Abstract
Amplified fragment length polymorphism (AFLP) was investigated for the differentiation of Corynebacterium diphtheriae isolates. Analysis using Taxotron revealed 10 distinct AFLP profiles among 57 isolates. Strains with ribotype patterns D1, D4, and D12 could not be distinguished; however, the technique discriminated isolates of ribotype patterns D3, D6, and D7 further. AFLP was rapid, fairly inexpensive, and reproducible and could be used as an alternative to ribotyping.
The diphtheria epidemic, which began in the Russian Federation in 1990, appears to be declining due to the implementation of vigorous control measures (4). In addition to the European region, countries in Africa, the Eastern Mediterranean, South America, Southeast Asia, and the Indian subcontinent also have evidence of substantial circulation of toxigenic Corynebacterium diphtheriae as manifested by outbreaks or large numbers of reported cases (6, 14). This situation poses a threat to individuals within and outside those countries who are susceptible to diphtheria (5, 7, 9) because of their low immunity levels. These reasons highlight the importance of rapid and reproducible molecular typing techniques for epidemiological characterization and monitoring of C. diphtheriae globally.
Various molecular methods to characterize strains of C. diphtheriae, such as ribotyping (3), pulsed-field gel electrophoresis (PFGE) (3), multilocus enzyme electrophoresis (10), and random amplification of polymorphic DNA (RAPD), have been described by several investigators. The current method agreed on by all members of the European Laboratory Working Group on Diphtheria (ELWGD) for typing C. diphtheriae is ribotyping (11). The technique is time-consuming, requires specialized equipment and technical expertise, and, therefore, cannot be performed in all laboratories. In contrast, the PCR-based method, RAPD analysis, which we recently described (2) is rapid; the technique requires a high degree of standardization to obtain reproducible results.
In this study, we report the use of a rapid PCR-based technique, amplified fragment length polymorphism (AFLP), for typing C. diphtheriae. The method is based on the selective PCR amplification of genomic restriction fragments of the whole genome (13) and has been shown to be rapid, reproducible, and highly discriminatory (1, 8). The AFLP method used was essentially that described by Valsangiacomo et al. (12). Ours is a simplified version of that technique, utilizing a one-step digestion-ligation reaction with one enzyme, and the PCR is performed using a single primer. This simplified version provides a small number of amplified bands, which can be separated by conventional agarose gel electrophoresis and visualized by staining in ethidium bromide.
A total of 57 C. diphtheriae isolates of nine distinct ribotypes (D1, D2, D3, D4, D5, D6, D7, D11, and D12) were analyzed (the previous nomenclature described by De Zoysa et al. in 1995 [3] has now been revised, and the prefixes “G” and “M” have been provisionally replaced by the prefix “D” for “diphtheria”). The isolates were from Russia, Finland, Estonia, Uzbekistan, Germany, Turkmenistan, Kyrgyzstan, Kazakhstan, Sweden, and Romania (Table 1). Four AFLP PstI primers (PstI-C, PstI-G, PstI-A, and PstI-T) were screened for their suitability to generate clear, definitive, and reproducible profiles which permitted good discrimination. Each primer was identical, 5′-GACTGCGTACATGCAGS-3′, except for the selective 3′-terminal base S, representing C, G, A, or T.
TABLE 1.
Summary of the 57 C. diphtheriae isolates analyzed by AFLP
| Ribotypea | No. of isolates | Country of isolation | Biotype | Toxigenicity | AFLP type |
|---|---|---|---|---|---|
| D1 | 18 | Russia, Finland, Estonia, Uzbekistan, Germany, Kazakhstan, Turkmenistan, Kyrgyzstan | Gravis | + | AP1 |
| D2 | 1 | Russia | Gravis | + | AP2 |
| D3 | 1 | Russia | Gravis | + | AP3 |
| D3 | 1 | Russia | Gravis | + | AP4 |
| D4 | 14 | Russia, Kazakhstan, Germany | Gravis | + | AP1 |
| D4 | 1 | Estonia | Gravis | − | AP1 |
| D5 | 1 | Russia | Gravis | + | AP5 |
| D6 | 1 | Russia | Gravis | + | AP6 |
| D6 | 2 | Russia | Gravis | + | AP7 |
| D7 | 6 | Russia, Kyrghystan | Mitis | + | AP8 |
| D7 | 2 | Russia | Mitis | + | AP9 |
| D11 | 8 | Russia, Sweden, Romania, Germany | Gravis | − | AP10 |
| D12 | 1 | Russia | Gravis | − | AP1 |
The previous nomenclature described by De Zoysa et al. in 1995 (3) has now been revised, and the previously used prefixes “G” and “M” have been replaced by the prefix “D” for “diphtheria” (provisional United Kingdom nomenclature).
The restriction-ligation reactions were performed as described previously (12). Briefly, the reaction was performed at 37°C for 3 h in a total volume of 20 μl. The reaction mixture consisted of 1.5 μg of genomic DNA, 0.2 μg of each adapter oligonucleotide (LG1, 5′-CTCGTAGACTGCGTACATGCA-3′, and LG2, 5′-TGTACGCAGTCTAC-3′ [Bioline]), 20 U of PstI (Boehringer), 1 U of T4 DNA ligase (Boehringer), and ligase buffer (1×, comprising 66 mM Tris [pH 7.5], 5 mM magnesium chloride, 1 mM dithiothreitol, and 1 mM ATP). The tagged fragments were precipitated using 7.5 M ammonium acetate and absolute ethanol, and the DNA was resuspended in 100 μl of TE buffer (10 mM Tris–0.5 mM EDTA [pH 8.0]) and diluted 1:100 for use.
The PCR was performed using PCR beads (Pharmacia Biotech). Each bead comprised 1.5 U of Taq polymerase, buffer (50 mM KCl–1.5 mM MgCl2 [pH 9.0]), 200 μM each deoxynucleoside triphosphate and stabilizers including bovine serum albumin. Reaction mixtures were prepared by adding 5 μl of diluted DNA (approximately 1 ng), 75 ng of primer AFLPPstI-G (5′-GACTGCGTACATGCAGG) (Bioline), and 1 mM MgCl2 (total MgCl2 concentration, 2.5 mM). (The AFLP primer PstI-G was chosen for AFLP typing because it produced clear, definitive, and reproducible AFLP fingerprints.) The reaction mixtures were overlaid with mineral oil (Sigma) and cycled through the following temperature profile: 94°C for 1 min, 60°C for 1 min, and 72°C for 2.5 min. Thermal cycling was performed on a Hybaid Omnigene thermal cycler, and amplified products were electrophoresed at 110 V for 6 h on 1.5% (wt/vol) agarose gels (Ultrapure; Life Technologies) in TBE buffer (0.089 M Tris–0.089 M boric acid–0.002 M EDTA [pH 8.0]). The bands were visualized by staining with ethidium bromide.
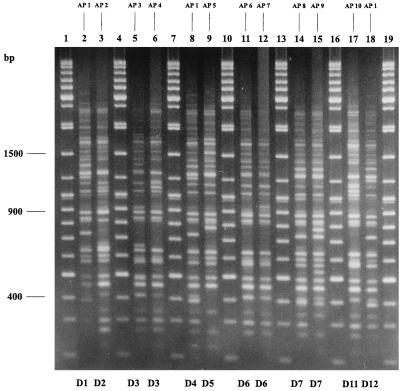
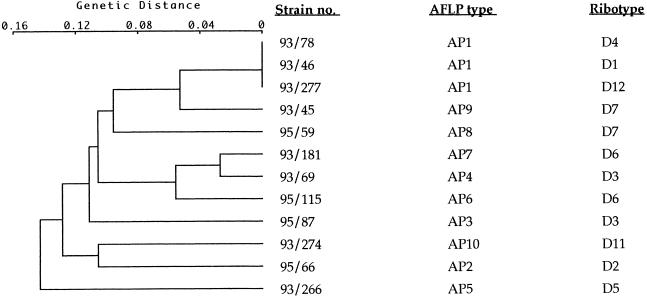
Approximately 25 to 33 AFLP fragments were generated when DNA was amplified with primer PstI-G. Fragment sizes ranged from 200 to 3,000 bp; fragments larger than 1,500 bp and fragments of less than 400 bp were excluded from the computer analysis of the gels due to inadequate resolution in these size ranges. Analysis of the gels using the Taxotron software package revealed 10 distinct AFLP profiles (designated AP1 to AP10) among the 57 C. diphtheriae isolates analyzed (Table 1; Fig. 1). Ribotype D1 (the predominant epidemic ribotype in the former Soviet Union) was not discriminated further by AFLP. Strains belonging to ribotype D3 (two strains) were distinguished further. Strains of ribotype D6 (three strains) were also further distinguished by AFLP. One D6 strain produced the AFLP type designated AP6, and the other two produced the pattern designated AP7. The eight strains belonging to ribotype D7 were also distinguished further by this technique. Six D7 strains produced the AFLP type designated AP8, and the remaining two D7 strains produced the pattern designated AP9. AFLP profiles of strains belonging to ribotypes D1, D4, and D12 were indistinguishable. The dendrogram (Fig. 2) represents the genetic relationships between the AFLP profiles produced by primer PstI-G, as determined using Taxotron.
FIG. 1.
AFLP profiles of C. diphtheriae produced by primer PstI-G. Lanes 1, 4, 7, 10, 13, 16, and 19, 100-bp molecular weight standard (with sizes indicated on the left). The remaining lanes show AFLP profiles of the following strains: C93/46 (lane 2), C95/66 (lane 3), C95/87 (lane 5), C93/69 (lane 6), C93/78 (lane 8), C93/266 (lane 9), C95/115 (lane 11), C93/181 (lane 12), C95/59 (lane 14), C93/45 (lane 15), C93/274 (lane 17), and C93/277 (lane 18). AFLP types are given above each lane, and ribotype designations are given at the bottom of each lane.
FIG. 2.
Cluster analysis of the AFLP profiles of the C. diphtheriae strains shown in Fig. 1. The strain numbers, AFLP types, and ribotype designations are given on the right. Analyses were performed with the Taxotron software package (Institut Pasteur). Patterns were clustered by the single-linkage method with a fixed tolerance of 4%. A genetic distance of zero is equal to 100% similarity.
The reproducibility of the technique was examined by performing duplicate AFLP runs for each isolate with two separate DNA extractions. Also, DNA from a single C. diphtheriae isolate was amplified in two different thermal cyclers (Hybaid Omnigene and Hybaid TouchDown). Under all these different conditions, the fragments for each AFLP profile were identical. However, variations in the intensities of some of the bands were observed with different PCR runs.
In conclusion, the 57 C. diphtheriae isolates of 9 distinct ribotypes were distinguished into 10 distinct AFLP profiles (AP1 to AP10). AFLP was not able to further discriminate the predominant epidemic ribotype in the former Soviet Union. However, the technique further discriminated strains belonging to ribotypes D3, D6, and D7. The two strains belonging to ribotype D3 were distinguished into two AFLP profiles. Profiles AP3 and AP4 have a three-band difference. Three strains belonging to ribotype D6 were analyzed, of which two produced the AP6 pattern and the other produced the AP7 pattern; these profiles have a single-band difference. Eight strains belonging to ribotype D7 were analyzed, of which six produced the AP8 profile and the remaining two produced the AP9 profile. Profiles AP8 and AP9 also have a single-band difference. Like PFGE (3), AFLP was not able to distinguish between strains belonging to ribotypes D1, D4, and D12. We have reported previously that ribotyping (3), PFGE (3), and RAPD (2) have shown a potential clonal relationship between strains of ribotypes D1, D4, and D12. The AFLP results obtained have provided further evidence for a potential clonal relationship between isolates of ribotypes D1, D4, and D12.
The AFLP technique appears to have several advantages in comparison to other molecular typing methods. It is easy to perform, rapid, discriminatory, and most importantly, highly reproducible. The technique analyzes the whole genome, requires only a small amount of DNA, and requires no prior sequence information about the target DNA. This simplified version also avoids the use of radioactive material for visualization of AFLP patterns as described in the original method.
AFLP appears to be an excellent tool for rapid and definitive analysis of outbreaks. The technique is, in many respects, easier and faster to perform than ribotyping (the current “gold standard” for typing of C. diphtheriae), as it allows the detection of restriction fragment length polymorphisms directly on agarose gels, eliminating the need for vacuum blotting and probe hybridizations. AFLP is also cheaper to perform than ribotyping (AFLP analysis on 57 isolates costs approximately $126.00, whereas ribotyping costs approximately $173.00). The method is also adaptable and therefore can be used as an alternative to ribotyping, especially in laboratories that have limited funding and equipment. AFLP has the potential to replace ribotyping as the “gold standard” within the ELWGD.
REFERENCES
- 1.Boumedine K S, Rodolakis A. AFLP allows the identification of genomic markers of ruminant Chlamydia psittaci strains useful for typing and epidemiological studies. Res Microbiol. 1998;149:735–744. doi: 10.1016/s0923-2508(99)80020-5. [DOI] [PubMed] [Google Scholar]
- 2.De Zoysa A S, Efstratiou A. PCR typing of Corynebacterium diphtheriae by random amplification of polymorphic DNA. J Med Microbiol. 1999;48:335–340. doi: 10.1099/00222615-48-4-335. [DOI] [PubMed] [Google Scholar]
- 3.De Zoysa A, Efstratiou A, George R C, Jahkola M, Vuopio-Varkila J, Deshevoi S, Tseneva G, Rikushin Y. Molecular epidemiology of Corynebacterium diphtheriae from northwestern Russia and surrounding countries studied by using ribotyping and pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:1080–1083. doi: 10.1128/jcm.33.5.1080-1083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dittmann S, Wharton M, Vitek C, Ciotti M, Galazka A, Guichard S, Hardy I, Kartoglu U, Koyama S, Kreysler J, Martin B, Mercer D, Ronne T, Roure C, Steinglass R, Strebel P. Successful control of epidemic diphtheria in the states of the former Union of Soviet Socialist Republics: lessons learned. J Infect Dis. 2000;181:S10–S22. doi: 10.1086/315534. [DOI] [PubMed] [Google Scholar]
- 5.Galazka A. The immunological basis for immunisation: diphtheria. Module 2, Expanded Programme on Immunisation. WHO/EP/GEN/93.12. Geneva, Switzerland: World Health Organization; 1993. [Google Scholar]
- 6.Galazka A M, Robertson S E. Diphtheria: changing patterns in the developing world and the industrialized world. Eur J Epidemiol. 1995;11:107–117. doi: 10.1007/BF01719955. [DOI] [PubMed] [Google Scholar]
- 7.Galazka A M, Robertson S E. Immunization against diphtheria with special emphasis on immunization of adults. Vaccine. 1996;14:845–857. doi: 10.1016/0264-410x(96)00021-7. [DOI] [PubMed] [Google Scholar]
- 8.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1993. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 9.Maple P A, Efstratiou A, George R C, Andrews N J, Sesardic D. Diphtheria immunity in UK blood donors. Lancet. 1995;345:963–965. doi: 10.1016/s0140-6736(95)90705-x. [DOI] [PubMed] [Google Scholar]
- 10.Popovic T, Kombarova S Y, Reeves M W, Nakao H, Mazurova I K, Wharton M, Wachsmuth I K, Wenger J D. Molecular epidemiology of diphtheria in Russia, 1985–1994. J Infect Dis. 1996;174:1064–1072. doi: 10.1093/infdis/174.5.1064. [DOI] [PubMed] [Google Scholar]
- 11.Popovic T, Mazurova I K, Efstratiou A, Vuopio-Varkila J, Reeves M W, De Zoysa A, Glushkevich T, Grimont P. Molecular epidemiology of diphtheria. J Infect Dis. 2000;181:S168–S177. doi: 10.1086/315556. [DOI] [PubMed] [Google Scholar]
- 12.Valsangiacomo C, Baggi F, Gaia V, Balmelli T, Peduzzi R, Piffaretti J. Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J Clin Microbiol. 1995;33:1716–1719. doi: 10.1128/jcm.33.7.1716-1719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. WHO Vaccine Preventable Diseases Monitoring System. Global summary, November 1999. WHO/V&B/99.17. Geneva, Switzerland: World Health Organization; 1999. [Google Scholar]