Abstract
Background:
Artificial intelligence (AI)-enabled analysis of 12-lead electrocardiograms (ECGs) may facilitate efficient estimation of incident atrial fibrillation (AF) risk. However, it remains unclear whether AI provides meaningful and generalizable improvement in predictive accuracy beyond clinical risk factors for AF.
Methods:
We trained a convolutional neural network (“ECG-AI”) to infer 5-year incident AF risk using 12-lead ECGs in patients receiving longitudinal primary care at Massachusetts General Hospital (MGH). We then fit three Cox proportional hazards models, each composed of: a) ECG-AI 5-year AF probability, b) the Cohorts for Heart and Aging in Genomic Epidemiology AF (CHARGE-AF) clinical risk score, and c) terms for both ECG-AI and CHARGE-AF (“CH-AI”). We assessed model performance by calculating discrimination (area under the receiver operating characteristic curve, AUROC) and calibration in an internal test set and two external test sets (Brigham and Women’s Hospital and UK Biobank). Models were recalibrated to estimate 2-year AF risk in the UK Biobank given limited available follow-up. We used saliency mapping to identify ECG features most influential on ECG-AI risk predictions and assessed correlation between ECG-AI and CHARGE-AF linear predictors.
Results:
The training set comprised 45,770 individuals (age 55±17 years, 53% women, 2,171 AF events), and the test sets comprised 83,162 individuals (age 59±13 years, 56% women, 2,424 AF events). AUROC was comparable using CHARGE-AF (MGH 0.802, 95% CI 0.767–0.836; BWH 0.752, 95% CI 0.741–0.763; UK Biobank 0.732, 95% CI 0.704–0.759) and ECG-AI (MGH 0.823, 95% CI 0.790–0.856; BWH 0.747, 95% CI 0.736–0.759; UK Biobank 0.705, 95% CI 0.673–0.737). AUROC was highest using CH-AI: MGH 0.838, 95% CI 0.807–0.869; BWH 0.777, 95% CI 0.766–0.788; UK Biobank 0.746, 95% CI 0.716–0.776). Calibration error was low using ECG-AI (MGH 0.0212; BWH 0.0129; UK Biobank 0.0035) and CH-AI (MGH 0.012; BWH 0.0108; UK Biobank 0.0001). In saliency analyses, the ECG P-wave had the greatest influence on AI model predictions. ECG-AI and CHARGE-AF linear predictors were correlated (Pearson r MGH 0.61, BWH 0.66, UK Biobank 0.41).
Conclusions:
AI-based analysis of 12-lead ECGs has similar predictive utility to a clinical risk factor model for incident AF and both approaches are complementary. ECG-AI may enable efficient quantification of future AF risk.
Keywords: atrial fibrillation, risk prediction, deep learning, electronic health records
Atrial fibrillation (AF) is a common and morbid arrhythmia.1–4 Identification of individuals at elevated AF risk is a clinical imperative since modifying lifestyle and behavioral factors may prevent AF,5,6 and since cardiac rhythm monitoring may identify individuals with undiagnosed AF thereby enabling prevention of strokes.7–9
Recent work highlights the potential for artificial intelligence (AI) to predict AF from a 12-lead electrocardiogram (ECG).10–12 Yet several important knowledge gaps exist. First, existing models have not explicitly incorporated event-free survival or censoring, which is important for accurately estimating absolute risk.11,12 Second, it is not clear whether AI complements, or extends, well-validated clinical risk factor models for AF that do not require ascertainment of an ECG for risk prediction such as the Cohorts for Heart and Aging Research in Genomic Epidemiology AF (CHARGE-AF) score.13–17 Third, it is unclear what ECG features influence AF risk estimates from ECG-based AI models, which is critical to assessing potential bias and promoting clinician confidence. Fourth, previous models are proprietary and have not been subjected to rigorous external validation.11,12
In the current study, we trained a convolutional neural network to explicitly predict time to incident AF (“ECG-AI”), within a sample of over 40,000 individuals receiving longitudinal primary care at Massachusetts General Hospital (MGH). We validated this model in over 80,000 individuals from three independent test sets including additional individuals from MGH, individuals receiving longitudinal primary care at a separate healthcare institution – the Brigham and Women’s Hospital (BWH), and a prospective cohort study – the UK Biobank. We then compared the predictive accuracy of ECG-AI to CHARGE-AF and examined the performance of a model including both ECG-AI and CHARGE-AF (“CH-AI”). We further examined what regions of the ECG most influenced the ECG-AI model performance for predicting AF.
Methods
Data availability
UK Biobank data are publicly available by application (www.ukbiobank.ac.uk). MGH and BWH data contain protected health information and cannot be shared publicly. Data processing scripts used to perform the analyses described herein are available at https://github.com/shaankhurshid/ecg_ai.
Study population
We trained and validated ECG-AI in the Community Care Cohort Project, a dataset comprising individuals aged 18–90 years that received longitudinal primary care within the Mass General Brigham (MGB) network between 2000 to 2019.18 Briefly, individuals were included if they received ≥2 primary care visits that occurred between 1–3 years apart at any of seven hospitals within the Mass General Brigham (MGB) network, all of which were linked to a common electronic health record (EHR) database.19 Follow-up started after the inclusion window and comprised data ascertained from the EHR through August 31, 2019. ECG-AI was trained among individuals with ≥1 ECG performed at MGH within three years prior to start of follow-up (see below). ECG-AI was then evaluated in independent test sets comprising 4,166 individuals from MGH (internal test set) and a separate set of 37,963 individuals with ≥1 ECG performed at BWH within the 3-year baseline period (Figure 1). There was no overlap between training and test sets.
Figure 1. Study overview.
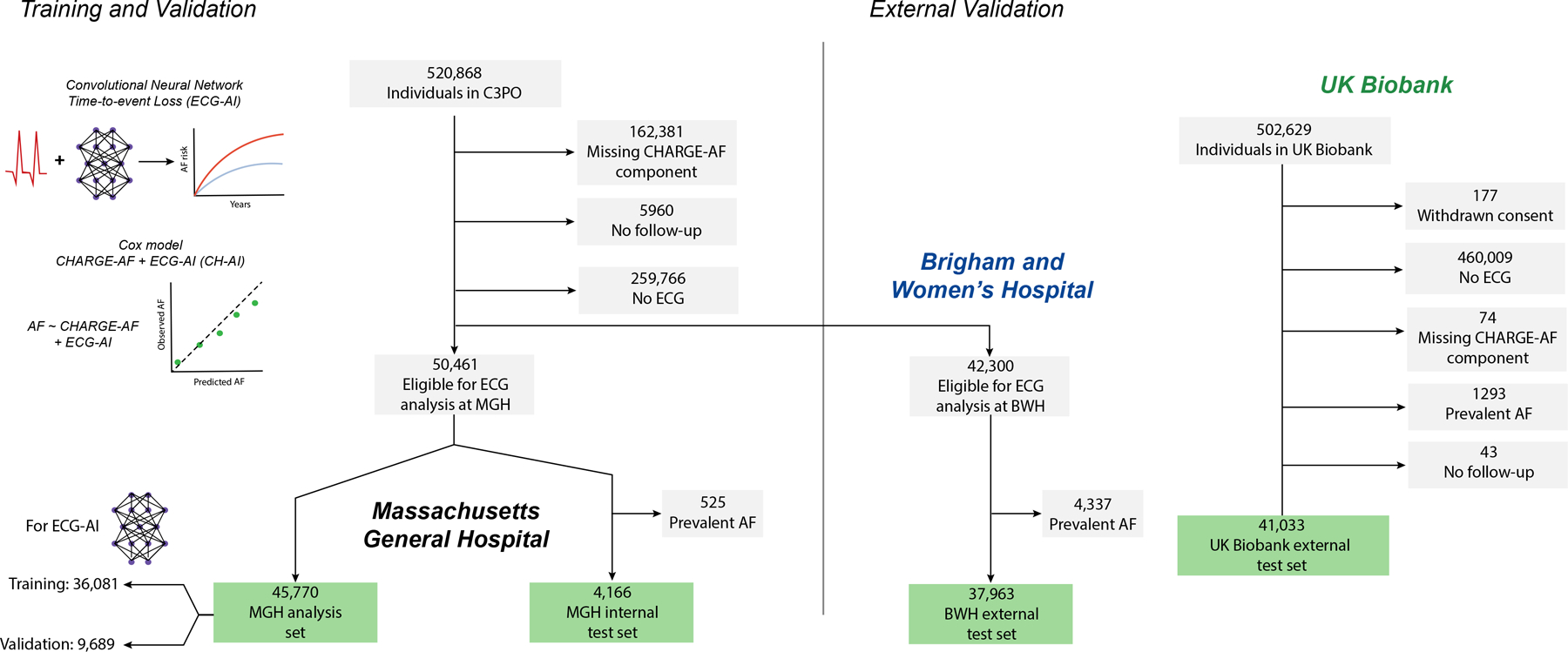
Depicted is an overview of the study. We trained a deep learning model to predict incident AF (ECG-AI) in Massachusetts General Hospital (MGH). We then developed a model combining ECG-AI and the CHARGE-AF clinical risk score (CH-AI) in the same training population. We then validated ECG-AI and CH-AI in three test sets: MGH, individuals from a separate hospital (Brigham and Women’s Hospital, BWH), and the UK Biobank prospective cohort study.
We performed additional external validation in the UK Biobank, a prospective cohort of 502,629 participants recruited between 2006–2010.20 Briefly, approximately 9.2 million individuals aged 40–69 living within 25 miles of the 22 assessment centers in England, Wales, and Scotland were invited, and 5.4% participated in the baseline assessment. Questionnaires, physical measures, and biological samples were collected at recruitment, with diagnostic tests in a large subset. Participants are followed for health outcomes using national datasets (updated through 02–28-2021). We analyzed all individuals who underwent a standardized study-based 12-lead ECG. An overview of the analysis samples is shown in Figure 1.
The UK Biobank was approved by the UK Biobank Research Ethics Committee (reference number 11/NW/0382). All UK Biobank participants provided written informed consent. Use of MGB and UK Biobank (application 7089) data were approved by the MGB Institutional Review Board.
ECG acquisition and ECG-AI training
ECG-AI is a convolutional neural network trained to predict 5-year AF-free survival. The input to ECG-AI is a single 12-lead ECG containing a time-series of 5,000 voltage measurements for each of 12 leads sampled at 500Hz and lasting 10 seconds in duration. A minority of ECGs having lower sampling rates were linearly resampled up to 500Hz, and ECGs having shorter durations were zero-padded to contain 5,000 measurements, resulting in a uniform input tensor of dimension 5,000 × 12. The raw waveform data as well as tabular metadata including date, time, machine type, sampling frequency, automated and physician reads were extracted from the MUSE Cardiology Information System (GE Healthcare, Chicago, IL), which stores data for all ECGs performed within MGB. All ECGs analyzed within MGH and BWH were performed for clinical purposes, whereas ECGs in the UK Biobank sample were performed prospectively as part of the study protocol. No ECGs were excluded from the training or test sets on the basis of particular findings.
Rather than binary classification,10,11 ECG-AI utilized an encoding and loss function21 accounting for both time to outcome (i.e., AF) and missingness introduced by censoring (defined as the earliest of death or loss to follow-up). To achieve this, the ECG-AI encoding divided time into discrete bins in which either an AF event or a censoring event could occur, and the loss function optimized the negative log likelihood of predicted AF occurring within each time bin. In this way, censored individuals do not contribute to the loss at time bins occurring after censoring (further detail in the Supplemental Methods). For training, ECG-AI was exposed to all 12-lead ECGs performed within 3 years prior to start of follow-up. For evaluation, ECG-AI was tested using only the most recent ECG prior to start of follow-up in MGH/BWH, and the single study visit ECG in the UK Biobank. Since sinus rhythm ECGs among individuals with a diagnosis of AF may contain useful training signal (i.e., they provide an example of AF-related changes in sinus rhythm)10 we included individuals with a history of AF in model training. However, we evaluated ECG-AI only among individuals without a history of AF. In addition to incident AF prediction, we trained ECG-AI to perform three related tasks (estimation of age, classification of sex, and identification of AF in the tracing) because we observed that the multi-task approach improved AF prediction performance compared to other model-building approaches we considered during model derivation (Table I in the Supplement). Further details of ECG-AI training are described in the Supplemental Methods. ECG-AI architecture is summarized in Figure I in the Supplement. Learning curves are shown in Figure II in the Supplement. Performance of ECG-AI for secondary tasks is shown in Figure III in the Supplement. ECG-AI performance varied modestly when training was performed utilizing alternative training and validation splits (Table II in the Supplement).
Clinical factors
We calculated CHARGE-AF, a validated AF prediction tool, for all individuals.13,14,17 Baseline age, sex, race, height, weight, and blood pressure values were obtained from the EHR.18 Anti-hypertensive use was determined using medication lists.14 Tobacco use was categorized as present or absent. Race was classified as white or non-white, as performed previously using CHARGE-AF.14,22 The presence of heart failure, diabetes, and myocardial infarction were ascertained using previously published sets of diagnostic codes and medications (e.g., anti-glygemics), having a reported positive predictive value for each diease of ≥85% in MGH and BWH.14,23 We utilized a complete case approach in all analyses (Figure 1).
Outcomes
The primary outcome was incident AF. Atrial flutter was considered equivalent to AF. In MGH and BWH, incident AF was defined using a previously published AF algorithm comprising combinations of diagnostic and procedural codes, and electrocardiogram reports (“Modified AF Algorithm”), with a reported positive predictive value of 92% for AF.24 In the UK Biobank, AF was defined using a previously published set of self-reported diagnoses and inpatient diagnostic and procedural codes. Although direct validation is not possible in the UK Biobank, the AF definition has been previously assessed in an external dataset with a reported positive predictive value of 92%.25 The details of each AF definition are shown in Table III in the Supplement.”
Statistical analysis
Consistent with prior work including the original CHARGE-AF derivation study,13 we defined the outcome of incident AF at 5 years in the MGH and BWH datasets. Due to limitations in available follow-up (median 2.8 years [quartile 1: 1.9, quartile 3: 4.3]), we could not assess 5-year AF in the UK Biobank and instead utilized a 2-year AF outcome. ECG-AI was trained to generate 5-year AF risk estimates. We calculated 5-year AF risk estimates for CHARGE-AF using the equation: 1 − 0.9718412736exp(score−12.58156),13 where score is the individual’s CHARGE-AF score. To compare ECG-AI to CHARGE-AF (a score derived as a Cox proportional hazards model) we fit Cox proportional hazards models in the MGH training set with incident AF as the outcome and a) ECG-AI probability, and b) ECG-AI probability and CHARGE-AF, as covariates (“CH-AI”). ECG-AI probability was logit-transformed to achieve an approximately linear relationship with the log hazard.12 We also fit an analogous model including only age and sex. Although ECG-AI and CHARGE-AF were correlated in the training set, the strength of correlation was only moderate (Pearson r=0.68, 95% CI 0.67–0.68). We utilized the CHARGE-AF score rather than fitting the individual score components given that the score has been validated across multiple settings.13,14,16,17 However in secondary analyses we also fit a version of CH-AI in which each CHARGE-AF component was included as an individual covariate. Given low death rates within the time window of interest (MGH training set: 4.6%, MGH test set 3.2%, BWH 2.8%, UK Biobank 0.4%), we did not account for death as a competing risk. We compared the age and sex, CHARGE-AF, ECG-AI and CH-AI models in each test set. The linear predictors of age and sex, ECG-AI, and CH-AI were converted to predicted probabilities of AF using the equation: where s0 is the average AF-free survival probability at the window of interest in the MGH training set, ∑βX is the individual’s score value, and ∑βY is the average score in the MGH training set. Values of s0 and ∑βY are provided in Table IV in the Supplement.
Within each test set, we assessed AF discrimination using the area under the time-dependent receiver operating characteristic curve (AUROC).26 Since AUROC may be misleading for relatively uncommon outcomes, we also assessed discrimination using time-dependent average precision (AP).27 Both AUROC and AP estimates were calculated using inverse probability of censoring weights to account for potential bias introduced by censoring.28,29 We calculated AUROC and AP in one year increments after ECG until the window of interest. Standard errors were estimated using 500-iteration bootstrapping, which were used to calculate 95% confidence intervals and perform pairwise Z-testing.
We assessed calibration using: 1) adaptive hazard regression30 curves of predicted versus observed AF risk, 2) calibration slopes, where a value of one indicates optimal calibration,31 and 3) integrated calibration index (ICI), the average prediction error weighted by the empirical risk distribution.30 Since initial 2-year AF estimates from ECG-AI and CH-AI substantially overestimated risk in the UK Biobank (Figure IV in the Supplement), and since the CHARGE-AF score was designed to predict 5-year AF,13 we recalibrated each score based on the observed 2-year AF incidence in the UK Biobank.32
We then quantified time-dependent net reclassification improvement (NRI).33 We assessed reclassification at standard risk thresholds (i.e., 5-year AF risk <2.5%, 2.5–5%, and ≥5% in MGH/BWH, and 2-year AF risk <0.5%, 0.5–1%, and ≥1% in the UK Biobank), at high risk thresholds (i.e., 5-year AF risk <20% versus ≥20% in MGH/BWH, and 2-year AF risk <2% versus ≥2% in UK Biobank), and using continuous risk values. Standard risk thresholds were chosen to mirror previous thresholds used when deriving the CHARGE-AF score,13 and high risk thresholds were chosen to reflect higher levels of AF risk which may be more clinically actionable. Prior to estimating NRI, models were recalibrated using the methods described above, with or without additional adjustment for the calibration slope as required32 to obtain well-calibrated scores (ICI range: 7.1×10−5 to 0.0129). We then plotted the cumulative risk of AF across high risk strata defined using ECG-AI and CHARGE-AF. To assess ECG-AI behavior, we produced saliency maps depicting areas of the ECG in which changes in voltage had the greatest influence on ECG-AI predicted risk estimates. We also plotted the median ECG waveforms for individuals at high estimated AF risk (≥5%) versus low estimated AF risk (<2.5%) for 1,000 randomly selected individuals within each stratum from the BWH test set, with cutoffs mirroring our standard risk thresholds.13 Further details of saliency map and median waveform analysis are described in the Supplemental Methods.
We performed several secondary analyses, described in detail in the Supplemental Methods. We considered two-sided p-values <0.05 statistically significant. Analyses were performed using Python v3.834 and R v4.0.35
Results
Training and Validation Samples
Overall, 45,770 individuals in the MGH sample were eligible for analysis (mean age 55, 53% female), and were divided into training (n=36,081) and validation (n=9,689) sets for ECG-AI. Characteristics of participants are displayed in Table 1. The training set included 100,954 12-lead ECGs (median ECGs per individual: 1 [quartile-1: 1, quartile-3: 3]). Following ECG-AI training, we fit Cox proportional hazards models in the full MGH sample of 45,770 individuals. When fitting CH-AI, both ECG-AI probability (hazard ratio [HR] 1.85 per 1-SD increase, 95% CI 1.75–1.95) and CHARGE-AF score (HR 1.97 per 1-SD increase, 95% CI 1.84–2.11) were associated with incident AF, without interaction (p=0.60).
Table 1.
Sample characteristics
| MGH training set (N=45,770)* | MGH test set (N=4,166) | BWH test set (N=37,963) | UK Biobank test set (N=41,033) | |
|---|---|---|---|---|
| Age (years) | 54.9 ± 16.6 | 53.5 ± 16.2 | 53.8 ± 15.3 | 64.5 ± 7.7 |
| Female | 24,047 (52.5%) | 2,300 (55.2%) | 23,211 (61.1%) | 21,426 (52.2%) |
| Race/Ethnicity | ||||
| White | 35,629 (77.8%) | 3,209 (77.0%) | 25,150 (66.2%) | 39,607 (96.5%) |
| Black | 2,959 (6.5%) | 273 (6.6%) | 5,283 (13.9%) | 300 (0.7%) |
| Hispanic or Latino | 1,897 (4.1%) | 197 (4.7%) | 3,240 (8.5%) | - |
| Asian or Pacific Islander | 2,155 (4.7%) | 228 (5.5%) | 1,082 (2.9%) | 587 (1.4%) |
| Mixed | 1 (0.002%) | 0 (0%) | 5 (0.01%) | 203 (0.5%) |
| Other | 1,953 (4.3%) | 177 (4.2%) | 1,673 (4.4%) | 226 (0.6%) |
| Unknown | 1,176 (2.6%) | 82 (2.0%) | 1,530 (4.0%) | 110 (0.3%) |
| Current smoker | 3,616 (7.9%) | 337 (8.1%) | 3,463 (9.1%) | 1,495 (3.6%) |
| Systolic blood pressure (mmHg) | 126 ± 17 | 126 ± 18 | 126 ± 18 | 138 ± 19 |
| Diastolic blood pressure (mmHg) | 76 ± 10 | 76 ± 11 | 76 ± 11 | 79 ± 10 |
| Anti-hypertensive medication use | 25,187 (55.0%) | 2,088 (50.1%) | 21,148 (55.7%) | 4,374 (10.7%) |
| Diabetes | 8,715 (19.0%) | 715 (17.2%) | 6,656 (17.5%) | 1,597 (3.9%) |
| Heart failure | 3,255 (7.1%) | 170 (4.1%) | 1,388 (3.7%) | 191 (0.5%) |
| Myocardial infarction | 3,574 (7.8%) | 245 (5.9%) | 2,643 (7.0%) | 933 (2.3%) |
Includes 5,183 individuals with prevalent AF who were used to train ECG-AI but not to fit CH-AI. Also includes 9,689 individuals in an internal validation set for ECG-AI who were used to fit CH-AI.
We assessed each model in three independent test sets: MGH (n=4,166), BWH (n=37,963), and UK Biobank (n=41,033). AF incidence rates were substantially higher in MGH (12.8 per 1,000 person-years, 95% CI 11.0–14.5) and BWH (12.9, 95% CI 12.3–13.4) as compared to the UK Biobank (4.2, 95% CI 3.7–4.7). Median follow-up for analysis was 5.0 years (quartile-1: 2.6, quartile 5.0) in MGH, 5.0 years (2.6, 5.0) in BWH, and 2.0 years (1.9, 2.0) in the UK Biobank. A sample overview is depicted in Figure 1 and baseline characteristics are shown in Table 1. Predicted AF risk distributions are shown in Figure V in the Supplement.
Discrimination
ECG-AI demonstrated moderate AF discrimination (AUROC MGH: 0.823, BWH 0.743, UK Biobank: 0.705), comparable to the full CHARGE-AF score (Table 2) (0.802, 0.752, 0.732). Compared to CHARGE-AF, CH-AI had consistently higher AUROC point estimates (0.838, 0.777, 0.746), although the difference was not statistically significant in the UK Biobank (p<0.05 for MGH and BWH, p=0.28 for UK Biobank) (Table 2). Improvements in discrimination were more prominent according to AP, where ECG-AI (0.27, 0.19, 0.06) and CH-AI (0.30, 0.21, 0.06) were favorable compared to CHARGE-AF (0.21, 0.17, 0.02) (ECG-AI: p=.06, p<0.05, p<0.05, respectively; CH-AI p<0.05 for all). Overall patterns in model discrimination were generally consistent for events occurring between 1 to 5 years, although discrimination for CH-AI and CHARGE-AF tended to increase with longer prediction windows while that using ECG-AI tended to remain constant (Figure 2 and Table V in the Supplement). When assessed at specific thresholds, ECG-AI and CH-AI tended to provide greater precision at higher specificity. For example, at 95% specificity, precision was substantially greater using ECG-AI (MGH: 17.3%, BWH: 12.6%, UK Biobank 4.12%) and CH-AI (17.9%, 14.6%, 4.85%) versus CHARGE-AF (11.0%, 12.0%, 3.28%) (Table VI in the Supplement).
Table 2.
Model performance for incident AF in test sets
|
Massachusetts General Hospital
(N=4,166) |
Brigham and Women’s Hospital
(N=37,963) |
UK Biobank
(N=41,033) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Hazard ratio (per 1-SD) | 5-year AUROC | 5-year average precision | Calibration slope | ICI‡ | Hazard ratio (per 1-SD) | 5-year AUROC | 5-year average precision | Calibration slope | ICI | Hazard ratio (per 1-SD) | 2-year AUROC | 2-year average precision | Calibration slope | ICI |
| Deep learning architectures | |||||||||||||||
| ECG-AI | - | 0.823* (0.790–0.856) |
0.27 (0.21–0.34) |
- | 0.0231 | - | 0.747* (0.736–0.759) |
0.19*† (0.17–0.20) |
- | 0.0124 | - | 0.705 (0.659–0.724) |
0.060* (0.043–0.087) |
- | 0.0768 |
| Cox proportional hazards models | |||||||||||||||
| Age and sex | 2.91 (2.44–3.47) |
0.768 (0.732–0.805) |
0.16 (0.13–0.20) |
1.05 (0.88–1.23) |
0.0074 | 2.48 (2.35–2.62) |
0.730 (0.717–0.743) |
0.14 (0.13–0.15) |
0.94 (0.88–1.00) |
0.0072 | 2.21 (1.96–2.50) |
0.728 (0.702–0.755) |
0.018 (0.015–0.024) |
1.48 (1.25–1.71) |
0.0019§ |
| CHARGE-AF | 3.36 (2.98–4.30) |
0.802* (0.767–0.836) |
0.21* (0.17–0.26) |
0.68 (0.58–0.77) |
0.0320 | 2.78 (2.63–2.94) |
0.752* (0.741–0.763) |
0.17* (0.15–0.18) |
0.57 (0.53–0.60) |
0.0344 | 2.26 (2.00–2.55) |
0.732 (0.704–0.759) |
0.020 (0.016–0.026) |
0.87 (0.75–1.00) |
0.0011§ |
| ECG-AI | 2.45 (2.23–2.69) |
0.823* (0.790–0.856) |
0.27* (0.21–0.34) |
1.06 (0.95–1.17) |
0.0212 | 2.05 (1.98–2.11) |
0.747* (0.736–0.759) |
0.19*† (0.17–0.20) |
0.81 (0.77–0.84) |
0.0129 | 2.01 (1.88–2.14) |
0.705 (0.673–0.737) |
0.060*† (0.044–0.090) |
0.75 (0.68–0.82) |
0.0035§ |
| CH-AI | 3.74 (3.24–4.33) |
0.838*† (0.807–0.869) |
0.30*† (0.24–0.38) |
1.13 (1.01–1.25) |
0.0120 | 2.76 (2.64–2.88) |
0.777*† (0.766–0.788) |
0.21*† (0.19–0.23) |
0.77 (0.74–0.81) |
0.0108 | 2.27 (2.11–2.44) |
0.746 (0.716–0.776) |
0.059*† (0.042–0.083) |
1.01 (0.92–1.10) |
0.0001§ |
p<0.05 for comparison against age and sex
p<0.05 for comparison against CHARGE-AF
Integrated calibration index (ICI), a quantitative measure of the average difference between predicted event risk and observed event incidence, weighted by the empirical distribution of event risk.30 Smaller values indicate better calibration.
Values reflect ICI after recalibration to the baseline 2-year AF risk in the UK Biobank
Difference in c-index for CH-AI vs ECG-AI: AUROC MGH p=NS, BWH p<0.05, UK Biobank p<0.05; average precision MGH p<0.05, BWH p<0.05, p=NS
AUROC = area under the receiver operating characteristic curve; ICI = integrated calibration index; SD = standard deviation
Figure 2. Discrimination of incident AF.
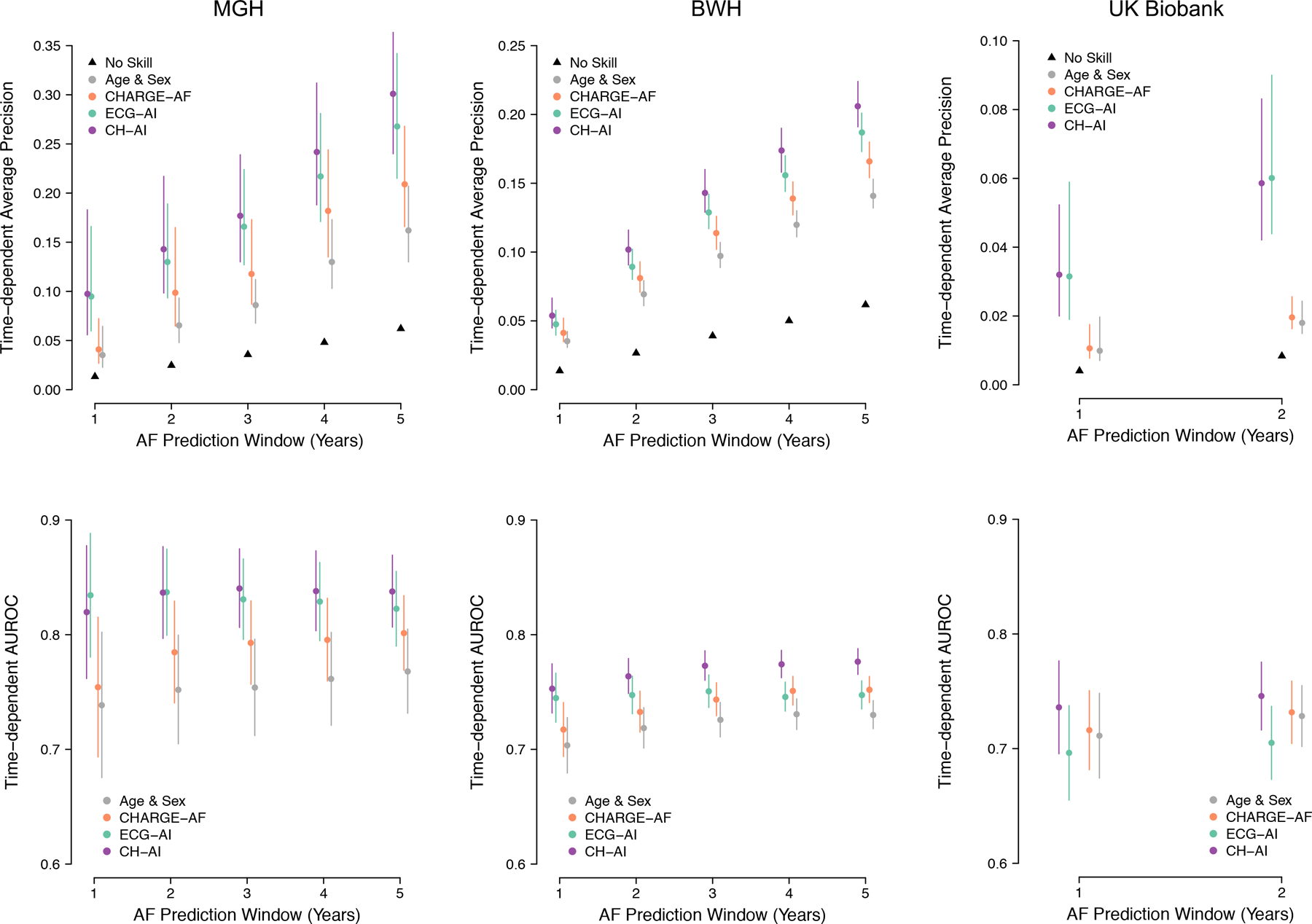
Depicted is discrimination of age and sex (gray), CHARGE-AF (orange), ECG-AI (green), and CH-AI (purple), in the MGH test set (left panels), BWH test set (middle panels), and UK Biobank test set (right panels). Top panels plot the average precision and bottom panels plot the area under the receiver operating characteristic curve (AUROC) across increasing length of the prediction window (x-axis). In the top panels, the black triangles represent the cumulative event rate (i.e., the precision of a randomly guessing model).
Calibration
CH-AI was well-calibrated in MGH (integrated calibration index 0.012) and BWH (0.019), but overestimated risk in the UK Biobank (0.068) (Figure IV in the Supplement). Given a near-optimal calibration slope (1.01, 95% CI 0.92–1.10), overestimation was likely attributable to a greater average 2-year AF risk in the MGH training set (1.48%) versus UK Biobank (0.56%). Accordingly, calibration of CH-AI in the UK Biobank was excellent after recalibration to the average 2-year AF hazard in the UK Biobank (integrated calibration index 7.1×10−5, Figure 3). Cumulative risk of AF was greatest among individuals classified as high risk using both CHARGE-AF and ECG-AI, lowest among individuals classified as low risk using both CHARGE-AF and ECG-AI, and intermediate among individuals classified as high risk using CHARGE-AF alone or ECG-AI alone (Figure 4). Cumulative risk of AF stratified by estimated risk using CH-AI is shown in Figure VI in the Supplement.
Figure 3. Calibration for incident AF.
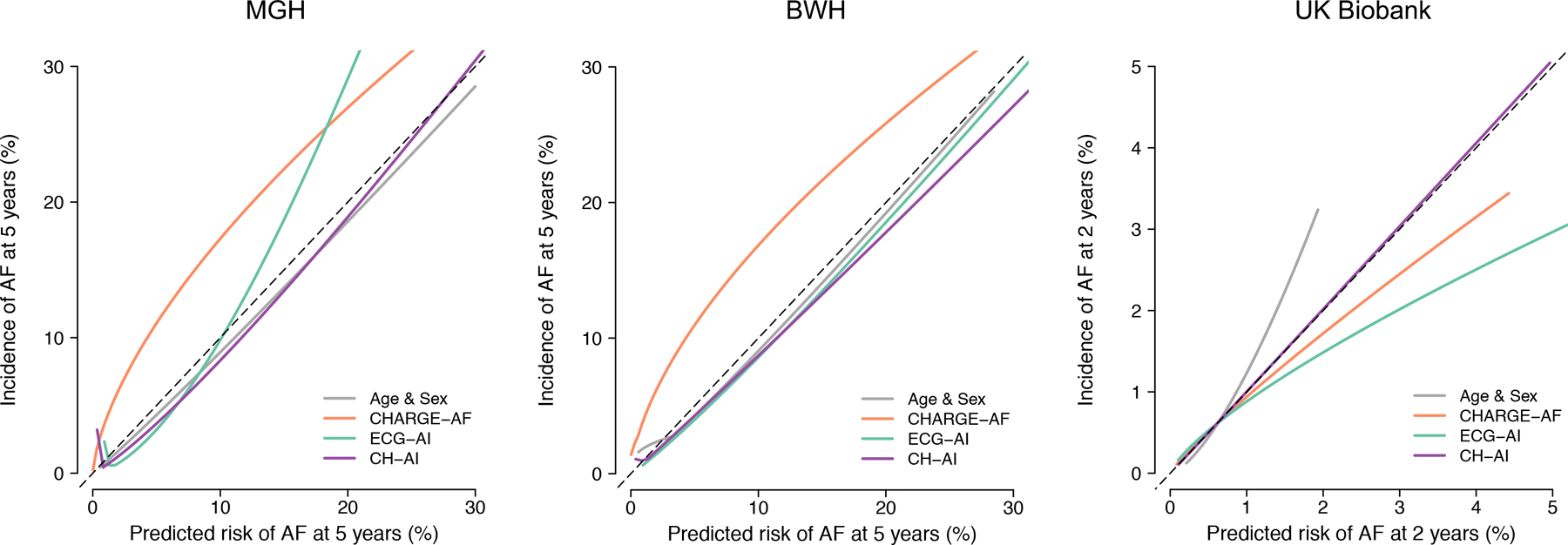
Depicted are fitted calibration curves demonstrating the relationship between predicted event risk (x-axis) and observed cumulative event incidence (y-axis) for and age and sex (gray), CHARGE-AF (orange), ECG-AI (green), and CH-AI (purple). Perfect calibration is indicated by the hashed diagonal line, denoting perfect correspondence between predicted and observed risk. Curves were obtained using adaptive hazard regression30 relating predicted risk and observed event risk.
Figure 4. Cumulative risk of AF stratified by predicted AF risk.
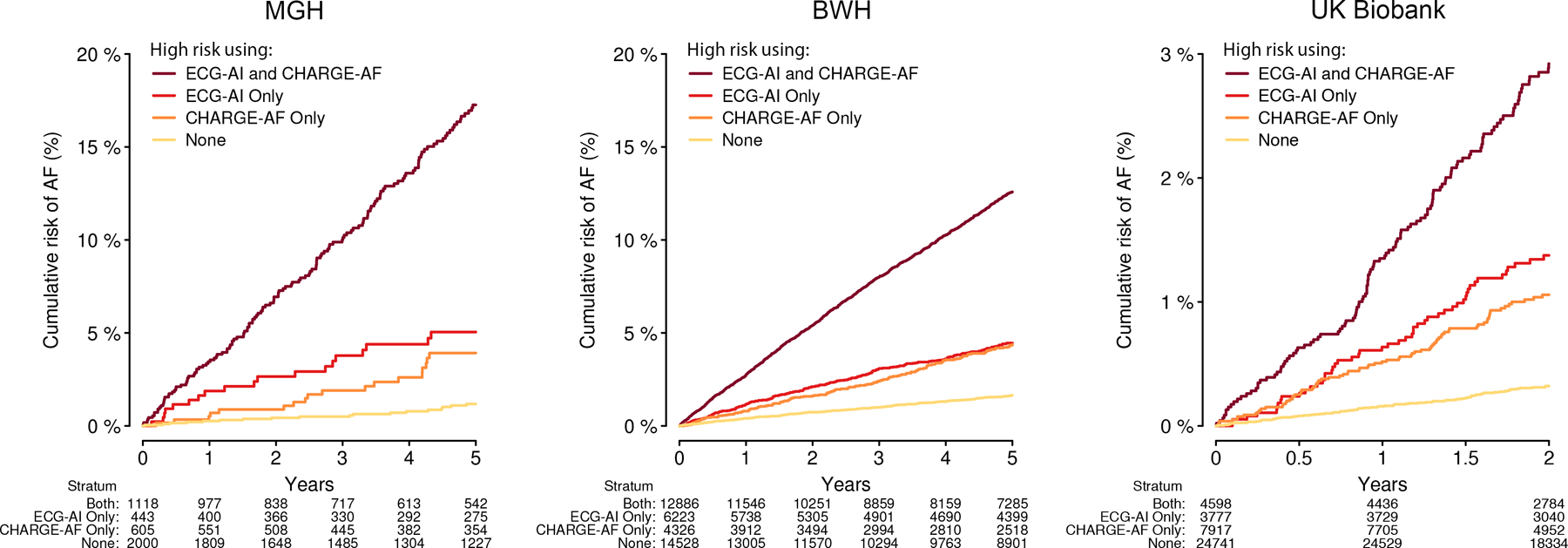
Depicted is the cumulative risk of AF stratified by high predicted risk of AF as determined using both ECG-AI and CHARGE-AF (dark red), ECG-AI only (red), CHARGE-AF only (orange), or neither model (yellow). High AF risk was defined as 5-year AF risk ≥5% in MGH and BWH (as performed in the original CHARGE-AF derivation study),13 and 2-year AF risk ≥1% in the UK Biobank (approximating the top tertile of risk). The number at risk across each stratum over time is depicted below each plot.
Model Behavior
Saliency maps demonstrated that the P wave and surrounding regions had the greatest impact on ECG-AI AF risk (Figure 5). Median waveform analysis demonstrated specifically that individuals with high estimated AF risk tended to have a longer P wave duration, as well as slightly wider QRS and a flatter ST segment (Figure 5).
Figure 5. Representations of ECG-AI behavior.
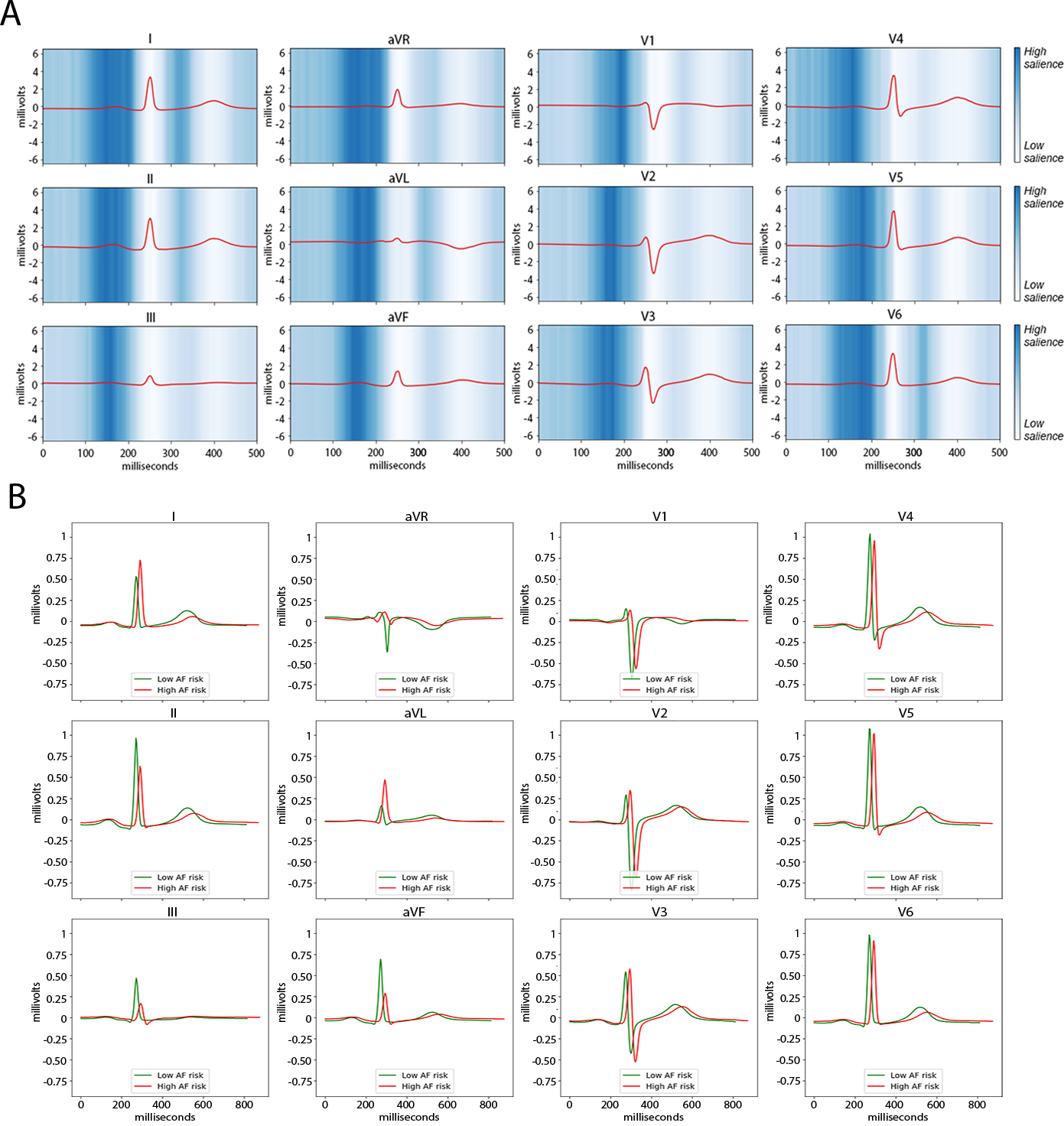
Depicted are two forms of visualizing the behavior of the ECG-AI deep learning model. Panel A is a saliency map of ECG-AI demarcating regions of the ECG waveform having the greatest influence on AF risk predictions. Blue shades depict the magnitude of the gradient of predicted AF risk with respect to the ECG waveform amplitude, where darker shades illustrate regions of the waveform exerting greater salience, or influence on AF risk predictions. Saliency was averaged over a random sample of 4,096 individuals in the BWH test set, and the red waveform depicts the median waveform in each lead among the 4,096 individuals. Panel B displays the median waveform of a random sample of 1,000 individuals in the BWH test set with low predicted AF risk (i.e., 5-year AF risk < 2.5%, green) versus the median waveform of a random sample of 1,000 individuals in the BWH test set with high predicted AF risk (i.e., 5-year AF risk > 5%, red).
Reclassification and Subgroup Analyses
Compared to CHARGE-AF, CH-AI demonstrated favorable NRI using standard risk thresholds, high risk thresholds, and continuous risk values (Tables VII–IX and Figure VII in the Supplement). Use of ECG-AI compared to CHARGE-AF did not result in favorable NRI using standard risk thresholds or continuous risk values, but did result in favorable reclassification at high risk thresholds (Tables VII–IX and Figure VII in the Supplement). Receiver operating characteristic and precision-recall curves are shown in Figure VIII in the Supplement.
Improvements in model performance using CH-AI versus CHARGE-AF were generally consistent among individuals with prevalent heart failure and stroke (Table X in the Supplement), and were more prominent within subgroups of age (Figure IX in the Supplement). Using models developed using only lead I, and separately lead II – vectors typical for single lead ECGs – CH-AI continued to provide greater discrimination than CHARGE-AF, although discrimination using ECG-AI was lower (Table XI in the Supplement). Performance of CH-AI and ECG-AI for 2-year AF risk in MGH and BWH were similar to that observed for 5-year AF risk (Table XII in the Supplement). When ECG-AI was fit sequentially along with covariate terms for sex and individual components of the CHARGE-AF score, we observed that inclusion of age and sex improved discrimination over ECG-AI alone, with even further improvement observed when additional CHARGE-AF components were added (Table XIII in the Supplement). A model fit using ECG-AI, sex, and each individual component of the CHARGE-AF score resulted in nearly identical discrimination to CH-AI, with worse calibration in the UK Biobank (Table XIII in the Supplement). Saliency maps across strata of CHARGE-AF and ECG-AI risk are shown in Figure X in the Supplement. The linear predictors of ECG-AI and CHARGE-AF were consistently correlated (Pearson r MGH 0.61, BWH 0.66, UK Biobank 0.41, p<0.01 for all).
Discussion
We developed ECG-AI, a deep learning model that explicitly predicts time to incident AF using 12-lead ECG data. ECG-AI was trained using roughly 100,000 ECGs from over 40,000 individuals within a primary care cohort. CH-AI, a model that combined both ECG-AI and CHARGE-AF demonstrated improved performance across multiple prognostic model metrics as compared to CHARGE-AF within three independent test sets including over 80,000 individuals whose clinical characteristics varied substantially. AF risk estimates from ECG-AI alone demonstrated comparable discrimination when compared to the 11-component CHARGE-AF score. We further observed that ECG-AI and CHARGE-AF were highly correlated, suggesting that much of the predictive utility of ECG-AI may reflect electrocardiographic manifestations of established clinical risk factors for AF. On balance, our findings suggest that deep learning-derived ECG-based risk signals provide comparable predictive utility to clinical risk models for prediction of incident AF, and that ECG-AI and clinical risk factors provide complementary information which augments risk prediction.
Attia et al. developed a deep learning model which was 80% accurate in the classification of AF status among individuals with established AF whose tracings showed normal sinus rhythm.10 Raghunath et al. subsequently developed a neural network to predict incident AF using 12-lead ECG, demonstrating good discrimination at one year, and modestly improved performance when compared to CHARGE-AF in a subset analysis.11 Our findings add substantively to previous work by introducing a deep learning model that explicitly incorporates survival time, and performing a rigorous epidemiologic assessment including quantification of discrimination, calibration, reclassification, and a broad external validation. We are unable to directly compare our approach to previous models given that the models are not available for application to our data. Even if previous models were available, important differences in model design (e.g., a standard c-statistic cannot be calculated for time-to-event models) would likely preclude direct comparison of our model metrics to previous models. Nevertheless, our results broadly support the notion that deep learning models utilizing 12-lead ECG provide important predictive utility for determining AF risk, and provide new evidence that risk estimates are generalizable, with predictive value maintained up to 5 years after an ECG is performed. Importantly, the ability to predict incident AF up to 5 years in the future may faciliate implementation of preventive interventions (e.g., alcohol cessation,5 achievement of healthy weight,6 control of high blood pressure36) designed to reduce risk of AF and associated complications.
Our findings demonstrate that deep learning models utilizing ECG to estimate AF risk are robust and valid across contrasting populations when assessed using rigorous epidemiologic metrics. Specifically, we assessed ECG-AI in test sets comprising independent individuals from: a) the same institution as the training set, b) a separate institution within the same healthcare network, and c) a prospective research cohort from a different continent in which AF risk was substantially lower. Consistent with prior findings,17 ECG-AI performed best in populations most closely resembling the training set, with decreasing discrimination across progressively different samples, underscoring the importance of widespread external validation of AI models for assessing clinical utility. Ultimately, we suspect that differences in discrimination may be related to differing sample characteristics (e.g., age, baseline AF risk) leading to varying relationships between specific ECG features and future AF risk. Nevertheless, CH-AI consistently outperformed CHARGE-AF, and ECG-AI alone consistently demonstrated at least moderate discrimination. Importantly, we observed substantial overestimation in AF risk estimates using CH-AI in the UK Biobank, a low risk sample. However, simple recalibration to the baseline hazard – a process commonly required for traditional prognostic models37 – resulted in excellent calibration. Notably, CHARGE-AF had worse calibration than CH-AI even after recalibration, suggesting that deep learned AF risk may contribute directly to more calibrated estimates.
We note two important implications of our results on the relations between deep learning-based ECG risk signals and traditional clinical risk factors for AF. First, clinical risk factors appear to manifest on the ECG in ways that are perceptible to deep learning models. Specifically, ECG-AI probability and CHARGE-AF score were moderately correlated within each test set. Furthermore, using saliency mapping and median waveform analysis, we observed that ECG-AI probability was critically influenced by the period of atrial depolarization and repolarization (i.e., P wave and surrounding period), a reflection of atrial structure and function which may be affected by age and chronic conditions such as hypertension.38,39
Second, deep learning models appear to extract elements of AF risk that are complementary to clinical risk factors. CH-AI – a model combining CHARGE-AF and ECG-AI – consistently demonstrated superior AF discrimination, calibration, and reclassification as compared to either ECG-AI or CHARGE-AF alone, suggesting meaningful improvement in predictive utility when combining clinical risk information with AI-enabled ECG risk stratification as compared to either alone. Similarly, AF risk was substantially higher among individuals classified as high risk according to both ECG-AI and CHARGE-AF as opposed to either model alone. Interestingly, discrimination using CH-AI and CHARGE-AF tended to increase with longer prediction windows, likely relating to the cumulative effect of clinical risk factors over time, whereas discrimination using ECG-AI tended to remain relatively constant, suggesting that the relative contribution of ECG-based AF risk may be greatest for predicting AF events in the shorter term. Future work is needed to better characterize the biological correlates of increased AF risk as indicated by deep learning on ECG independent of clinical risk factors.
The convenience and external validity of estimating AF risk using a single modality rather than a complex clinical score suggests that deep learning models may be useful for clinical application. Although clinical factors are increasingly available, risk score functions remain challenging to implement given requirements for user interaction and susceptibility to misclassification of inputs. In contrast, models like ECG-AI may enable instantaneous AF risk estimation, which may facilitate rapid identification of individuals at elevated AF risk to guide preventive efforts, and increase the efficiency of AF screening by targeting individuals most likely to have AF identified with diagnostic testing.40 To this end, ECG-AI retained predictive utility in two populations of particular clinical interest – HF and stroke – in which AF risk assessment may have particular relevance considering the associated morbidity and high risk of stroke associated with AF. Furthermore, ECG-AI also had stable performance in models utilizing only a single ECG lead, suggesting that deep learning may facilitate AF risk estimation using wearable devices, which are commonly equipped with single-lead ECG capability. Future work is warranted to validate and prospectively assess the predictive utility of ECG-AI in different clinical settings and as applied to wearable ECGs.
Our study should be interpreted in the context of design. First, we trained ECG-AI on individuals with at least one ECG performed for clinical purposes. We also required that individuals have each component of the CHARGE-AF score available at baseline. Both of these requirements introduce potential selection bias. However, we submit that analyzing an EHR sample comprising individuals receiving longitudinal primary care is likely to reduce bias,18 and note our models continued to discriminate AF in a completely independent prospective cohort study. Second, our training set represented individuals from a single institution. Training on larger samples across multiple institutions may lead to more accurate and generalizable models. Third, given limited follow-up in the UK Biobank, we assessed a shorter prediction window of two years, as opposed to five years in MGH and BWH. Fourth, ECG-AI is a black box model. However, in contrast to previous AF prediction models10–12 we use saliency maps and median waveform analysis to demonstrate that biologically plausible ECG changes (e.g., longer p wave duration) had greatest influence on AF risk estimates. Fifth, a 5 year prediction window may represent AF risk that is less immediately actionable. However, we note that our models had consistent discrimination across shorter time windows as well. We cannot exclude that ECG-AI identifies individuals with preexisting undiagnosed AF. Sixth, ECG-AI and CH-AI required recalibration in the UK Biobank. Recalibration is frequently necessary when transferring prognostic models across populations,37 and simple recalibration to the observed AF incidence in the UK Biobank resulted in very well-calibrated estimates, suggesting that initial miscalibration was fully attributable to a baseline AF incidence in the UK Biobank that was roughly one-third that in MGH and BWH. Furthermore, CH-AI had better calibration than CHARGE-AF even though CHARGE-AF was similarly recalibrated in the UK Biobank.
In summary, across three independent test sets spanning over 80,000 individuals, ECG-AI, a deep learning model that explicitly predicts time to incident AF using 12-lead ECG, offers comparable discrimination of 5-year AF risk when compared to the 11-component CHARGE-AF clinical risk score. CH-AI – a model integrating CHARGE-AF and ECG-AI – offers consistently superior discrimination, calibration, and reclassification. Deep learned ECG-based AF risk signals have the potential for broad deployment to provide accurate and generalizable absolute AF risk estimates up to several years after an ECG is performed.
Supplementary Material
What Is New?
Artificial intelligence (AI)-based analysis of 12-lead ECGs has similar predictive utility to an established clinical risk factor model for incident AF and both are complementary.
An ECG-AI model for AF had predictive utility across independent study samples, discriminated risk in patients with heart failure and stroke, and was applicable to single-lead ECG tracings.
What Are the Clinical Implications?
AI-based AF risk prediction models utilizing 12-lead ECGs may enable efficient quantification of future AF risk.
Prediction of AF can be performed using clinical risk factors or AI-based analysis of ECGs, but the combination of both provides greatest predictive accuracy.
Funding Sources
This work was supported by NIH grants R01HL139731 (Lubitz), R01 HL134893 (Ho), R01 HL140224 (Ho), K24 HL153669 (Ho), 2R01HL092577 (Ellinor) and K24HL105780 (Ellinor); T32HL007208 (Khurshid); American Heart Association (Dallas, Texas) 18SFRN34250007 (Lubitz, Anderson) and 18SFRN34110082 (Ellinor); Doris Duke Foundation Clinical Scientist Development Award 2014105 (Lubitz); and Fondation Leducq 14CVD01 (Ellinor).
Disclosures
Dr. Lubitz receives sponsored research support from Bristol Myers Squibb / Pfizer, Bayer AG, Boehringer Ingelheim, Fitbit, and IBM, and has consulted for Bristol Myers Squibb / Pfizer, Bayer AG, and Blackstone Life Sciences. Dr. Ellinor receives sponsored research support from Bayer AG and IBM, and has consulted for Novartis, MyoKardia and Bayer AG. Dr. Ho has received sponsored research support from Bayer AG and research supplies from EcoNugenics, Inc. Dr. Batra receives sponsored research support from Bayer AG and IBM, and has consulted for Novartis and Prometheus Biosciences. Dr. Anderson receives sponsored research support from Bayer AG and has consulted for ApoPharma.
Non-standard Abbreviations and Acronyms
- AF
atrial fibrillation
- AI
artificial intelligence
- AP
average precision
- AUROC
area under the receiver operating characteristic curve
- BWH
Brigham and Women’s Hospital
- CH-AI
CHARGE-AF and ECG-AI
- CHARGE-AF
Cohorts for Aging Research and Genomic Epidemiology Atrial Fibrillation
- ECG
Electrocardiogram
- ECG-AI
Electrocardiogram Artificial Intelligence
- EHR
Electronic health record
- ICI
Integrated calibration index
- MGB
Mass General Brigham
- MGH
Massachusetts General Hospital
- NRI
Net reclassification improvement
Footnotes
Supplemental Materials
References 41–46
References
- 1.Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation: a major contributor to stroke in the elderly. The Framingham Study. Arch Intern Med. 1987;147:1561–1564. [PubMed] [Google Scholar]
- 2.Corley SD, Epstein AE, DiMarco JP, Domanski MJ, Geller N, Greene HL, Josephson RA, Kellen JC, Klein RC, Krahn AD, et al. Relationships between sinus rhythm, treatment, and survival in the Atrial Fibrillation Follow-Up Investigation of Rhythm Management (AFFIRM) Study. Circulation. 2004;109:1509–1513. [DOI] [PubMed] [Google Scholar]
- 3.Carlisle MA, Fudim M, DeVore AD, Piccini JP. Heart Failure and Atrial Fibrillation, Like Fire and Fury. JACC Heart Fail. 2019;7:447–456. [DOI] [PubMed] [Google Scholar]
- 4.Diener H-C, Hart RG, Koudstaal PJ, Lane DA, Lip GYH. Atrial Fibrillation and Cognitive Function: JACC Review Topic of the Week. J Am Coll Cardiol. 2019;73:612–619. [DOI] [PubMed] [Google Scholar]
- 5.Voskoboinik A, Kalman JM, De Silva A, Nicholls T, Costello B, Nanayakkara S, Prabhu S, Stub D, Azzopardi S, Vizi D, et al. Alcohol Abstinence in Drinkers with Atrial Fibrillation. New England Journal of Medicine. 2020;382:20–28. [DOI] [PubMed] [Google Scholar]
- 6.Middeldorp ME, Pathak RK, Meredith M, Mehta AB, Elliott AD, Mahajan R, Twomey D, Gallagher C, Hendriks JML, Linz D, et al. PREVEntion and regReSsive Effect of weight-loss and risk factor modification on Atrial Fibrillation: the REVERSE-AF study. Europace. 2018;20:1929–1935. [DOI] [PubMed] [Google Scholar]
- 7.Lip GYH, Nieuwlaat R, Pisters R, Lane DA, Crijns HJGM. Refining clinical risk stratification for predicting stroke and thromboembolism in atrial fibrillation using a novel risk factor-based approach: the euro heart survey on atrial fibrillation. Chest. 2010;137:263–272. [DOI] [PubMed] [Google Scholar]
- 8.Ruff CT, Giugliano RP, Braunwald E, Hoffman EB, Deenadayalu N, Ezekowitz MD, Camm AJ, Weitz JI, Lewis BS, Parkhomenko A, et al. Comparison of the efficacy and safety of new oral anticoagulants with warfarin in patients with atrial fibrillation: a meta-analysis of randomised trials. Lancet. 2014;383:955–962. [DOI] [PubMed] [Google Scholar]
- 9.Stroke Prevention in Atrial Fibrillation Study. Final results. Circulation. 1991;84:527–539. [DOI] [PubMed] [Google Scholar]
- 10.Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, et al. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet. 2019;394:861–867. [DOI] [PubMed] [Google Scholar]
- 11.Raghunath S, Pfeifer JM, Ulloa-Cerna AE, Nemani A, Carbonati T, Jing L, vanMaanen DP, Hartzel DN, Ruhl JA, Lagerman BF, et al. Deep Neural Networks Can Predict New-Onset Atrial Fibrillation From the 12-Lead Electrocardiogram and Help Identify Those at Risk of AF-Related Stroke. Circulation. 2021;143:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christopoulos G, Graff-Radford J, Lopez CL, Yao X, Attia ZI, Rabinstein AA, Petersen RC, Knopman DS, Mielke MM, Kremers W, et al. Artificial Intelligence-Electrocardiography to Predict Incident Atrial Fibrillation: A Population-Based Study. Circ Arrhythm Electrophysiol. 2020;13:1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonso A, Krijthe BP, Aspelund T, Stepas KA, Pencina MJ, Moser CB, Sinner MF, Sotoodehnia N, Fontes JD, Janssens ACJW, et al. Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the CHARGE-AF consortium. J Am Heart Assoc. 2013;2:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulme OL, Khurshid S, Weng L-C, Anderson CD, Wang EY, Ashburner JM, Ko D, McManus DD, Benjamin EJ, Ellinor PT, et al. Development and Validation of a Prediction Model for Atrial Fibrillation Using Electronic Health Records. JACC Clin Electrophysiol. 2019;5:1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y-G, Pastori D, Farcomeni A, Yang P-S, Jang E, Joung B, Wang Y-T, Guo Y-T, Lip GYH. A Simple Clinical Risk Score (C2HEST) for Predicting Incident Atrial Fibrillation in Asian Subjects: Derivation in 471,446 Chinese Subjects, With Internal Validation and External Application in 451,199 Korean Subjects. Chest. 2019;155:510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khurshid S, Kartoun U, Ashburner JM, Trinquart L, Philippakis A, Khera AV, Ellinor PT, Ng K, Lubitz SA. Performance of Atrial Fibrillation Risk Prediction Models in Over 4 Million Individuals. Circ Arrhythm Electrophysiol. 2021;14:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christophersen IE, Yin X, Larson MG, Lubitz SA, Magnani JW, McManus DD, Ellinor PT, Benjamin EJ. A comparison of the CHARGE-AF and the CHA2DS2-VASc risk scores for prediction of atrial fibrillation in the Framingham Heart Study. Am Heart J. 2016;178:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khurshid S, Reeder C, Harrington LX, Singh P, Sarma G, Friedman SF, Achille PD, Diamant N, Cunningham JW, Turner AC, et al. Cohort Design and Natural Language Processing to Reduce Bias in Electronic Health Records Research: The Community Care Cohort Project. medRxiv. Preprint posted online May 30, 2021. doi: 10.1101/2021.05.26.21257872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nalichowski R, Keogh D, Chueh HC, Murphy SN. Calculating the benefits of a Research Patient Data Repository. AMIA Annu Symp Proc. 2006;1044. [PMC free article] [PubMed] [Google Scholar]
- 20.Littlejohns TJ, Sudlow C, Allen NE, Collins R. UK Biobank: opportunities for cardiovascular research. Eur Heart J. 2019;40:1158–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gensheimer MF, Narasimhan B. A scalable discrete-time survival model for neural networks. PeerJ. 2019;7:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alonso A, Roetker NS, Soliman EZ, Chen LY, Greenland P, Heckbert SR. Prediction of Atrial Fibrillation in a Racially Diverse Cohort: The Multi-Ethnic Study of Atherosclerosis (MESA). J Am Heart Assoc. 2016;5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang EY, Hulme OL, Khurshid S, Weng L-C, Choi SH, Walkey AJ, Ashburner JM, McManus DD, Singer DE, Atlas SJ, et al. Initial Precipitants and Recurrence of Atrial Fibrillation. Circ Arrhythm Electrophysiol. 2020;13:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khurshid S, Keaney J, Ellinor PT, Lubitz SA. A Simple and Portable Algorithm for Identifying Atrial Fibrillation in the Electronic Medical Record. Am J Cardiol. 2016;117:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khurshid S, Choi SH, Weng L-C, Wang EY, Trinquart L, Benjamin EJ, Ellinor PT, Lubitz SA. Frequency of Cardiac Rhythm Abnormalities in a Half Million Adults. Circ Arrhythm Electrophysiol. 2018;11:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uno H, Tian L, Cai T, Kohane IS, Wei LJ. A unified inference procedure for a class of measures to assess improvement in risk prediction systems with survival data. Stat Med. 2013;32:2430–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Y, Zhou QM, Li B, Cai H, Chow EJ, Armstrong GT. A threshold-free summary index of prediction accuracy for censored time to event data. Stat Med. 2018;37:1671–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung H, Chiang C-T. Estimation methods for time-dependent AUC models with survival data. Can J Statistics. 2009;38:8–26. [Google Scholar]
- 29.Uno H, Cai T, Pencina MJ, D’Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011;30:1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Austin PC, Harrell FE, Klaveren D. Graphical calibration curves and the integrated calibration index (ICI) for survival models. Statistics in Medicine. 2020;39:2714–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens RJ, Poppe KK. Validation of clinical prediction models: what does the “calibration slope” really measure? J Clin Epidemiol. 2020;118:93–99. [DOI] [PubMed] [Google Scholar]
- 32.Demler OV, Paynter NP, Cook NR. Tests of calibration and goodness-of-fit in the survival setting. Stat Med. 2015;34:1659–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pencina MJ, D’Agostino RB, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Python Core Team. (2015). Python: A dynamic, open source programming language. Python Software Foundation. https://www.python.org/. [Google Scholar]
- 35.R Core Team (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- 36.Dzeshka MS, Shantsila A, Shantsila E, Lip GYH. Atrial Fibrillation and Hypertension. Hypertension. 2017;70:854–861. [DOI] [PubMed] [Google Scholar]
- 37.D’Agostino RB, Grundy S, Sullivan LM, Wilson P, CHD Risk Prediction Group. Validation of the Framingham coronary heart disease prediction scores: results of a multiple ethnic groups investigation. JAMA. 2001;286:180–187. [DOI] [PubMed] [Google Scholar]
- 38.Dagli N, Karaca I, Yavuzkir M, Balin M, Arslan N. Are maximum P wave duration and P wave dispersion a marker of target organ damage in the hypertensive population? Clin Res Cardiol. 2008;97:98–104. [DOI] [PubMed] [Google Scholar]
- 39.Havmoller R, Carlson J, Holmqvist F, Herreros A, Meurling CJ, Olsson B, Platonov P. Age-related changes in P wave morphology in healthy subjects. BMC Cardiovasc Disord. 2007;7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khurshid S, Mars N, Haggerty CM, Huang Q, Weng L-C, Hartzel DN, Lunetta KL, Ashburner JM, Anderson CD, Benjamin EJ, et al. Predictive Accuracy of a Clinical and Genetic Risk Model for Atrial Fibrillation. Circ Genom Precis Med. 2021;14:561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snoek J, Larochelle H, Adams RP. Practical Bayesian Optimization of Machine Learning Algorithms. Advances in Neural Information Processing Systems. 2012;25:1–9. [Google Scholar]
- 42.Lu Y, Gould S, Ajanthan T. Bidirectionally Self-Normalizing Neural Networks. arXiv. Preprint posted online May 18, 2021. doi:arXiv.2006.12169v4. [Google Scholar]
- 43.Liu L, Jiang H, He P, Chen W, Liu X, Gao J, Han J. On the Variance of the Adaptive Learning Rate and Beyond. arXiv. Preprint posted online April 17, 2020. doi:arXiv.1908:03265v3 [Google Scholar]
- 44.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. Journal of Machine Learning Research. 2014;15:1929–1958. [Google Scholar]
- 45.Khurshid S, Healey JS, McIntyre WF, Lubitz SA. Population-Based Screening for Atrial Fibrillation. Circ Res. 2020;127:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are publicly available by application (www.ukbiobank.ac.uk). MGH and BWH data contain protected health information and cannot be shared publicly. Data processing scripts used to perform the analyses described herein are available at https://github.com/shaankhurshid/ecg_ai.


