato Hairy Roots. Journal of Visualized Experiments, (110). 10.3791/53843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabelka EA, Carlson SR, & Diers BW (2005). Localization of Two Loci that Confer Resistance to Soybean Cyst Nematode from PI 468916. Crop Science, 45(6), 2473. 10.2135/cropsci2005.0027 [DOI] [Google Scholar]
- Kabelka EA, Carlson SR, & Diers BW (2006). Glycine soja PI 468916 SCN Resistance Loci’s Associated Effects on Soybean Seed Yield and Other Agronomic Traits. Crop Science, 46(2), 622. 10.2135/cropsci2005.06-0131 [DOI] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, & Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 10(6), 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ki D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 14(4), R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, & Diers BW (2013). Fine Mapping of the SCN Resistance QTL and from PI 468916. Crop Science, 53(3), 775. 10.2135/cropsci2012.07.0425 [DOI] [Google Scholar]
- Liao Y, Smyth GK, Shi W, (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu A, Xiao Z, Li M-W, Wong F-L, Yung W-S, Ku Y-S, … Lam H-M (2018). Transcriptomic reprogramming in soybean seedlings under salt stress: Soybean transcriptome under salt stress. Plant, Cell & Environment. 10.1111/pce.13186 [DOI] [PubMed] [Google Scholar]
- Liu H, Ding Y, Zhou Y, Jin W, Xie K, & Chen L-L (2017). CRISPR-P 2.0: An Improved CRISPR-Cas9 Tool for Genome Editing in Plants. Molecular Plant, 10(3), 530–532. 10.1016/j.molp.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, … Meksem K (2012). A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature, 256–260. 10.1038/nature11651 [DOI] [PubMed] [Google Scholar]
- McCarville MT, Marett CC, Mullaney MP, Gebhart GD, & Tylka GL (2017). Increase in Soybean Cyst Nematode Virulence and Reproduction on Resistant Soybean Varieties in Iowa From 2001 to 2015 and the Effects on Soybean Yields. Plant Health Progress. 10.1094/PHP-RS-16-0062 [DOI] [Google Scholar]
- Melito S, Heuberger AL, Cook D, Diers BW, MacGuidwin AE, & Bent AF (2010). A nematode demographics assay in transgenic roots reveals no significant impacts of the Rhg1 locus LRR-Kinase on soybean cyst nematode resistance. BMC Plant Biology, 10(1), 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Wrather JA, Heinz RD, Shannon JG, & Danekas G (2007). Variability in Distribution and Virulence Phenotypes of Heterodera glycines in Missouri During 2005. Plant Disease, 91(11), 1473–1476. 10.1094/PDIS-91-11-1473 [DOI] [PubMed] [Google Scholar]
- Niblack TL, et al. , (2008). Shift in Virulence of Soybean Cyst Nematode is Associated with Use of Resistance from PI 88788. Plant Health Progress. 10.1094/PHP-2008-0118-01-RS [DOI] [Google Scholar]
- Niblack TL, Arelli PR, Noel GR, Opperman CH, Orf JH, Schmitt DP, … Tylka GL (2002). A Revised Classification Scheme for Genetically Diverse Populations of Heterodera glycines. Journal of Nematology, 34(4), 279–288. [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2013). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from http://www.R-project.org [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK, (2010). EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1) 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincker K, Cary T, & Diers BW (2017). Impact of Soybean Cyst Nematode Resistance on Soybean Yield. Crop Science, 57(3), 1373. 10.2135/cropsci2016.07.0628 [DOI] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, … Jackson SA (2010). Genome sequence of the palaeopolyploid soybean. Nature, 463(7278), 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Siddique S, Matera C, Radakovic ZS, Hasan MS, Gutbrod P, Rozanska E, Sobczak M, Torres MA, Grundler FMW (2014). Parasitic worms stimulate host NADPH oxidases to produce reactive oxygen species that limit plant cell death and promote infection. Sci. Signal 7, ra33. 10.1126/scisignal.2004777 [DOI] [PubMed] [Google Scholar]
- Sottosanto JB, Gelli A, & Blumwald E (2004). DNA array analyses of Arabidopsis thaliana lacking a vacuolar Na+/H+ antiporter: impact of AtNHX1 on gene expression: Impact of nhx1 on gene expression. The Plant Journal, 40(5), 752–771. 10.1111/j.1365-313X.2004.02253.x [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, & Birchler JA (2008). Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends in Genetics, 24(8), 390–397. 10.1016/j.tig.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Verma A, Lee C, Morriss S, Odu F, Kenning C, Rizzo N, … Mitchum MG (2018). The novel cyst nematode effector protein 30D08 targets host nuclear functions to alter gene expression in feeding sites. New Phytologist, 219(2), 697–713. 10.1111/nph.15179 [DOI] [PubMed] [Google Scholar]
- Wang D, Arelli PR, Shoemaker RC, Diers BW (2001) Loci underlying resistance to Race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor Appl Genet 103:561–566. [Google Scholar]
- Wang P, Zhou Z, Hu A, Ponte de Albuquerque C, Zhou Y, Hong L, … Fu X-D (2014). Both Decreased and Increased SRPK1 Levels Promote Cancer by Interfering with PHLPP-Mediated Dephosphorylation of Akt. Molecular Cell, 54(3), 378–391. 10.1016/j.molcel.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart SW, Griff IC, Brunner M, Clary DO, Mayer T, Buhrow SA, & Rothman JE (1993). SNAP family of NSF attachment proteins includes a brain-specific isoform. Nature, 362(6418), 353–355. 10.1038/362353a0 [DOI] [PubMed] [Google Scholar]
- Yu N, & Diers BW (2017). Fine mapping of the SCN resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Euphytica, 213(2). 10.1007/s10681-016-1791-2 [DOI] [Google Scholar]
- Yuan M, Chu Z, Li X, Xu C, & Wang S (2009). Pathogen-Induced Expressional Loss of Function is the Key Factor in Race-Specific Bacterial Resistance Conferred by a Recessive R Gene xa13 in Rice. Plant and Cell Physiology, 50(5), 947–955. 10.1093/pcp/pcp046 [DOI] [PubMed] [Google Scholar]
- Zhang C, Hou Y, Hao Q, Chen H, Chen L, Yuan S, … Huang W (2015). Genome-Wide Survey of the Soybean GATA Transcription Factor Gene Family and Expression Analysis under Low Nitrogen Stress. PLOS ONE, 10(4), e0125174. 10.1371/journal.pone.0125174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Xu Y, Yu S, Lu L, Ding M, Cheng J, … Tian Y (2015). An Efficient Genotyping Method for Genome-modified Animals and Human Cells Generated with CRISPR/Cas9 System. Scientific Reports, 4(1). 10.1038/srep06420 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.