Abstract
Amplification of a specific, 500-bp fragment from Mycobacterium bovis isolates and use of the fragment to differentiate between Mycobacterium tuberculosis and M. bovis was previously reported (J. G. Rodriguez, G. A. Meja, P. Del Portillo, M. E. Patarroyo, and L. A. Murillo, Microbiology 141:2131–2138, 1995). In the present study, 30 M. bovis isolates from Sardinian cattle were examined for the presence of this 500-bp fragment; 4 of the 30 isolates lacked the fragment. This result indicates that identification of M. bovis strains by amplification of the 500-bp sequence may lead to false-negative results.
Mycobacterium tuberculosis is the most common cause of tuberculosis in humans, but an unknown proportion of human tuberculosis is caused by Mycobacterium bovis (2, 6, 9, 14, 17). Human M. bovis infections are more common in areas of the world where bovine tuberculosis is uncontrolled and pasteurization of milk is not universal (2, 9). It is difficult to discriminate between M. tuberculosis and M. bovis with conventional methods (3, 11, 16). Recently, different molecular techniques have been proposed to rapidly identify and differentiate M. bovis, even directly in biological samples (6, 8, 12, 19, 20). The PCR assay reported by Rodríguez et al. (10, 11) is a simple and accurate method to identify and differentiate M. bovis.
In a surveillance study in Sardinia, Italy, we isolated 30 M. bovis strains from cattle. All isolates were confirmed to belong to the M. tuberculosis complex by PCR-restriction fragment polymorphism analysis (PRA) of the gene for the 65-kDa heat shock protein (hsp65) as previously described by Telenti et al. (18). The isolates were further identified as M. bovis by biochemical methods. PCR ribotyping and DNA fingerprinting with IS6110, enterobacterial repetitive intergenic consensus (ERIC)-PCR, and GTG5-PCR have already been reported for 22 of these isolates (14). In this study, the isolates were evaluated by the PCR assay described by Rodríguez et al. (10).
Thirty strains of M. bovis were isolated from cattle in various regions of Sardinia from 1996 to 1999. M. bovis ATCC 27290 was used as the positive control; M. tuberculosis strain H37Rv (American Type Culture Collection) and 20 M. tuberculosis clinical isolates were used as negative controls. All strains used in this study were identified as M. bovis by the following biochemical properties: scarce growth in glycerol, lack of detectable niacin production (kit from Difco Laboratories, Detroit, Mich.), negative results in tests for catalase and pyrazinamidase, and susceptibility to thiophene carboxylic hydrazide (8) (Sigma Chemical Co.). Isolates were maintained on Lowenstein-Jensen slants until subjected to further analysis.
Mycobacterial strains were grown in 10 ml of 7H9 medium with albumin dextrose catalase, and genomic DNA was extracted and analyzed as described previously (15).
All DNA amplifications were performed in a DNA thermal cycler (Hybaid, model TR3CM220; Omnigene, Teddington, United Kingdom). Strains were identified as belonging to the M. tuberculosis complex by PRA of the hsp65 gene (22). PRA was performed with primers tb11 (5′-ACCAACGATGGTGTGTCCAT) and tb12 (5′-CTTGTCGAACCGCATACCCT). Twenty microliters of the PCR product was digested by BstEII (Promega, Madison, Wis.) and HaeIII (New England Biolabs, Inc., Beverly, Mass.), and 15 μl of the restriction digest was loaded on a 2% (wt/vol) Metaphor agarose gel (FMC Bioproducts, Rockland, Maine) and visualized by staining with ethidium bromide.
The species-specific PCR proposed by Rodríguez et al. (10) was used to identify M. bovis strains. Primers JB21 (5′-TCGTCCGCTGATGCAAGTGC) and JB22 (5′-CGTCCGCTGACCTCAAGAAG) were used at a concentration of 20 pmol each, and the reaction was performed in a total volume of 50 μl containing 2.5 U of Taq polymerase, 20 mM Tris (pH 8.3), 50 mM KCl, 1.5 mM MgCl, and 200 μM deoxynucleoside triphosphate (Gibco BRL Life Technology, Paisley, United Kingdom). Reaction mixtures were overlaid with 1 drop of paraffin oil and then incubated for 2 min at 94°C, followed by 35 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 10 min and a final extension at 70°C for 20 min. The amplification products were visualized after electrophoresis at 90 V for 90 min in a 1.8% Metaphor agarose gel and staining with ethidium bromide.
Sequencing of the 500-bp fragment was performed using the dideoxy chain termination method of Sanger et al. (13) with Sequenase (United States Biochemical Corporation, Cleveland, Ohio). A GenBank search (1) of the 500-bp sequence of strain 621-11 revealed identity with the M. bovis BCG 500-bp region sequenced by Gordon et al. (5). The 500-bp fragment is part of a 4,999-bp fragment that is not present in the M. tuberculosis H37Rv chromosome (1, 5). It is located at the 3′ end of a putative gene called RvD1-Rv2031c.
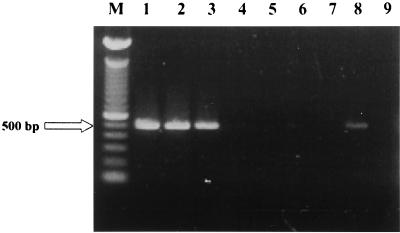
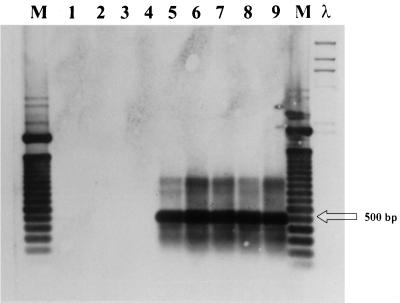
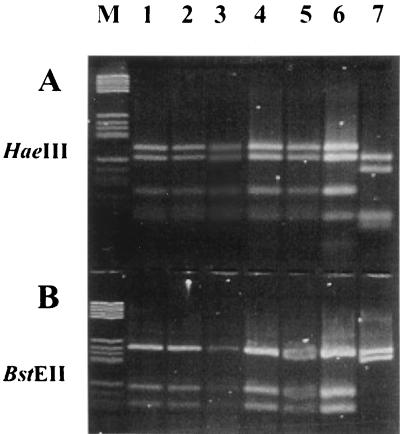
Each of the isolates from Sardinian cattle was identified as M. bovis by biochemical methods. However, 4 of the 30 strains did not generate the 500-bp fragment (strains 868-30, 621-1, 621-2, and 621-10) (Fig. 1, lanes 4, 5, 6, and 7, respectively) when evaluated in the M. bovis PCR assay described by Rodríguez et al. (10) and assessed by ethidium bromide staining. The 500-bp sequence also was not detected in chromosomal DNA from these four strains as determined by hybridization (Fig. 2, lanes 1, 2, 3, and 4). These strains were confirmed as belonging to the M. tuberculosis complex by PRA of hsp65 (Fig. 3, lanes 1, 2, 3, and 4).
FIG. 1.
Agarose gel electrophoresis (Metaphor 1.8%) of DNA amplifications of the 500-bp fragment of different M. bovis clinical isolates. Lane M, molecular marker consisting of a 100-bp ladder (Gibco Life Science); lane 1, strain 621-11; lane 2, strain 621-13; lane 3, strain 621-15; lane 4, strain 868-30; lane 5, strain 621-1; lane 6, strain 621-2; lane 7, strain 621-10; lane 8, M. bovis ATCC 27290; lane 9, negative control.
FIG. 2.
Southern blot of chromosomal DNA of different M. bovis isolates showing hybridization with the labeled 500-bp fragment. Lane 1, strain 868-30; lane 2, strain 621-1; lane 3, strain 621-2; lane 4, strain 621-10; lane 5, strain 621-11; lane 6, strain 621-13; lane 7, strain 621-15; lane 8, strain 621-16; lane 9, M. bovis ATCC 27290; lane M, molecular marker consisting of a 100-bp ladder; λ, molecular marker consisting of HindIII-digested DNA.
FIG. 3.
Agarose gel electrophoresis (Metaphor, 1.8%) of amplified DNA of the hsp65 gene digested with HaeIII and BstEII. Lane M, molecular marker V (Boehringer Mannheim, Monza, Italy); lane 1, strain 868-30; lane 2, strain 621-1; lane 3, strain 621-2; lane 4, strain 621-10; lane 5, strain 621-11; lane 6, M. bovis ATCC 27290; lane 7, M. avium as a negative control.
The results of other molecular characterizations of these four strains (14) were reviewed to determine if there were genetic differences between isolates lacking the 500-bp fragment. The four strains produced two different IS6110 fingerprinting patterns: strains 621-10 and 868-30 did not hybridize with the probe, whereas strains 621-1 and 621-2 produced a band of hybridization at 5.5 kb (14). The strains were also differentiated with two other previously published PCR-based fingerprinting methods, GTG5-PCR and PCR ribotyping (14). Strain 621-1 (which generated a 5.5-kb band of hybridization with IS6110 probe) and strain 621-10 (generating no bands with IS6110 fingerprinting) could not be differentiated by PCR ribotyping, whereas strains 868-30 and 621-2 produced different patterns after endonuclease digestion. GTG5-PCR discriminated strains 621-1 and 621-10 from strains 621-2 and 868-30 (14). Strains 621-1, 621-2, and 621-10 were isolated from different cattle of the same herd.
Our results have implications for the application of PCR in laboratory diagnosis of M. bovis infections. PCR has the potential to provide a more rapid, sensitive, and specific diagnostic assay than conventional methods for M. bovis identification in clinical specimens. Rodríguez et al. showed with PCR the presence of the 500-bp fragment in the chromosomes of 121 M. bovis isolates and 4 M. tuberculosis strains isolated from sea lions (10). The present study demonstrates that not all M. bovis strains can be detected with primers directed toward the 500-bp fragment. Use of such primers can lead to false-negative results. Other PCR techniques, such as PCRs based on amplification of IS6110 (7), also can lead to false-negative results if used alone; we have found five M. bovis strains with no IS6110 insertions in their genomes (14). Strains 868-30 and 621-10 were negative in both the 500-bp fragment PCR and the IS6110 hybridization. Only the PRA of the hsp65 gene identified these strains as belonging to the M. tuberculosis complex. Thus, even the combined molecular tests (in this case, assays for IS6110 and the 500-bp fragment) can give false-negative results, and only the use of multiple molecular techniques and biochemical methods will ensure the proper identification of isolates.
Previous molecular data on the four isolates lacking the 500-bp fragment were reviewed (14). Two of the strains negative for the 500-bp fragment (868-30 and 621-10) generated the same pattern with GTG5-PCR and also with PCR ribotyping after HaeIII digestion (14), but they showed slightly different patterns with ERIC-PCR and PCR ribotyping after PvuII digestion (14). The GTG5-PCR pattern of these isolates was not found elsewhere among the Sardinian isolates examined (14). This indicates that the isolates were very similar; the herds from which these isolates originated are very close geographically and a common progenitor may have infected both herds.
Acknowledgments
This work was supported in part by the first and second national project “Tubercolosi” of the “Istituto Superiore di Sanità,” Rome, Italy, and by grant 9806297296 from MURST.
We thank E. Manca and D. Delogu for technical help.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cosivi O, Grange J M, Daborn C J, Raviglione M C, Fujikura T, Cousins D, Robinson R A, Huchzermeyer H F A K, de Kantor I, Meslin F-X. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg Infect Dis. 1998;4:1–15. doi: 10.3201/eid0401.980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cousins D, Williams S, Liébana E, Aranaz A, Bunschoten A, Van Embden J, Ellis T. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J Clin Microbiol. 1998;36:168–178. doi: 10.1128/jcm.36.1.168-178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fadda G, Roe S L. Recovery and susceptibility testing of Mycobacterium tuberculosis from extrapulmonary specimens by the BACTEC radiometric method. J Clin Microbiol. 1984;19:720–721. doi: 10.1128/jcm.19.5.720-721.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon S V, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole S T. Identification of variable regions in the genomes of tubercle bacilli using bacterial chromosome arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 6.Gutiérrez M, Samper S, Jiménez M S, van Embden J D A, Marín J F G, Martín C. Identification by spoligotyping of a caprine genotype in Mycobacterium bovis strains causing human tuberculosis. J Clin Microbiol. 1997;35:3328–3330. doi: 10.1128/jcm.35.12.3328-3330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liébana E, Aranaz A, Dominguez L, Mateos A, González Llamazares O, Rodriguez-Ferri E F, Domingo M, Vidal D, Cousins D. The insertion element IS6110 is a useful tool for DNA fingerprinting of Mycobacterium bovis isolates from cattle and goats in Spain. Vet Microbiol. 1997;54:223–233. doi: 10.1016/s0378-1135(96)01282-5. [DOI] [PubMed] [Google Scholar]
- 8.Metchock B G, Nolte F S, Wallace R J., Jr . Mycobacterium. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 399–437. [Google Scholar]
- 9.O'Reilly L M, Daborn C J. The epidemiology of Mycobacterium bovis infection in animals and man: a review. Tuberc Lung Dis. 1995;76:1–46. doi: 10.1016/0962-8479(95)90591-x. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez J G, Fissanoti J C, Del Portillo P, Patarroyo M E, Romano M I, Cataldi A. Amplification of a 500-base-pair fragment from cultured isolates of Mycobacterium bovis. J Clin Microbiol. 1999;37:2330–2332. doi: 10.1128/jcm.37.7.2330-2332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez J G, Mejia G A, Del Portillo P, Patarroyo M E, Murillo L A. Species-specific identification of Mycobacterium bovis by PCR. Microbiology. 1995;141:2131–2138. doi: 10.1099/13500872-141-9-2131. [DOI] [PubMed] [Google Scholar]
- 12.Romero R E, Garzon D L, Mejia G A, Monroy W, Patarroyo M E, Murillo L A. Identification of Mycobacterium bovis in bovine clinical samples by PCR species-specific primers. Can J Vet Res. 1999;63:101–106. [PMC free article] [PubMed] [Google Scholar]
- 13.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sechi L A, Leori G, Lollai S A, Duprè I, Molicotti P, Fadda G, Zanetti S. Different strategies for molecular differentiation of Mycobacterium bovis strains isolated in Sardinia, Italy. Appl Environ Microbiol. 1999;65:1781–1785. doi: 10.1128/aem.65.4.1781-1785.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sechi L A, Zanetti S, Duprè I, Delogu G, Fadda G. Enterobacterial repetitive intergenic consensus sequences as molecular targets for typing of Mycobacterium tuberculosis strains. J Clin Microbiol. 1998;36:128–132. doi: 10.1128/jcm.36.1.128-132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stonebrink B. The use of a pyruvate containing egg medium in the culture of isoniazid resistant strains of Mycobacterium tuberculosis var. Hominis Acta Tuberc Scand. 1958;35:67–80. [Google Scholar]
- 17.Szewzyk R, Svenson S B, Hoffner S E, Bölske G, Wahlström H, Englund L, Engvall A, Källenius G. Molecular epidemiological studies of Mycobacterium bovis infections in humans and animals in Sweden. J Clin Microbiol. 1995;33:3183–3185. doi: 10.1128/jcm.33.12.3183-3185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whipple D L, Clarke P R, Jaragin J L, Payeur J B. Restriction fragment length polymorphism analysis of Mycobacterium bovis isolates from captive and free-ranging animals. J Vet Diagn Investig. 1997;9:381–386. doi: 10.1177/104063879700900407. [DOI] [PubMed] [Google Scholar]
- 20.Wilson S M, Goss S, Drobniewski F. Evaluation of strategies for molecular fingerprinting for use in the routine work of a mycobacterium reference unit. J Clin Microbiol. 1998;36:3385–3388. doi: 10.1128/jcm.36.11.3385-3388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]