Abstract
Owing to its great threat to human health and environment, Pb2+ pollution has been recognized as a major public problem by the World Health Organization (WHO). Many DNA aptamers have been utilized in the development of Pb2+-detection sensors, but the underlying mechanisms remain elusive. Here, we report three Pb2+-complexed structures of the thrombin binding aptamer (TBA). These high-resolution crystal structures showed that TBA forms intramolecular G-quadruplex and Pb2+ is bound by the two G-tetrads in the center. Compared to K+-stabilized G-quadruplexes, the coordinating distance between Pb2+ and the G-tetrads are much shorter. The T3T4 and T12T13 linkers play important roles in dimerization and crystallization of TBA, but they are changeable for Pb2+-binding. In combination with mutagenesis and CD spectra, the G8C mutant structure unraveled that the T7G8T9 linker of TBA is also variable. In addition to expansion of the Pb2+-binding aptamer sequences, our study also set up one great example for quick and rational development of other aptamers with similar or optimized binding activity.
Subject terms: X-ray crystallography, Analytical biochemistry, DNA, Biophysical chemistry
Introduction
Lead (Pb) is one of the most toxic heavy metals known to date. Through mimicking of essential elements such as iron, zinc and calcium, Pb can enter and accumulate in the human body1, impairing the functions of multiple organs including kidney, heart and brain2. The direct relationships between Pb poisoning and growth3, delayed puberty4 and hematological changes5 have been well documented. Human can be exposed to Pb under diverse circumstances such as battery6,7 and automotive manufacturing8, refining and smelting9. Contaminated drinking water is another important source of Pb exposure10 and has occurred in many areas11,12.
Owing to its great threat to human health, Pb poisoning has been recognized as one major public problem. To detect the Pb present in human body as well as in the surrounding environment, various methods have been developed. Many methods rely on the traditional analytical techniques such as atomic absorption spectroscopy, anodic stripping voltammetry and reverse-phase high-performance liquid chromatography. Although these methods are sensitive, they require sophisticated equipments. The requirement of complicated sample treatment is another major limitation of these methods. To develop faster and simpler methods for Pb detection, scientists have performed extensive trials and found that some DNA sequences possess high Pb ion (Pb2+) binding affinity and specificity. These sequences were termed as Pb2+-binding DNA aptamers and have been extensively utilized in Pb2+ detector development. Compared to the traditional Pb2+ detecting methods, these aptamer-based methods are easier in handling and less expensive.
To date, various Pb2+-binding DNA aptamers have been identified13. Exampled by PS2.M14, PW17, T30695 and the thrombin binding aptamer (TBA), many Pb2+-binding DNA aptamers are rich in guanines. Guanine-rich sequences are often observed at the ends of chromosomal telomeres or at the promoter regions of human genes15. Interestingly, these sequences are also present in the genomes of many viruses such as Papilloma Virus and Human Immunodeficiency Virus (HIV), playing important regulatory roles in viral replication and gene expression. Guanine-rich sequences are capable of forming quadruplexes16. Quadruplexes are normally stabilized by the stacking of multiple Hoogsteen hydrogen-bonded G-tetrads and the electrostatic interactions between the guanines and the cations (commonly K+ or Na+) residing in the center of the tetrads.
Circular dichroism (CD), UV and/or NMR spectra indicated that Pb2+ can cause large conformational changes and induce the quadruplex formation of many DNA aptamers such as PS2.M, PW17 and T3069513,17. Compared to K+ and Na+, Pb2+ is more efficient in quadruplex formation of these aptamers. Although the structural basis is not very clear, Pb2+-binding aptamers have been utilized in the design of Pb2+-controlled DNA gate13 and other nanodevices. Assisted by other technologies, such as Fluorescence Anisotropy (FA)18, Fluorescent Metal Nanocluster (NCs)19 and Forster Resonance Energy Transfer (FRET)20, these Pb2+-binding aptamers have also been used to develop ultrasensitive Pb2+ sensors. Due to their intrinsic difficulty in crystallization, the Pb2+-DNA aptamer complex structures are barely available, which hindered our understanding on Pb2+-binding mechanisms by DNA aptamers. Here, we report three complex structures of Pb2+-TBA, which revealed the detailed basis for Pb2+ binding by TBA. In combination with site-specific mutations and CD analyses, our studies revealed the detailed mechanistic insights into the Pb2+-TBA interactions and also shed great light for the development of novel Pb2+-binding aptamers.
Results
Overall structure of TBA-Pb2+ complex
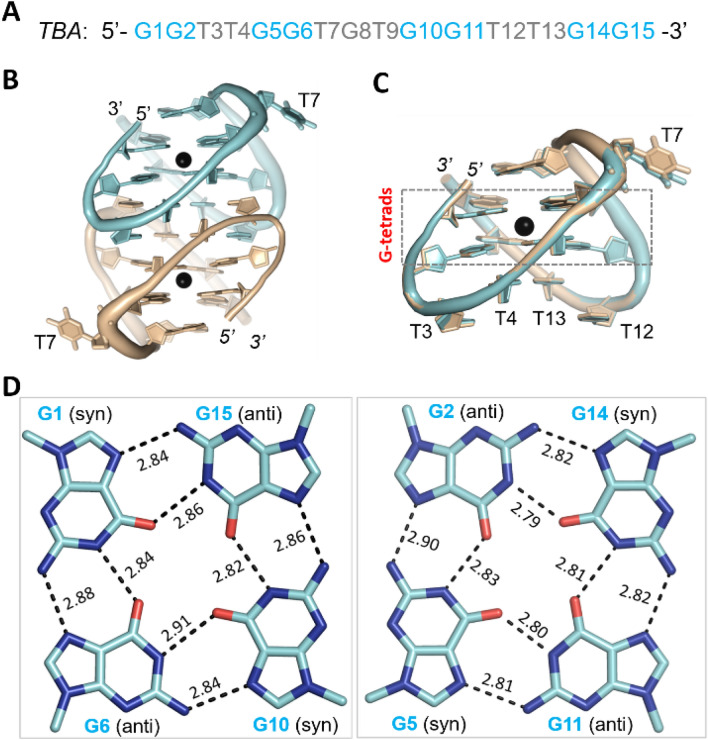
To reveal the underlying basis of Pb2+-binding by DNA aptamers, we performed extensive crystallization trials. Although no crystals were obtained for PS2.M, PW17 or T30695, we solved two TBA-Pb2+ complex structures with high-resolution in both A-form and B-form (Table 1). As calculated by the Matthews Coefficient program, the solvent content (44.7%) of the A-form structure is higher than that (40.0%) of the B-form structure. The A-form structure belongs to the tetragonal space group P41212; it contains two TBA-Pb2+ complexes per asymmetric unit (Fig. 1B). The B-form structure belongs to the orthorhombic space group P212121; per asymmetric unit contains six TBA-Pb2+ complexes. Due to their different space groups, packing of the TBA-Pb2+ complexes are very different in the A- and B-form structures (Fig. S1A,B). However, the overall folding of the eight TBA-Pb2+ complexes are similar. The root mean square deviations value (RMSD) between the two TBA-Pb2+ complexes is only 0.15 Å within the A-form structure; and, it ranges from 0.3 to 0.5 Å between the complexes of the A- and B-form structures.
Table 1.
Data collection and refinement statistics.
| Structure (PDB ID) |
TBA-Pb2+ complex (A-form) 7D31 |
TBA-Pb2+ complex (B-form) 7D32 |
G8C-Pb2+ complex 7D33 |
|---|---|---|---|
| Data collectiona | |||
| Space group | P41212 | P212121 | C2 |
| Cell parameter | |||
| a (Å) | 40.0 | 27.3 | 51.4 |
| b (Å) | 40.0 | 53.1 | 78.0 |
| c (Å) | 99.9 | 152.5 | 101.7 |
| α (°) | 90.0 | 90.0 | 90.0 |
| β (°) | 90.0 | 90.0 | 113.6 |
| ρ (°) | 90.0 | 90.0 | 90.0 |
| Wavelength (Å) | 0.9793 | 0.9793 | 0.9793 |
| Resolution (Å) | 30.0–1.4 | 30.0–1.7 | 37.4–2.1 |
| Last shell (Å) | 1.45–1.4 | 1.76–1.70 | 2.23–2.1 |
| Completeness (%) | 97.9 (89.6) | 96.0 (84.4) | 96.5 (98.5) |
| Redundancy | 14.4 (3.8) | 5.7( 2.6) | 6.7 (6.7) |
| I/σ (I) | 44.9 (3.3) | 9.6 (1.9) | 6.7 (2.4) |
| Rmerge (%) | 10.9 (38.1) | 14.6 (43.2) | 22.5 (93.9) |
| Refinement | |||
| Resolution (Å) | 27.2–1.4 | 26.4–1.7 | 25.3–2.1 |
| Rwork (%)/Rfree (%) | 12.6/14.8 | 18.9/21.5 | 21.2/24.0 |
| No. of atoms | |||
| DNA | 630 | 1890 | 1560 |
| Pb2+ | 2 | 6 | 5 |
| Water | 57 | 85 | 70 |
| R.m.s. deviations | |||
| Bond length (Å) | 0.007 | 0.006 | 0.005 |
| Bond angle (°) | 0.997 | 0.918 | 0.751 |
aValues in parentheses are for the last resolution shell.
Figure 1.
TBA-Pb2+ complex structure. (A) Sequence of TBA aptamer. (B) Overall folding of the A-form TBA-Pb2+ complex structure. (C) Superposition of the two TBA-Pb2+ complexes. (D) The G-tetrads observed in the A-form TBA-Pb2+ complex structure. The detailed Hoogsteen H-bond distances are labelled with numbers (Å). In (B,C), TBA molecules A and B are shown as cartoons in cyan and wheat, respectively. Pb2+ ions are shown as black spheres. In (D), the C-atoms are colored in cyan for the G-tetrads of TBA molecule A, respectively. Figures were displayed using PyMOL (http://www.pymol.org/).
Compared to the B-form structure, the resolution (1.4 Å) of the A-form TBA-Pb2+ complex structure is higher, therefore it was used for further structural analysis. TBA is composed of 15 nucleotides. As depicted in Fig. 1C, each TBA molecule folds into an antiparallel chair-like quadruplex, which can be divided into four regions: two TT linkers (T3T4 and T12T13), one TGT linker (T7G8T9) and the central G-tetrads. The central G-tetrads are formed by eight G nucleotides, in which G1, G6, G10 and G15 arranged in one layer and G2, G5, G11 and G14 arranged in a second layer (Fig. S2). In both layers, two G nucleotides adopt syn conformation and the other two adopt anti conformation. Interestingly, although the conformations of the G nucleotides varied within the two G-tetrads, the interactions mediated by the individual G nucleotide conserved. As depicted in Fig. 1D, each G nucleotide forms four Hoogsteen hydrogen bond interactions. The average in-plane N1–O6 and N2–N7 distances are 2.83 Å and 2.85 Å, respectively. Compared to other G-quadruplex structures, both the N1-O6 and N2-N7 distances are shorter in the TBA-Pb2+ complex structure (Table 2). The two G-tetrads of TBA-Pb2+ complex form stable hydrophobic stacking interactions, the average between-plane O6–O6 distance is only 2.98 Å, which is significantly shorter than other quadruplexes.
Table 2.
Mean distances (Å) observed in the A-form TBA-Pb2+ complex and reported quadruplex structures.
| A-form TBA-Pb2+(PDB:7D31) |
thrombin-TBA complex (PDB: 4DII) | oxytricha nova telomeric quadruplex (PDB: 3NZ7) | |
|---|---|---|---|
| Pb2+–O6 | 2.65 | ||
| K+–O6 | 2.82 | 2.86 | |
| O6–O6 (between G-tetrads) | 2.98 | 3.31 | 3.44 |
| O6–O6 (within G-tetrads) | 3.12 | 3.25 | 3.28 |
| N1–O6 H-bond | 2.83 | 2.83 | 2.88 |
| N2–N7 H-bond | 2.85 | 2.89 | 2.92 |
Pb2+ forms tight coordination with TBA
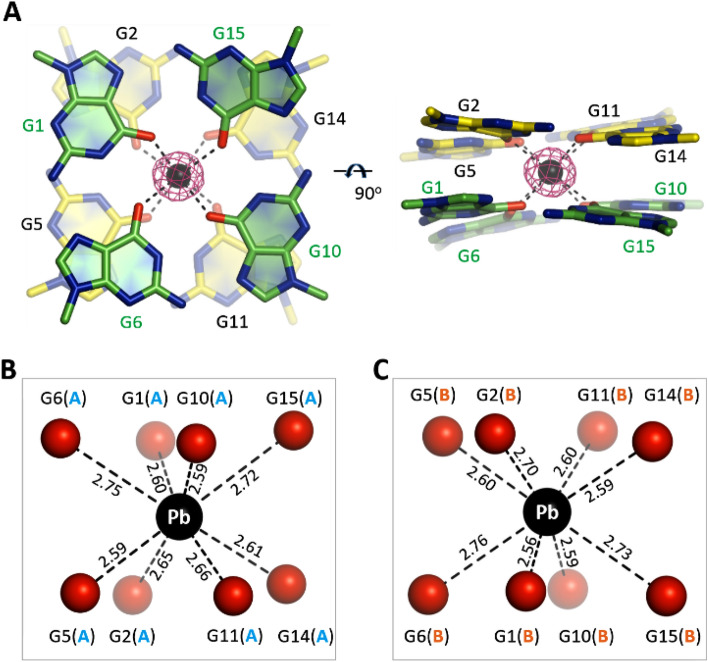
TBA is one of the earliest identified Pb2+-binding aptamers. Although it is not preferred by many other quadruplexes, Pb2+ is effective in folding and stabilizing the structures of TBA. In the A-form TBA-Pb2+ complex structure, each TBA molecule captures one Pb2+ ion. The Pb2+ ion resides in-between the two G-tetrads (Fig. 2A), coordinating with the eight O6 atoms of the G-tetrads (Fig. 2B,C). The shortest coordination (2.56 Å) forms between Pb2+ and the G1 nucleotide of TBA molecule B. In addition, Pb2+ forms several short coordination (~ 2.60 Å) with other G nucleotides. The average Pb2+–O6 coordinating distance is only 2.65 Å in the A-form TBA-Pb2+ complex structure (Table 2). Similar Pb2+–O6 coordination and coordinating distance are also observed in the B-form TBA-Pb2+ complex structure.
Figure 2.
Pb2+ coordination by TBA. (A) Electron density and coordination of the Pb2+ ions. The two G-tetrads are shown as sticks. Pb2+ ions are shown as black spheres. The 2Fo − Fc electron density maps of Pb2+ are contoured at 10 σ level. (B,C) The detailed coordinating geometry and coordinating distances (Å) of the Pb2+ ions. Pb2+ and the O6 atoms of the G-tetrads are shown as spheres in black and red, respectively. Figures were displayed using PyMOL (http://www.pymol.org/).
Compared to Pb2+, monovalent cations (especially K+ and Na+) are more common in stabilizing quadruplex structures. Due to its great potential in inhibiting HIV replication, TBA received much more attentions than other DNA aptamers21–23. CD and NMR studies confirmed that K+ can induce quadruplex formation of TBA, but the quadruplex is very dynamic and requires high concentration of K+. We were able to obtain the crystals for the TBA-Pb2+ complex. However, in consistent with its intrinsic dynamic, no crystal of TBA-K+ complex could be obtained in previous and our studies. Although it is not preferred by other quadruplexes, these observations indicated that Pb2+ is more powerful than K+ in TBA quadruplex folding and stabilization.
No crystal structure of TBA-K+ complex is available to date, but one crystal structure of TBA-thrombin complex (PDB_ID: 4DII)24 has been reported, which captured one K+ in-between the two G-tetrads. The high-resolution crystal structures of many other K+-stabilized quadruplexes have also been reported, including oxytricha nova telomeric quadruplex (PDB_ID: 3NZ7)25. To reveal the basis for the strong TBA-stabilizing ability of Pb2+, we analyzed the TBA-Pb2+ complex, the TBA-thrombin complex and the oxytricha nova telomeric quadruplex. We found that the coordination geometries of Pb2+ and K+ are actually very similar in the three structures. However, compared to the average Pb2+–O6 distance, the average K+–O6 coordinating distance is about 0.21 Å longer (Table 2). In addition, the average within-plane and between-plane O6–O6 distances are also longer in the K+-stabilized quadruplexes. The ionic radius of Pb2+ is 1.29 Å, which is smaller than that (1.51 Å) of K+. However, compared to K+, Pb2+ is more positive in charge. Most likely, the smaller ionic radius and stronger electropositivity of Pb2+ play critical roles in the tighter coordination, the more compacted G-tetrad conformations and the higher TBA-Pb2+ stability.
Dimerization is important for TBA-Pb2+ crystallization
As depicted in Fig. 1B, the two TBA-Pb2+ complexes assembled into one dimer in the A-form structure. In fact, the six TBA-Pb2+ complexes within the asymmetric unit of the B-form structure also assembled into dimers: dimer AB, CD and EF (Fig. S3A). The overall fold of the AB and CD dimers is very similar to that of the A-form structure (Fig. S3B,C). The overall fold of the EF dimer is also very similar to other dimers, but the relative orientation of the EF dimer is different from others. As depicted in Fig. 3A, when the EF dimer is superimposed with the AB dimer of the A- or B-form structure, there is one 180° rotation (along the central vertical axis) between the F and B molecules.
Figure 3.
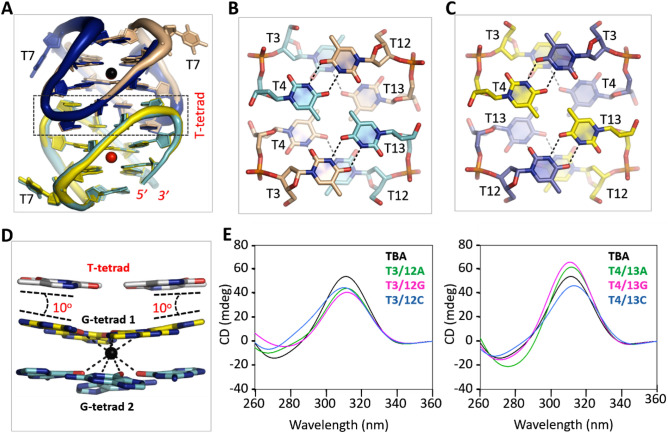
Functional characterization of the TT linkers. (A) Structural superposition of the A-form complex with the EF dimer of the B-form complex, which is colored in yellow, blue, and red. The A-form complex is colored as in Fig. 1A. (B) Dimerization mediated by the TT linkers of the A-form structure. (C) Dimerization mediated by the TT linkers of the EF dimer of the B-form structure. (D) Stacking with the pseudo T-tetrad and tilting of the G-tetrad 1. (E) CD spectra analysis of the native and TT linker mutated TBAs. Except the mutations listed in the figure, the sequences of the mutants are identical to that of TBA.TBA (native or mutant) and Pb2+ concentrations are 10 μM and 50 μM, respectively. Structural figures were displayed using PyMOL (http://www.pymol.org/).
Dimerization of the TBA-Pb2+ complexes is mediated by the two TT linkers. For the AB (or CD) dimers in the A- and B-form structures (Fig. 3B), T3 and T4 arrange in two layers; similar conformations were also adopted by T12 and T13. However, instead of T12 or T13 from the same molecule, T3 and T4 form hydrogen (H) bond interactions with T13 and T12 from the partner molecule, respectively (Fig. S4A,B). T3, T4, T12 and T13 also arrange in two layers in the EF dimer of the B-form structure (Fig. 3C). However, due to the rotation of the F molecule, T3 and T4 of molecule F interact with T4 and T3 of molecule E, respectively. Likewise, T12 and T13 of the two molecules interact with each other (Fig. S4C,D).
To investigate the functional importance of the dimerization mediated by the TT linkers, we designed six TBA mutants with TT linker mutation, including T3/12A, T3/12G, T3/12C, T4/13A, T4/13G, and T4/13C (Table 3). Like native TBA, we performed extensive crystallization trials for all the TT linker mutated sequences. However, none of these sequences could produce crystal. These observations indicated that the TT linkers-mediated dimerization is critical for the crystal growth of TBA-Pb2+ complex.
Table 3.
Sequences of native and mutated TBAs.
| Name | Sequence (from 5′ to 3′) |
|---|---|
| TBA | GGTTGGTGTGGTTGG |
| T3/12A | GGATGGTGTGGATGG |
| T3/12G | GGGTGGTGTGGGTGG |
| T3/12C | GGCTGGTGTGGCTGG |
| T4/13A | GGTAGGTGTGGTAGG |
| T4/13G | GGTGGGTGTGGTGGG |
| T4/13C | GGTCGGTGTGGTCGG |
| T7A | GGTTGGAGTGGTTGG |
| T7G | GGTTGGGGTGGTTGG |
| T7C | GGTTGGCGTGGTTGG |
| G8A | GGTTGGTATGGTTGG |
| G8T | GGTTGGTTTGGTTGG |
| G8C | GGTTGGTCTGGTTGG |
| T9A | GGTTGGTGAGGTTGG |
| T9G | GGTTGGTGGGGTTGG |
| T9C | GGTTGGTGCGGTTGG |
Significant letters are given in bold.
TT linkers are changeable for Pb2+-binding by TBA
For all TBA-Pb2+ dimers observed in the A- and B-form structures, the eight thymidine residues of the TT linkers formed two pseudo T-tetrads (Fig. 3B,C). Maybe due to the presence of 5-methyl groups at the center, no cation could be bound in-between the two T-tetrads or in-between the T- and G-tetrads (Fig. 3D). The pseudo T-tetrad stacks against G-tetrad 1, the G-tetrad formed by G2, G5, G11 and G14. However, compared to the stacking interactions formed between regular G-tetrads, the stacking interactions between the pseudo T-tetrad and the G-tetrad is weaker, which allows ~ 10° tilt of the nucleobases of the G-tetrad and leads to the tight coordination with Pb2+.
The TT linker-mediated dimerization is important for TBA-Pb2+ complex crystallization. However, in the TBA-thrombin complex structure, TBA exists as a monomer. Superposition with the TBA-Pb2+ complex structures showed that T4 and T12 adopt conserved conformations, but the conformation of T3 and T13 are very different (Fig. S5A). Together with its weaker interaction with the G-tetrad and the variable dimerization manner, we speculated that the pseudo T-tetrads are actually very dynamic and may be not critical for Pb2+ binding. To investigate this possibility, we performed CD spectra analysis for the native and mutated TBA sequences (Fig. 3E). Upon addition of Pb2+, native TBA showed one strong positive peak near 312 nm, which is the most characteristic peak of Pb2+-stabilized quadruplexes. Besides TBA-Pb2+, this peak was also observed for PS2.M-Pb2+ and PW17-Pb2+ complexes previously12,17. Like native TBA, all the TBA mutants showed positive peak near 312 nm. Compared to that of native sequence, the heights of the 312-nm peaks are slightly lower for the T3/12A, T3/12G, T3/12C and T4/13C mutants, whereas they are increased for the T4/13A and T4/13G mutants.
TGT linker helps the stabilization of TBA
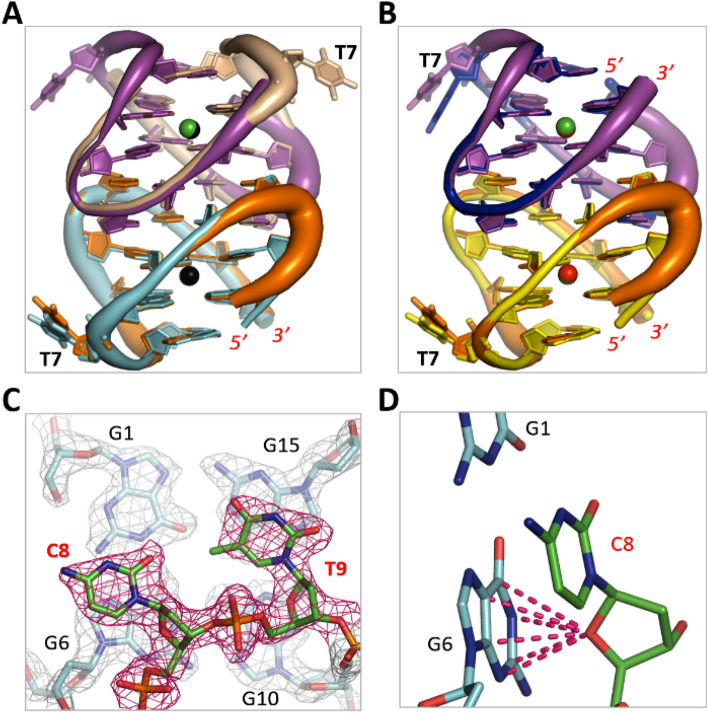
Unlike the T3T4 and T12T13 linkers, no nucleotide of the T7G8T9 linker forms tetrads in the A- and B-form structures. Instead, the T7G8T9 linker adopts one loop-like conformation (Fig. 4A). The nucleobase of T7 points away from the main body of TBA and does not participate in Pb2+ binding. The phosphate backbone of T7 is well organized. However, as indicated by the weak electron densities and high B-factors, the conformation of T7 nucleobase is very flexible. The flexibility of T7 nucleobase could also be further supported by the conformational differences between the A- and B-form structures (Fig. S3B).
Figure 4.
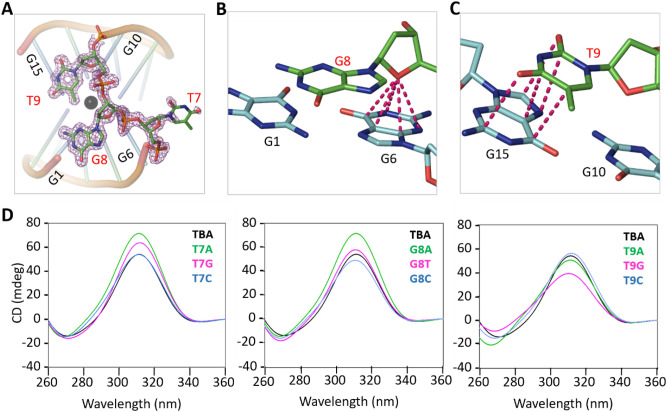
Functional characterization of the TGT linker. (A) Detailed conformation of the TGT linker, which are shown as sticks. The 2Fo − Fc electron density maps are contoured at 1.0 σ level. (B) O–π interactions formed between the sugar pucker of G8 and the nucleobase of G6. (C) Stacking interactions formed between the nucleobases of T9 and G15. (D) CD spectra analysis of the native TBA and the TGT linker mutants. Except the mutations listed in the figure, the sequences of the mutants are identical to that of TBA. TBA (native or mutant) and Pb2+ concentrations are 10 μM ad 50 μM, respectively. Structural figures were displayed using PyMOL (http://www.pymol.org/).
G8 and T9 lie on the top of G-tetrad 2, the G-tetrad formed by G1, G6, G10 and G15 (Fig. 4A, Supplementary Fig. S5B). Different from T7, the backbones and nucleobases of G8 and T9 are well defined. The sugar pucker of G8 is perpendicular to the G-tetrad; via its O4′ atom, G8 forms stable O–π interactions with the six-member ring of G6 nucleobase (Fig. 4B). In the A-form structure, the N1 atom of G8 nucleobase forms H-bond interaction with the O5′ atom of G1 (Fig. S6A). Although such interactions were observed for some TBA molecules in the B-form structure, the G8 N1 atoms of other TBA molecules interact with water (Fig. S6B). Similar to G8, the sugar pucker of T9 is also perpendicular to the G-tetrad, but it did not form stable O–π interaction. Instead, T9 forms stacking interaction with G15 (Fig. 4C).
The T7G8T9 linker also adopts loop-like conformation in the TBA-thrombin structure. However, structural superposition showed that the detailed conformations of the T7G8T9 linker is very different from those in the TBA-Pb2+ complex (Fig. S5B). May be due to its interactions with symmetry-related thrombin molecules, T9 points away from the main body of TBA in the TBA-thrombin complex. As a compensation, the nucleobase of T7 folds back and forms stacking interactions with G6. Like T7, the nucleobase of G8 also lies on the top of the G-tetrad 2, forming stacking interactions with G10. Although they could be very different in conformations, both TBA-thrombin complex and our TBA-Pb2+ complex structures indicated that the T7G8T9 linker interacts with the central G-tetrad and helps the stabilization of TBA.
TGT linker is variable for Pb2+-binding
The structural observations suggested that T7, G8, and T9 might be important for the folding of TBA, whereas they are not directly involved in Pb2+-binding. To further clarify the functional roles of the T7G8T9 linker, we synthesized several TBA single-point mutants (Table 3) and performed CD analysis (Fig. 4D). In agree with our structural analysis, substitution of T7 by other nucleotides did not inhibit Pb2+-binding by TBA. In fact, the T7A and T7G mutants might be more efficient in Pb2+-binding, suggested by the higher 312-nm peaks. Substitution of G8 by C8 (for the G8C mutant) or T8 (for the G8T mutant) has no obvious impact on Pb2+-binding; whereas A8 substitution (for the G8A mutant) could lead to stronger Pb2+-binding. When T9 was substituted by either A9 or C9 (for the T9A and the T9C mutants), no clear difference on the CD spectra was observed. In contrast to other T7G8T9 linker mutants, the Pb2+-binding affinity of T9G mutant is weaker, indicated by the lower peak near 312 nm.
Similar to native TBA, we also carried out crystallographic studies for the T7G8T9 linker mutants and successfully solved one complex structure of G8C-Pb2+ at 2.1 Å resolution (Table 1). The structure belongs to the monoclinic space group C2 and it contains five G8C-Pb2+ complexes per asymmetric unit. Four of the complexes formed two intermolecular dimers, whereas the last one formed dimer with symmetric-related molecule. Other than the A-form structure, the overall folding and orientation of the G8C dimers are more similar to dimer EF in the B-form structure (Fig. 5A,B). Due to their different cell units and space groups, packing of G8C-Pb2+ complexes is very different from the A-form and B-form structures (Fig. S1C). However, the Pb2+-coordinating geometry is almost identical in the three complex structures. C8 is well ordered in the G8C-Pb2+ complex structure (Fig. 5C). Similar to G8 in the A- and B-form structures, the sugar pucker of C8 also forms stable O–π interaction with G6 (Fig. 5D), which may lead to the similar CD spectra of G8C mutant and native TBA.
Figure 5.
G8C-Pb2+ complex structure. (A) Superposition of G8C-Pb2+ and the A-form TBA-Pb2+ structure. (B) Superposition of G8C-Pb2+ and the EF dimer of the B-form TBA-Pb2+ structure. (C) Detailed conformation and the 2Fo − Fc electron density map of C8. The maps are contoured at 1.2 σ level. (D) O–π interactions formed between the sugar pucker of C8 and the nucleobase of G6. The G8C-Pb2+ complexes are colored in orange and magenta in both (A,B). Figures were displayed using PyMOL (http://www.pymol.org/).
Discussion
In addition to Pb2+-binding DNA aptamers, several DNA enzymes (DNAzymes, such as GR5 and 8-17E) with Pb2+ binding ability and specificity have also been discovered26. GR5 and 8-17E can cleave RNA in a sequence-specific manner. Owing to their great potential in gene silencing and therapeutics, GR5 and 8-17E have been extensively studied27,28. However, due to their intrinsic difficulties in crystallization, the structural information of the Pb2+-binding DNAzymes is very limited. The only 8–17 DNAzyme structure was determined by us in 2017, which was facilitated by one protein molecule29. As revealed by the crystal structure, Pb2+-binding with 8–17 DNAzyme is actually very dynamic, indicated by the relatively low occupancy (~ 50%) of Pb2+ in the structure. Besides the enzyme strands, folding of the Pb2+-binding DNAzymes also needs RNAs. The RNA strands function as substrates and are degraded during the analysis, which makes the DNAzyme-based Pb2+-detection method expensive and difficult in preparation.
Unlike Pb2+-binding DNAzymes, the Pb2+-binding DNA aptamers are all composed of single stable DNA strands. Compared to the DNAzyme-based methods, the aptamer-based methods are much easier to handle and less expensive. TBA is one of the earliest discovered Pb2+-binding DNA aptamers. Like TBA, many other DNA sequences can also assemble into intramolecular quadruplex. Owing to their functional roles in DNA recombination, replication, and transcription, quadruplex structures have attracted significant attentions in the past30,31. However, the TBA sequence is very different from the conventional G3-L1-G3-L2-G3-L3-G3 sequence units for identifying potential G-quadruplex structures in bioinformatic studies32. Most likely, the unique sequence provides TBA with weak K+ binding affinity. The Pb2+-binding affinity of TBA is very high and the Pb2+-complexed TBA-quadruplex is very stable, which allows us to obtain the crystals and solve three TBA structures at atomic resolution, including two native and one G8C mutant structures. In addition to the detailed coordination, these structures also revealed the detailed basis for the tight Pb2+ binding by TBA, such as tilting of the G-tetrad (Fig. 3D).
DNAs are very stable biomolecules. Although not as common as duplexes, DNA quadruplexes also have great potential in nanodevice development. Previous, the Pb2+-binding aptamers PS2.M, PW17 and T30695 have been utilized to create K+–Pb2+-switched DNA gate13. The Pb2+-stabilized TBA structure is more stable than that stabilized by K+. Besides Pb2+ binding, TBA can also function as potential inhibitor for HIV replication. Therefore, TBA received much more attentions than other counterpart aptamers33. Whereas, the possibility to develop TBA-based nanodevices has not been fully explored. During this study, we noticed that the 5′- and 3′-OH groups are not involved in Pb2+-binding by TBA (Fig. S6). Theoretically, florescent groups or additional DNA sequences could be attached to the 5′- or 3′-end of TBA, which will enable the development of novel types of Pb2+-sensors or DNA nanodevices.
Guided by the structural observations, we did systematic mutation. CD spectra analysis confirmed that all the TT linker or the TGT linker mutants could bind with Pb2+. Compared to native TBA, some mutants might have higher Pb2+-binding abilities, such as T4/13A, T4/13G, T7A, T7G and G8A mutants. These results significantly expanded the sequence for the Pb2+-binding aptamers. Like Pb2+-binding aptamers, many other aptamers have also been reported. Aptamer identification usually involves multiple cycles of in vitro selection and DNA sequencing, which are costy and time-consuming. Our study suggested that structure-based design may be an ideal alternative method to resolve these problems and contribute to the development of new aptamers.
Methods
Chemicals
All chemicals and buffers used are of analytical reagent grade. Sodium chloride, lead chloride, lead nitrate, Tris acetate, and acetic acid were purchased from Sigma-Aldrich. Spermine tetra-HCl, sodium cacodylate, (±)-2-Methyl-2,4-pentanediol (MPD) were purchased from Hampton Research. Native TBA and TBA mutants were purchased from Integrated DNA Technologies (IDT), Shanghai Generay company or synthesized and purified in house.
Crystallization of TBA-Pb2+ complexes
Lead chloride, native TBA (5′-GGTTGGTGTGGTTGG-3′, Fig. 1A) and TBA mutants were all dissolved in ultra-pure water. Mix native TBA (or mutants) and lead chloride with a final concentration of 1.0 mM and 2.0 mM, respectively. Incubate the sample at room temperature for 30 min prior to crystallization. The initial crystallization conditions were identified using the Gryphon crystallization robot system from Art Robbins Instrument and crystallization kits from Hampton Research. The sitting-drop vapor diffusion method was used during the initial screening at 16 °C, whereas all the crystallization optimization procedure were performed at 18 °C using the hanging-drop vapor diffusion method.
Well solutions are composed of 30% v/v MPD for all the crystals. The drops contain 1.6 μl TBA-Pb2+ sample and 0.4 μl crystallization buffer (10% v/v MPD, 40 mM sodium cacodylate pH 7.0, 12 mM spermine tetra-HCl, and 80 mM sodium chloride) for the A-form crystals, whereas they are composed of equal volume (1.0 μl) of sample and crystallization buffer for the B-form crystals. The drops are composed of 0.7 μl sample and 0.7 μl crystallization buffer (10% v/v MPD, 40 mM sodium cacodylate pH 7.0, 28 mM spermine tetra-HCl, and 80 mM sodium chloride) for the G8C-Pb2+ complex crystals. Growth of the crystals are slow. They normally appeared 2 weeks after the crystallization and reached the full sizes in another 2 weeks.
Diffraction data collection
All the crystals were cryoprotected using 30% v/v MPD and flash-frozen by quickly dipping into liquid nitrogen. The X-ray diffraction data were collected on beamline BL17U1, BL18U1, and BL19U1 at Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China) at cryogenic temperature, maintained with cryogenic system. One single crystal was used for each structure; data processing was carried out using the HKL2000 or HKL3000 programs34. The data collection and processing statistics were summarized in Table 1.
Structure determination and refinement
The A-form TBA-Pb2+ complex structure was solved by the SAD (single anomalous diffraction) method35 using the anomalous signals of Pb2+. The Autosol program embedded in the phenix suite36 could identify two Pb2+ ions with figure-of-merit (FAM) value of 0.741. The resulting electron density maps were used as guide for manually model building by Coot program37. The model were then refined by the Refmac5 program embedded in the CCP4i suite38. 5% randomly selected data was set aside for free R-factor cross validation calculations during the refinement. Both the B-form TBA-Pb2+ complex structure and the G8C-Pb2+ complex structure were solved by the molecular replacement method using the phaser program39, the A-form structure was used as the search model. The final refinement of the three structures were all done using the phenix.refine program of phenix. The detailed refinement statistics were summarized in Table 1.
Circular dichroism (CD) spectroscopy
All native and mutant TBAs (Table 3) utilized in the circular dichroism (CD) spectra analysis were synthesized by solid phase synthesizer and purified by High Performance Liquid Chromatography (HPLC) in the laboratory. All samples were prepared by mixing TBA (or mutants) and lead nitrate that were dissolved in 10 mM Tris acetate buffer (pH 7.2). The final TBA and lead nitrate concentrations are 10 μM and 50 μM, respectively. All solutions were heated to 85 °C for 3 min, then cooled slowly to room temperature and stored at 4 °C for overnight. CD studies were carried out in utilizing a Jasco-815 CD spectrometer in a quartz cell with a 10-mm path length. CD spectra were collected from 360 to 260 nm and with a scanning speed of 100 nm/min. The bandwidth was set to 1.0 nm, and the digital integration time was 1.0 s. All CD spectra were base line corrected against the blank buffer.
Supplementary Information
Acknowledgements
We thank the staff of the BL17U1 beamline at the Shanghai Synchrotron Radiation Facility (SSRF) and the staff of BL18U1, BL19U1 beamlines of the National Facility for Protein Science in Shanghai for help during data collection. This work is supported by the National Natural Science Foundation of China (32171197 and 31870721) and US National Science Foundation (NSF CHE-1845486).
Author contributions
J.S. and J.H.G. conceived the principal idea and designed the experiments. H.H.L. and Y.Q.G. performed the crystallographic studies. J.M. and F.S.S. performed the CD analysis. X.C., Y.Y.L., Z.W.S., Y.X.Z. and Q.Y.S. helped with X-ray diffraction data collection. H.H.L., J.S. and J.H.G. wrote the manuscript. All the authors discussed the results and commented on the manuscript.
Code availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB, www.pdb.org) under accession ID codes 7D31, 7D32, and 7D33 for the A-form TBA-Pb2+ complex, the B-form TBA-Pb2+ complex and the G8C-Pb2+ complex, respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hehua Liu, Yanqing Gao and Johnsi Mathivanan.
Contributor Information
Jia Sheng, Email: jsheng@albany.edu.
Jianhua Gan, Email: ganjhh@fudan.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-04243-2.
References
- 1.Bridges CC, Zalups RK. Molecular and ionic mimicry and the transport of toxic metals. Toxicol. Appl. Pharmacol. 2005;204:274–308. doi: 10.1016/j.taap.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yassa HA. Autism: A form of lead and mercury toxicity. Environ. Toxicol. Pharmacol. 2014;38:1016–1024. doi: 10.1016/j.etap.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Burns JS, et al. Peripubertal blood lead levels and growth among Russian boys. Environ. Int. 2017;106:53–59. doi: 10.1016/j.envint.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah F, et al. Exposures of lead to adolescent workers in battery recycling workshops and surrounding communities. J. Expo. Sci. Environ. Epidemiol. 2012;22:649–653. doi: 10.1038/jes.2012.64. [DOI] [PubMed] [Google Scholar]
- 5.Mazumdar I, Goswami K, Ali MS. Status of serum calcium, vitamin D and parathyroid hormone and hematological indices among lead exposed jewelry workers in Dhaka, Bangladesh. Indian J. Clin. Biochem. 2017;32:110–116. doi: 10.1007/s12291-016-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qasim SF, Baloch M. Lead toxicity in battery workers. J. Coll. Phys. Surg. Pak. 2014;24(Suppl 3):S284–S286. [PubMed] [Google Scholar]
- 7.Basit S, Karim N, Munshi AB. Occupational lead toxicity in battery workers. Pak. J. Med. Sci. 2015;31:775–780. doi: 10.12669/pjms.314.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurtado CM, Gutierrez M, Echeverry J. Clinical manifestations of lead levels in children exposed to automobile battery recycling processes in Soacha and Bogota, D.C. Biomedica. 2008;28:116–125. doi: 10.7705/biomedica.v28i1.114. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez-Ortega N, Caballero-Gallardo K, Olivero-Verbel J. Low blood lead levels impair intellectual and hematological function in children from Cartagena, Caribbean coast of Colombia. J. Trace Elem. Med. Biol. 2017;44:233–240. doi: 10.1016/j.jtemb.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Gallacher J, Harris L, Elwood PC. Lead toxicity from water. Nature. 1983;305:280. doi: 10.1038/305280a0. [DOI] [PubMed] [Google Scholar]
- 11.Bellinger DC. Lead contamination in flint—An abject failure to protect public health. N. Engl. J. Med. 2016;374:1101–1103. doi: 10.1056/NEJMp1601013. [DOI] [PubMed] [Google Scholar]
- 12.Hanna-Attisha M, LaChance J, Sadler RC, Champney Schnepp A. Elevated blood lead levels in children associated with the flint drinking water crisis: A spatial analysis of risk and public health response. Am. J. Public Health. 2016;106:283. doi: 10.2105/AJPH.2015.303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, Wang E, Dong S. Potassium-lead-switched G-quadruplexes: A new class of DNA logic gates. J. Am. Chem. Soc. 2009;131:15082–15083. doi: 10.1021/ja9051075. [DOI] [PubMed] [Google Scholar]
- 14.Majhi PR, Shafer RH. Characterization of an unusual folding pattern in a catalytically active guanine quadruplex structure. Biopolymers. 2006;82:558–569. doi: 10.1002/bip.20507. [DOI] [PubMed] [Google Scholar]
- 15.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smirnov I, Shafer RH. Lead is unusually effective in sequence-specific folding of DNA. J. Mol. Biol. 2000;296:1–5. doi: 10.1006/jmbi.1999.3441. [DOI] [PubMed] [Google Scholar]
- 18.Zhang D, et al. A sensitive fluorescence anisotropy method for detection of lead (II) ion by a G-quadruplex-inducible DNA aptamer. Anal. Chim. Acta. 2014;812:161–167. doi: 10.1016/j.aca.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 19.Zhang B, Wei C. Highly sensitive and selective detection of Pb(2+) using a turn-on fluorescent aptamer DNA silver nanoclusters sensor. Talanta. 2018;182:125–130. doi: 10.1016/j.talanta.2018.01.061. [DOI] [PubMed] [Google Scholar]
- 20.Khoshbin Z, Housaindokht MR, Izadyar M, Verdian A, Bozorgmehr MR. A simple paper-based aptasensor for ultrasensitive detection of lead (II) ion. Anal. Chim. Acta. 2019;1071:70–77. doi: 10.1016/j.aca.2019.04.049. [DOI] [PubMed] [Google Scholar]
- 21.Musumeci D, Montesarchio D. Polyvalent nucleic acid aptamers and modulation of their activity: A focus on the thrombin binding aptamer. Pharmacol. Ther. 2012;136:202–215. doi: 10.1016/j.pharmthera.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 23.Roxo C, Kotkowiak W, Pasternak A. G-quadruplex-forming aptamers-characteristics, applications, and perspectives. Molecules. 2019 doi: 10.3390/molecules24203781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo Krauss I, et al. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012;40:8119–8128. doi: 10.1093/nar/gks512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell NH, Smith DL, Reszka AP, Neidle S, O'Hagan D. Fluorine in medicinal chemistry: Beta-fluorination of peripheral pyrrolidines attached to acridine ligands affects their interactions with G-quadruplex DNA. Org. Biomol. Chem. 2011;9:1328–1331. doi: 10.1039/c0ob00886a. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XB, Kong RM, Lu Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekesan S, York DM. Dynamical ensemble of the active state and transition state mimic for the RNA-cleaving 8–17 DNAzyme in solution. Nucleic Acids Res. 2019;47:10282–10295. doi: 10.1093/nar/gkz773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HK, et al. Metal-dependent global folding and activity of the 8–17 DNAzyme studied by fluorescence resonance energy transfer. J. Am. Chem. Soc. 2007;129:6896–6902. doi: 10.1021/ja0712625. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, et al. Crystal structure of an RNA-cleaving DNAzyme. Nat. Commun. 2017;8:2006. doi: 10.1038/s41467-017-02203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopes J, et al. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, et al. Kinetics and mechanism of conformational changes in a G-quadruplex of thrombin-binding aptamer induced by Pb2+ J. Phys. Chem. B. 2011;115:13051–13056. doi: 10.1021/jp2074489. [DOI] [PubMed] [Google Scholar]
- 34.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 35.Giacovazzo C, Siliqi D. Phasing via SAD/MAD data: The method of the joint probability distribution functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:73–82. doi: 10.1107/s0907444903022406. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ. Solving structures of protein complexes by molecular replacement with phaser. Acta Crystallogr. D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB, www.pdb.org) under accession ID codes 7D31, 7D32, and 7D33 for the A-form TBA-Pb2+ complex, the B-form TBA-Pb2+ complex and the G8C-Pb2+ complex, respectively.