Abstract
Ninety-six plasma and whole blood specimens from nine selected patients were analyzed for the presence of Aspergillus DNA. Nineteen specimens from three patients with proven aspergillosis were PCR positive in both materials, whereas an additional 22 were PCR positive in whole blood only. All 36 samples from six patients without signs of aspergillosis were negative in both assays. We conclude that although plasma and whole blood spiked with Aspergillus conidia showed an identical lower detection limit (10 CFU), the sensitivity of plasma PCR was lower than that of PCR performed on whole blood samples.
Invasive aspergillosis has become a major cause of infection-related morbidity and mortality in neutropenic cancer patients and recipients of allogeneic bone marrow transplants. In the last decade, the incidence of invasive aspergillosis has increased steadily (2). Because of a lack of sensitive and specific conventional tests that allow early diagnosis of invasive aspergillosis (8), different protocols based on the amplification of fungal DNA in blood samples have been published recently (14, 16). In selected groups of patients, PCR-based assays showed a promising sensitivity and specificity, indicating the potential for early diagnosis of invasive fungal infections.
Our previously published PCR assay (3) amplifies a wide range of fungal DNA using primers binding to highly conserved regions of the 18S rRNA gene. Specificity for various fungal pathogens is achieved by species-specific oligonucleotide hybridization.
Plasma samples have been successfully used for the detection of bacterial, protozoal, and viral DNA (5, 7, 15). However, only limited data are available for the detection of fungal DNA in plasma samples.
In order to establish a method for extracting DNA from plasma, 5-ml EDTA-anticoagulated blood samples from healthy volunteer donors were centrifuged for 10 min at 1,500 × g. Then, plasma (1 ml) was spiked with Aspergillus conidia (106 to 100 CFU/ml, in serial dilutions) or Aspergillus DNA (100 pg to 10 fg, in serial dilutions), which had been extracted previously. DNA extraction was performed immediately. Spiked plasma was also stored at room temperature for 24, 48, or 72 h and at 4°C, −20°C, and −80°C for 1, 7, 28, and 84 days. Fungi for spiking experiments were obtained from the German Collection of Microorganisms and cultured on Sabouraud-glucose-agar for 72 h (Aspergillus fumigatus, DSM 790) at 30°C. Serial dilutions of conidia were prepared with sterile saline suspensions, adjusted to a McFarland standard of 0.5 (= 106 conidia/ml).
In addition, 96 plasma and whole blood samples from nine selected patients receiving an allogeneic bone marrow transplant were analyzed. Two 5-ml blood specimens were prospectively collected from all patients twice weekly, starting at admission to the transplant unit until day 100 posttransplantation or death. Three patients developed proven aspergillosis, which was documented histopathologically at autopsy, and six patients showed no signs of aspergillosis (halo sign or air crescent sign in computerized tomography scan). Two of the six patients received amphotericin B therapy because of fever of unknown origin refractory to broad-spectrum antibiotics. Both patients had culture-proven bacteremia.
All patients were treated in laminar air flow rooms until recovery of neutrophils to above 1,000/μl was documented. For antifungal prophylaxis, fluconazole (400 mg/day) was administered. The PCR assay was run by an investigator blinded to clinical and microbiologic data. PCR results were not known to physicians and thus were not used in the management of the patients.
Samples were processed within 2 h after collection. One tube was centrifuged immediately (1,500 × g, 10 min) to obtain the plasma. One milliliter of the plasma was aliquoted and stored at −80°C as a backup sample. DNA from an additional 1 ml of plasma was extracted immediately. In parallel, DNA extraction was performed from the whole blood specimens (1 ml from the second blood tube). To monitor for contamination, aliquots of saline or DNA from healthy control persons were prepared concurrently. Thirty samples from five healthy individuals were analyzed as controls in order to screen nonneutropenic plasma samples for the presence of Aspergillus DNA.
DNA from whole blood specimens was extracted as described previously (3). For DNA extraction from plasma, 1 ml of plasma was centrifuged for 10 min at 15,000 × g. Then, 900 μl was decanted, and 500 μl of white cell lysis buffer (10 mM Tris, 10 mM EDTA, 50 mM sodium chloride, 0.2% sodium dodecyl sulfate, 200 μg of proteinase K per ml) was added to the remaining volume (incubation for 45 min at 65°C), followed by a repeated incubation in liquid nitrogen and at 95°C in a heating block. For protein precipitation, 500 μl of double-distilled water and 500 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added and then centrifuged for 10 min at 15,000 × g. The supernatant was pipetted into a new tube. DNA precipitation was performed using 900 μl of ice-cold isopropanol. Samples were stored at −80°C for 10 min, followed by centrifugation for 10 min at 15,000 × g. DNA was purified once with 500 μl of 70% ethanol. After lyophilization, 40 μl of double-distilled water was added for resuspension. Amplification of Aspergillus DNA was performed under standard conditions (3). Primers (5′-ATTGGAGGGCAAGTCTGGTG and 5′-CCGATCCCTAGTCGGCATAG; Roth, Karlsruhe, Germany) bind to conserved regions of the 18S rRNA gene. Thirty-four cycles of repeated denaturation (30 s at 94°C), annealing (1 min at 62°C), and enzymatic chain extension (2 min at 72°C) were performed in a PE 2400 thermocycler (Perkin Elmer, Dreieich, Germany). In order to exclude the presence of PCR inhibitors in plasma samples, all specimens were analyzed twice in the same PCR assay (the original clinical sample [α-sample] as well as another sample from the same patient to which 1 pg of Aspergillus DNA was added [γ-sample]). In order to control the sensitivity of each assay, genomic Aspergillus DNA from dilution series (105 to 100 cells) was amplified in each assay. DNA extraction, PCR, and amplicon detection were performed in separate rooms. Those pipetting the PCR mixtures wore one-way gowns, sterile gloves, and face masks.
Amplicons were hybridized in a PCR-enzyme-linked immunosorbent assay (Roche, Mannheim, Germany) according to the manufacturer's manual using 2 pmol of biotin-labeled oligonucleotide specific for A. fumigatus (5′-TGGGGAACCTCATGGCCTTCACTGGCTGTG; Roth) per ml (10).
Plasma was spiked with Aspergillus conidia (106 to 100/ml) or Aspergillus DNA (100 pg to 10 fg) in order to determine the sensitivity of the assay. A sensitivity of 10 CFU/ml and 100 fg of Aspergillus DNA was documented. All γ-samples were positive in the hybridization assay, excluding the presence of PCR inhibitors in the plasma samples. These results corresponded to whole blood assays that we obtained previously (10). In addition, the sensitivity was tested for samples kept at 4°C, −20°C, and −80°C for 1, 7, 28, and 84 days. The sensitivity was identical for samples frozen at −20°C or −80°C for up to 84 days. However, in samples stored at 4°C, slightly higher optical densities were observed. After a 24-h incubation at room temperature, the sensitivity for 100 fg of DNA was unchanged, whereas after 48 and 72 h, the sensitivity decreased to 1 pg after hybridization, indicating a loss of sensitivity by enzymatic degradation.
All 30 samples from the five healthy individuals as well as the 36 samples from the six patients without clinical signs of invasive fungal infection were negative in both assays. From three patients with histologically proven invasive aspergillosis, 19 specimens were positive in both assays, whereas an additional 22 samples were positive in the PCR from whole blood only (Fig. 1).
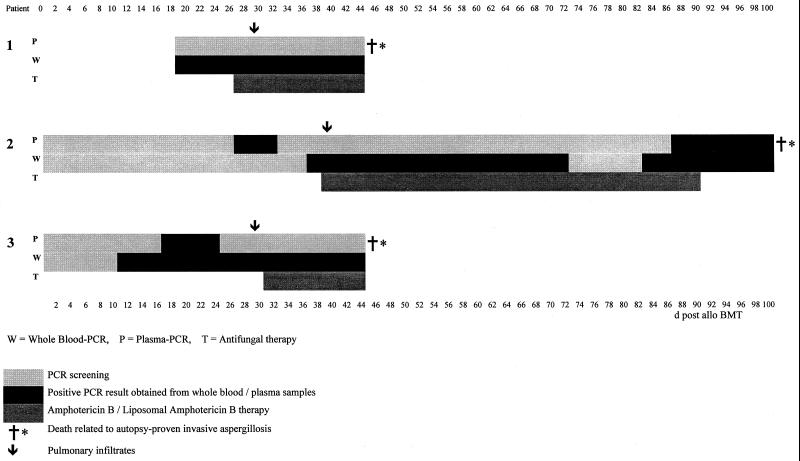
FIG. 1.
PCR results for three patients with histologically proven invasive aspergillosis. Numbers above and below indicate days postallogeneic bone marrow transplant (d post allo BMT).
Patient 1 was positive in the whole blood PCR from day 18 until day 45 but constantly negative in the plasma PCR. He developed pulmonary infiltrates on day 29 and died on day 45 despite therapy with liposomal amphotericin B, which was initiated on day 27. Patient 2 showed 19 positive samples in the whole blood PCR but only 9 in the PCR performed with plasma samples. The first positive sample could be detected at day 36 posttransplantation in the whole blood PCR, whereas plasma PCR remained negative until day 86. Pulmonary infiltrates were seen on day 39. On the same day, liposomal amphotericin B therapy was initiated (until day 93). He died of cerebral aspergillosis (parahippocampus) in the later posttransplant period. Patient 3 was positive in the plasma PCR only from days 16 until 24 but in the whole blood PCR from days 10 until 36. Pulmonary infiltrates were documented on day 28. She received conventional amphotericin B therapy from day 30. The patient died on day 45 of invasive aspergillosis.
Fungal DNA was amplified successfully from whole blood (6), serum (1), and bronchoalveolar lavage (4) samples. Until now, the sensitivity of a PCR assay has not been compared for different clinical specimens from the same patient. Thus, we analyzed 96 plasma and whole blood specimens from nine immunocompromised patients. Studies using spiked plasma and blood samples showed a comparable sensitivity for both materials (16).
Spin columns are not recommended for DNA extraction from plasma, as there might be a high loss of target DNA. As strong PCR inhibitors like albumin and globulins might be present in the plasma, an extraction procedure focusing on complete protein precipitation is essential for amplifying DNA obtained from plasma samples. Therefore, phenol-chloroform-isoamyl alcohol was used for protein precipitation.
In addition, we analyzed the sensitivity of samples stored at different temperatures. The sensitivity was comparable (10 CFU) for samples frozen at −20°C or −80°C for 7, 28, and 84 days. If plasma samples are frozen prior to DNA extraction, overnight shipment of samples to other laboratories is possible. Moreover, we could demonstrate that conidia and hyphae spiked into plasma samples might grow at 4°C. This circumstance might disturb quantification when samples are stored in the refrigerator.
In whole blood samples, fungal cells floating in the blood or found within human leukocytes are pelleted by centrifugation at 1,500 × g without releasing their DNA. Incubation with white cell lysis buffer containing proteinase K results in lysis of the leukocytes. Consequently, fungal cells are released. After centrifugation, cell debris and free DNA are decanted with the supernatant, whereas fungal DNA remains within the fungal pathogen. In contrast, in serum and plasma samples, only free DNA may be detected, as after centrifugation at 1,500 × g, hyphal cells are mainly found in the pellet together with the leukocytes. Free-floating Aspergillus DNA might have been released from nonviable hyphae into the blood. This has been reported for cancer patients in whom mutant DNA from cancer cells was detected in plasma specimens (11). Two decades ago, in patients with chronic autoimmune disorders, increased quantities of DNA were demonstrated to circulate in the plasma (9). Thus, depending on the type of clinical material (whole blood or plasma), the sensitivity of the PCR assay may vary.
Bougnoux et al. (1) showed that purified fungal DNA injected into rabbits at a dose of 200 μg was still detectable in serum samples for at least 120 min postinjection, demonstrating free DNA to be stable in blood specimens. In contrast, probably due to DNA-degrading enzymes like DNase and other nucleases present in serum samples, Rumore et al. (13) reported a half-life of circulating DNA of less than 5 min, indicating that the ability to detect free fungal DNA is dose dependent. Prince et al. (12) demonstrated that a minimum concentration of DNase in serum was required for degradation of human chromatin. In addition, it has been shown that EDTA abolishes DNase activity, enabling successful amplification of DNA after storage of EDTA-anticoagulated blood samples.
We conclude from this study that, despite the benefit that plasma can be shipped frozen without loss of sensitivity, DNA extraction from plasma is complex, and the sensitivity of the plasma PCR is lower than that of the PCR performed on whole blood samples.
REFERENCES
- 1.Bougnoux M E, Dupont C, Mateo J, Saulnier P, Faivre V, Payen D, Nicolas-Chanoine M H. Serum is more suitable than whole blood for diagnosis of systemic candidiasis by nested PCR. J Clin Microbiol. 1999;37:925–929. doi: 10.1128/jcm.37.4.925-930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denning D W. Invasive aspergillosis. Clin Infect Dis. 1998;26:781–805. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 3.Einsele H, Hebart H, Roller G, Löffler J, Rothenhöfer I, Müller C A, Bowden R A, van Burik J A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Einsele H, Quabeck K, Müller K D, Hebart H, Rothenhöfer I, Löffler J, Schaefer U W. Colonization of the lower respiratory tract by Aspergillus species at the time of transplant predicts invasive pulmonary aspergillosis in marrow graft recipients. Lancet. 1998;352:1443. doi: 10.1016/s0140-6736(05)61265-2. [DOI] [PubMed] [Google Scholar]
- 5.El-Zaatari F A, Naser S A, Markesich D C, Kalter D C, Engstand L, Graham D Y. Identification of Mycobacterium avium complex in sarcoidosis. J Clin Microbiol. 1996;34:2240–2245. doi: 10.1128/jcm.34.9.2240-2245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flahaut M, Sanglard D, Monod M, Bille J, Rossier M. Rapid detection of Candida albicans in clinical samples by DNA amplification of common regions from C. albicans-secreted aspartic proteinase genes. J Clin Microbiol. 1998;36:395–401. doi: 10.1128/jcm.36.2.395-401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomes M L, Macedo A M, Vago A R, Pena S D, Galvao L M, Chiari E. Trypanosoma cruzi: optimization of polymerase chain reaction for detection in human blood. Exp Parasitol. 1998;88:28–33. doi: 10.1006/expr.1998.4191. [DOI] [PubMed] [Google Scholar]
- 8.Latgé J P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leon S A, Shapiro B, Slaroff D M, Yaros M J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1997;37:646–650. [PubMed] [Google Scholar]
- 10.Löffler J, Hebart H, Sepe S, Schumacher U, Klingebiel T, Einsele H. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med Mycol. 1998;36:275–279. doi: 10.1080/02681219880000441. [DOI] [PubMed] [Google Scholar]
- 11.Mulcahy H E, Croke D T, Farthing M J. Cancer and mutant DNA in blood plasma. Lancet. 1996;348:628. doi: 10.1016/S0140-6736(05)65067-2. [DOI] [PubMed] [Google Scholar]
- 12.Prince W S, Baker D, Dodge A H, Ahmet A R, Chestnut R W, Sinicropi D V. Pharmacodynamics of recombinant human DNAse I in serum. Clin Exp Immunol. 1998;113:289–296. doi: 10.1046/j.1365-2249.1998.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rumore P, Muralidhar B, Lin M, Lai C, Steinman C R. Haemodialysis as a model for studying endogenous plasma DNA: oligonucleotide-like structure and clearance. Clin Exp Immunol. 1992;90:56–62. doi: 10.1111/j.1365-2249.1992.tb05831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin J H, Notle F S, Morrison C J. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J Clin Microbiol. 1997;35:1454–1459. doi: 10.1128/jcm.35.6.1454-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun R, Ku J, Jayakar H, Kuo J C, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–2969. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Burik J A, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]