Abstract
The quantification of supramolecular host–guest interactions is important for finely modulating supramolecular systems. Previously, most host–guest interactions quantified using force spectroscopic techniques have been reported in force units. However, accurately evaluating the adhesion energies of host–guest pairs remains challenging. Herein, using a surface forces apparatus, we directly quantify the interaction energies between cyclodextrin (CD)-modified surfaces and ditopic adamantane (DAd) molecules in water as a function of the DAd concentration and the CD cavity size. The adhesion energy of the β-CD–DAd complex drastically increased with increasing DAd concentration and reached saturation. Moreover, the molecular adhesion energy of a single host–guest inclusion complex was evaluated to be ~9.51 kBT. This approach has potential for quantifying fundamental information toward furthering the understanding of supramolecular chemistry and its applications, such as molecular actuators, underwater adhesives, and biosensors, which require precise tuning of specific host–guest interactions.
Subject terms: Molecular self-assembly, Self-assembly, Organic molecules in materials science
Quantification of supramolecular host–guest interactions is important for modulating supramolecular systems but accurately evaluating the adhesion energies of host–guest pairs remains challenging. Here, the authors directly quantify the interaction energies between cyclodextrin modified surfaces and ditopic adamantane using a surface force apparatus.
Introduction
Biological systems are driven by various physical interactions, called noncovalent interactions, including H-bonding, van der Waals forces, electrostatic interactions, hydrophobic interactions, and metal–ligand coordination1. Complex combinations of these interactions lead to specific binding interactions, more commonly referred to as ligand–receptor, complementary, lock-and-key, or host–guest interactions2,3. The reversible nature of these interactions has inspired the development of supramolecular chemistry toward understanding biological processes such as DNA replication/transcription4–6 and enzyme activity7, which involve repeated assembly and disassembly through the highly selective recognition of specific target molecules. In particular, within supramolecular chemistry, the host–guest interaction is a crucial component for developing fundamental molecular recognition principles (e.g., the lock and key model)8,9. Thus, host–guest interactions have been widely investigated, from understanding molecular functions to biological applications such as hydrogels10,11, bioadhesives12,13, sensors14,15, and drug delivery systems16. Recent studies have reported that desirable properties in host–guest materials (e.g., self-healing, adhesion, and stability) can be affected by the binding affinities of host–guest inclusion complexes17–19.
The binding affinities of host–guest interactions have been commonly investigated using thermodynamic parameters, including the Gibbs free energy (ΔG) and the association constant (Ka), as determined using nuclear magnetic resonance (NMR)20, isothermal titration calorimetry (ITC)21, and surface plasmon resonance (SPR)22. The ΔG and Ka values of various host–guest inclusion complexes under thermodynamic equilibrium conditions have been used as relative indicators to develop new host–guest-interaction-based materials. However, the correlation between these thermodynamic parameters and the mechanical properties of host–guest materials remains ambiguous. Thus, for furthering the understanding of host–guest interactions and expanding practical applications, measurements of the direct interaction forces and energies of host–guest inclusion complexes are essential.
The interaction forces of host–guest inclusion complexes have been investigated using single-molecule force spectroscopy (SMFS) (e.g., magnetic/optical tweezers and atomic force microscopy (AFM))23, which can provide the individual rupture forces of host–guest complexes at the single-molecule level24–28. In particular, the Vancso group studied the individual rupture forces of host–guest inclusion complexes using a β-cyclodextrin (β-CD)-modified surface as the host and a guest-immobilized AFM tip. They reported that the individual rupture force (55 pN for ferrocene–β-CD) was independent of the loading rate, the spacer chain length, and the host–guest complex concentration, which showed that the designed system was under thermodynamic equilibrium29,30. Furthermore, they found that the individual rupture forces (39–102 pN) measured for several types of guest molecule on the β-CD-modified surface followed the same trend as the ΔG values determined by ITC or SPR31. Similarly, Blass et al. investigated molecular kinetics and cooperative effects for host–guest complexes in terms of the rupture and friction forces measured by AFM32–34. In addition, they designed an energy potential model to estimate ΔG values from the measured forces33. However, the absolute ΔG values derived from the measured forces were significantly different from those determined by ITC or SPR owing to technical limitations such as inaccurate rupture distances, multiple interactions, and the absence of a precise and systematic model31,33. Thus, a different technical approach is needed for directly and accurately converting measured forces into the energies under thermodynamic equilibrium conditions.
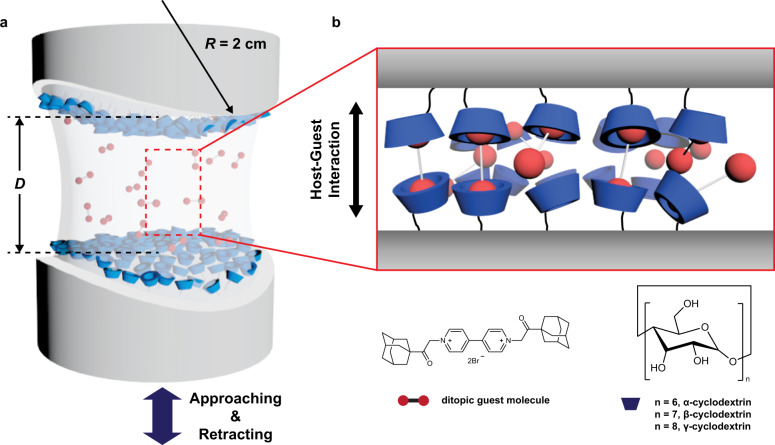
Herein, we measured the host–guest interaction forces between β-CD and adamantane (Ad), which were selected as a representative host–guest pair with an association constant of ~104 M−1 21, using a surface forces apparatus (SFA). The SFA has been widely used to measure the absolute distances and interaction forces between macroscopic surfaces35–37. Compared with other SMFS techniques, the SFA has lower force resolution (~10 nN); however, in terms of accuracy and resolution for interaction energies, the SFA outperforms other force spectroscopic techniques. As the SFA utilizes a molecularly smooth surface (RMS roughness: ~0.42 Å)38, roughness effects can be ignored. Moreover, the SFA measures interaction forces between macroscopic surfaces with a curvature of ~2 cm, which is much larger than the working distance of the SFA (D < 1 μm) (Fig. 1a). As a result, the Derjaguin approximation and the Johnson–Kendall–Roberts model, which are models for converting a force between two curved surfaces to an energy per unit area, are extremely accurate39. Furthermore, the SFA measures the absolute distance between substrates rather than the relative displacement based on a (steric) hard wall; thus, the exact thickness of molecules at a specific applied force can be evaluated. To measure the adhesion forces between CD and Ad using the SFA, symmetric CD-modified surfaces and a ditopic adamantane (DAd) guest molecule were designed, which are expected to form CD–DAd–CD inclusion complexes in water (Fig. 1b). The formation of host–guest inclusion complexes was confirmed using 2D ROESY NMR measurements. Moreover, the surface density of grafted CD molecules was determined using a quartz crystal microbalance with dissipation monitoring (QCM-D), and the molecular interaction energy per host–guest inclusion complex was calculated.
Fig. 1. Experimental scheme.
a Surface forces apparatus (SFA) setup for interaction force measurements. b Graphical illustration showing the host–guest pairs between symmetric host (CD)-modified surfaces and ditopic guest molecules (DAd).
Results
Synthesis of DAd as a ditopic guest molecule
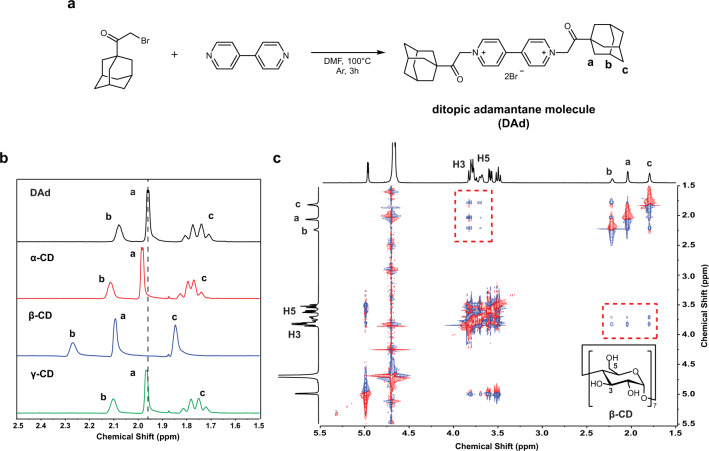
To measure the host–guest interaction forces, a ditopic guest molecule was adopted to provide versatility in controlling the solution conditions between symmetric host surfaces (Fig. 1b). As a ditopic guest molecule, DAd was synthesized from 4,4′-bipyridine and 1-adamantyl bromomethyl ketone (Fig. 2a). The pyridinium salt structure of DAd ensured sufficient solubility of the hydrophobic guest molecules in water. Additionally, a short and stiff connector was required between guest moieties to achieve accurate 1:1 host–guest interactions between the opposing CD-modified surfaces. This connector prevented DAd from forming inclusion complexes with CDs within the same surface (Supplementary Fig. 1 and Supplementary Note 1). In our work, a pyridinium connector was utilized to decrease unintended intrasurface host–guest interactions40. The adamantly group was used as a guest moiety owing to its strong binding affinity with β-CD (Ka ~104 M−1)21. The characterization of the synthesized DAd is shown in Supplementary Fig. 2.
Fig. 2. Formation of the host–guest inclusion complex in water.
a Scheme of the synthesis of DAd. b 1H NMR spectra of the 1:2 mixtures of 1.0 mM DAd with α-, β-, and γ-CD in D2O. c 2D ROESY NMR spectrum of the 1:2 mixture of 1.0 mM DAd with β-CD in D2O.
The CD–DAd host–guest interaction was confirmed to be dependent on the size the CD cavity using NMR spectroscopy. Figure 2b shows the 1H NMR spectra of mixtures of DAd with α-, β-, and γ-CD. The downfield shifts observed for the methylene and methine protons of the adamantly group in the mixture of DAd with β-CD indicated the formation of a complex between the adamantly group and β-CD. The ROESY NMR spectrum of the mixture of DAd with β-CD showed correlation signals for the methylene and methine protons with H-3 and H-5 in β-CD (Fig. 2c). These results reveal that the adamantyl groups of DAd can strongly penetrate the cavity of β-CD but not those of α- and γ-CD owing to size compatibility.
Characterization of CD surfaces
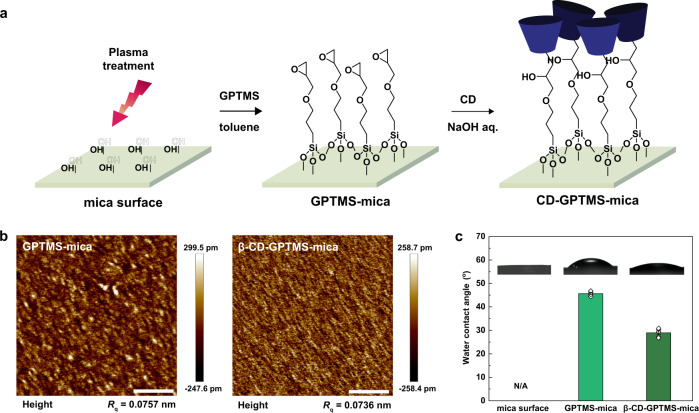
To prepare the CD-modified surfaces, a molecularly smooth mica surface was functionalized with (3-glycidyloxypropyl)trimethoxysilane (GPTMS), followed by deposition of a CD layer (Fig. 3a). Topographic images of the GPTMS- and CD–GPTMS-modified surfaces were acquired using AFM tapping mode (Fig. 3b and Supplementary Fig. 3a). AFM topography analysis gave an RMS roughness of 0.074–0.092 nm for both modified surfaces, indicating the formation of homogeneous and smooth surfaces without any aggregation.
Fig. 3. Preparation and characterization of CD-modified surfaces.
a Schematic illustration of the procedure for preparing CD-modified mica surfaces. b Topographic AFM images (scale bar, 200 nm). c WCA measurements (error bars represent the standard error of the mean (s.e.m.), where n = 5). Source data are provided as a Source Data file.
In addition, water contact angle (WCA) measurements were performed to confirm the successful modification of the surfaces with GPTMS and CD. The GPTMS-modified mica surface showed a higher WCA (45.7°) than the plasma-treated mica surface (~0°) because of the relatively hydrophobic GPTMS groups on the surface41. After chemically attaching CD to the surface, the WCA decreased owing to the hydrophilic hydroxyl groups of the CD molecule (Fig. 3c and Supplementary Fig. 3b). These WCA trends were comparable to those observed for GPTMS and CD monolayers in previous studies42,43.
Direct interaction force measurements
We measured the force–distance profiles for two opposing CD-coated surfaces as a function of DAd concentration using the SFA. The successful grafting of CD was further confirmed by the steric wall thickness (Dsw, defined as D at F/R = 40 mN m−1). The thickness of β-CD–GPTMS was estimated to be ~2 nm, which approximately corresponds to the sum of the GPTMS (8–10 Å) and β-CD (7.8–15.3 Å) molecular lengths44. Under pure distilled water (DI water) conditions, a Dsw value of ~4.4 nm was obtained, confirming that β-CD–GPTMS was well grafted to both surfaces.
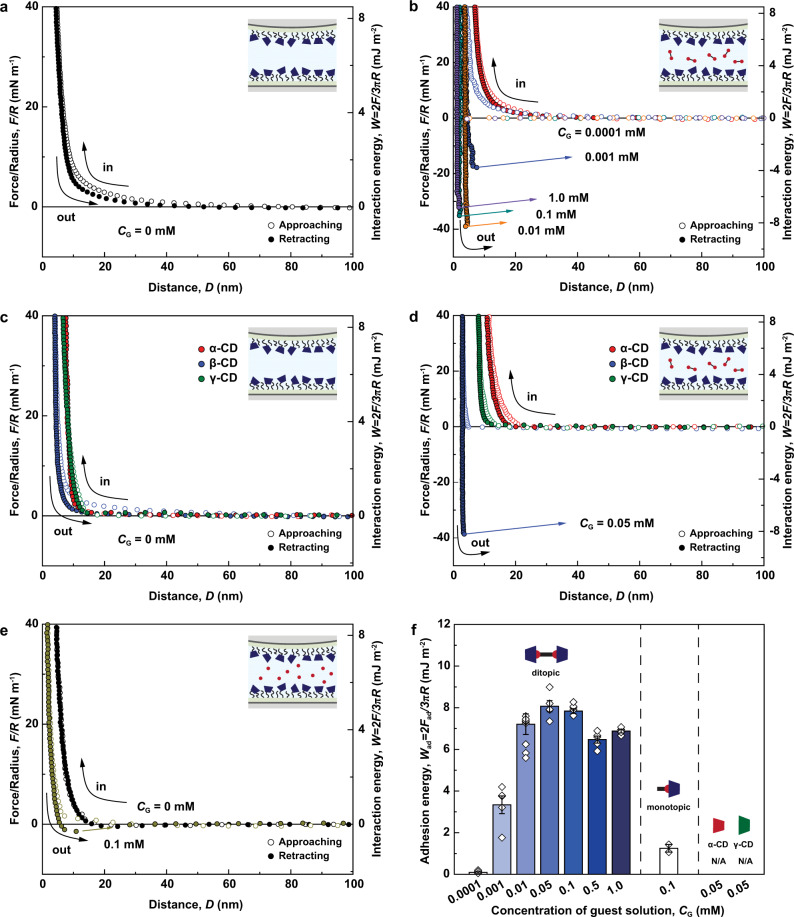
Figure 4b shows the force–distance profiles for symmetric β-CD-coated surfaces as a function of DAd concentration (CG = 0, 0.0001, 0.001, 0.01, 0.1, and 1.0 mM). Without DAd, a purely repulsive force profile was observed for the β-CD coated surfaces (Fig. 4a). The decay length of the repulsive force (λ−1) was ~15.19 nm (Supplementary Fig. 4), indicating the electrostatic repulsion between (vacant) negatively charged mica–mica surfaces upon approach. When D was less than 15 nm, λ−1 was ~2.14 nm, which originated from hydration repulsion45 and steric repulsion. In addition, no adhesion force (Fad) was observed at low CG (<0.001 mM), indicating the existence of a critical association concentration (CAC) for DAd. Above the CAC (CG ≥ 0.001 mM), Fad abruptly increased with increasing CG until reaching a maximum (Fad/R = 38.02 mN m−1, Wad = 8.07 mJ m−2) at CG = 0.05 mM, which followed the Langmuir isotherm model (Ka = 2.66 × 105 M−1) (Supplementary Fig. 5 and Supplementary Note 2). When CG was increased further (>0.05 mM), the adhesion plateaued or slightly decreased (Fig. 4f). This behavior indicates that all β-CD in this system was fully occupied with DAd molecules at a CG of ~0.05 mM (Fig. 4f).
Fig. 4. Force versus distance profiles for symmetric CD-modified surfaces and guest molecules.
Symmetric β-CD-modified surfaces a without guest molecules and b with DAd at various concentrations (0.0001–1.0 mM). Symmetric CD-modified surfaces with various cavity sizes (α-, β-, and γ-CD) c without guest molecules and d with 0.05 mM DAd. e Symmetric β-CD-modified surface without guest molecules and with 0.1 mM MAd. f Overall adhesion energies (Wad) (error bars represent the s.e.m., where n ≥ 3). Source data are provided as a Source Data file.
At high CG (>0.05 mM), when the two surfaces are far apart from each other, all the β-CDs are expected to be occupied by DAd molecules with one adamantyl moiety dangling in the bulk solution (β-CD–DAd inclusion complex) (Supplementary Fig. 6a). Upon approach, this arrangement could lead to the screening of vacant β-CD sites on the opposing surface, resulting in an increase in steric repulsion between DAd molecules and a steep decrease in the bridging of DAd molecules between the two β-CD-modified surfaces (Supplementary Fig. 6b). However, the measured interactions showed only a slight decrease in adhesion (~15%) and no significant steric repulsion was observed, even when CG was increased 20-fold to 1 mM (Fig. 4f). This discrepancy indicates that the bridging of DAd molecules between two opposing β-CD-modified surfaces (β-CD–DAd–β-CD inclusion complex) (Supplementary Fig. 6c) is more thermodynamically favorable than the complex with one dangling free end of DAd (Supplementary Fig. 6b). Nevertheless, the slight decrease in adhesion at high CG implies that the bridging interaction was hindered by a small number of dangling adamantyl moieties.
In addition, to confirm that the CD–DAd–CD inclusion complex was in thermodynamic equilibrium, we performed adhesion force measurements as a function of contact time (tc) (Supplementary Fig. 7) and loading rate (Supplementary Fig. 8). Changes in tc or loading velocity did not affect the bridging force of the CD–DAd–CD complex, indicating that the system reached thermodynamic equilibrium within 2 min. This result is in contrast to previous studies46–48 on the adhesive properties of biomacromolecules, which require long times to equilibrate and reach thermodynamic equilibrium, showing a significant increase in adhesion force and decrease in film thickness as tc increases.
We further measured the adhesion between 1-adamantylamine (monotopic adamantane; MAd) and β-CD-modified surfaces to verify the ability of DAd molecules to bridge two β-CD-modified surfaces. For this experiment, 0.1 mM of MAd, which has the same number of adamantyl groups as 0.05 mM DAd, was injected between two β-CD-modified surfaces (Fig. 4e). As expected, only a small adhesion energy was measured (Wad = 0.95 mJ m−2; less than 12% of the highest energy with DAd), suggesting that the β-CD–MAd inclusion complexes were unable to bridge two opposing surfaces.
Finally, to confirm the size dependency of the host–guest interaction between CD and Ad, force–distance profiles were obtained using surfaces modified with different types of CDs (α-, β-, and γ-CD). α-, β-, and γ-CDs have been used in various host–guest studies to evaluate cavity compatibility. At CG = 0 mM, the measured Dsw values confirmed that the α- and γ-CD monolayers had thicknesses of ~5 nm, and the force profiles were purely repulsive, similar to that of β-CD (Fig. 4c). However, for the α- and γ-CD-modified surfaces, significant adhesion forces were not measured in the presence of DAd (CG = 0.05 mM). Thus, the bridging interactions were maximized in the β-CD system (Fig. 4d), as expected from the CD cavity diameters (α-CD: 4.7–5.3 Å, β-CD: 6.0–6.5 Å, and γ-CD: 7.5–8.3 Å)49 and the size of Ad (~6.5 Å)50. In the case of α-CD, the DAd molecules are too large to be inserted into α-CD cavities to form bridging interactions (Supplementary Fig. 9). Instead, the DAd molecules seemed to adsorb weakly to the exterior of α-CD, inducing an increase in Dsw and a repulsive force upon compression. In contrast, although DAd can easily penetrate the large cavity of γ-CD, the cavity is too large to form stable CD–Ad inclusion complexes (Supplementary Fig. 9). Previous studies have reported that the Ka value for Ad binding to β-CD (Ka ~104–105 M−1) is more than 100 times larger than those for Ad binding to α-CD (Ka ~102 M−1) and γ-CD (Ka ~102–104 M−1)21,51.
Moy et al. reported the correlation between adhesion energy (Wad) and Ka for the biotin–avidin specific interaction52.
| 1 |
| 2 |
where NR and NL are the receptor and ligand densities, respectively, η is a proportionality constant, and Kd (or 1/Ka) is the dissociation constant of the complex. They showed that in the high binding affinity regime (Ka > 106 M−1), Wad scales with the logarithm of Ka (Eq. (1)), whereas in the low binding affinity regime (Ka < 106 M−1), Wad is linearly proportional to Ka (Eq. (2)). As the Ka values for CD–Ad complexes are smaller than 106 M−1, Ka is expected to be linearly proportional to Wad. This linear relationship is consistent with the SFA results, where the adhesion energies of α- and γ-CD with Ad are negligible compared to that of β-CD with Ad.
Surface density measurements using QCM-D
We further investigated the adsorption of DAd on β-CD-modified surfaces using a QCM-D (Fig. 5). β-CD was introduced onto SiO2-coated quartz sensor chips through the same modification procedure as for the mica surfaces. In the QCM-D experiments, the frequency response of the β-CD-modified surface was equilibrated using DI water (I). Then, a 0.05 mM DAd aqueous solution, corresponding to the concentration that showed the highest interaction forces in the SFA experiment, was injected for 4 h (II). Finally, the β-CD-modified surface was rinsed using DI water to remove loosely bound DAd molecules (III). After injecting the DAd solution, a frequency shift of −1.30 Hz was measured, indicating the formation of the β-CD–DAd inclusion complex on the surface.
Fig. 5. QCM-D measurement of the β-CD-modified surface.
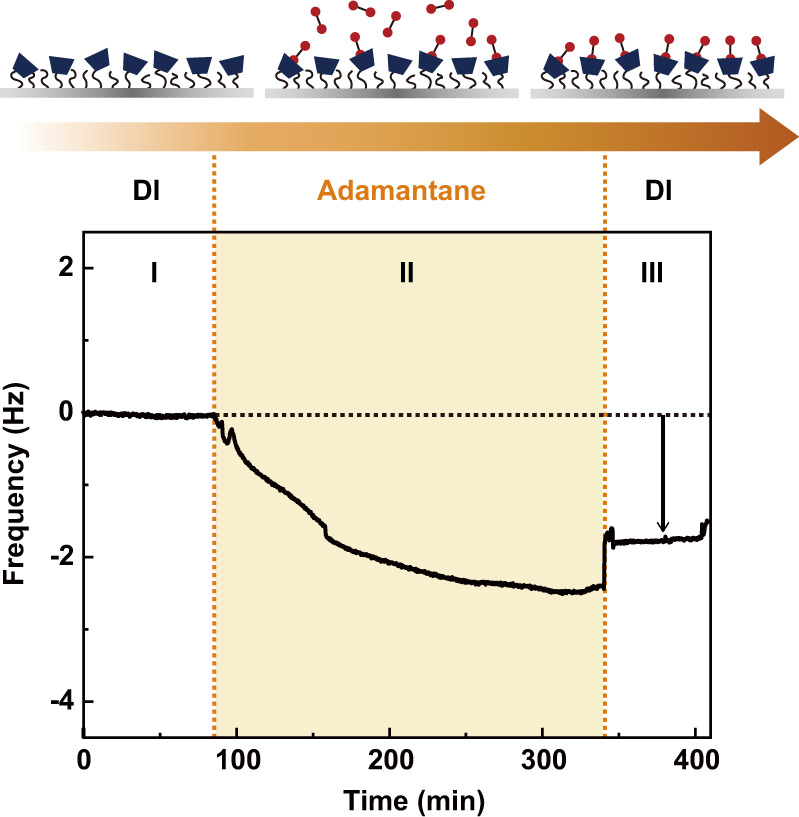
Real-time frequency shift (∆f) signal for a β-CD-modified QCM sensor surface in response to flowing 0.05 mM DAd aqueous solution.
The QCM-D results showed that ∆f and the dissipation change (∆D) overlapped on all harmonics with low dispersion and that ∆D because almost negligible as the frequency decreased, which indicates that the surface with host–guest inclusion complexes could be considered as a rigid surface. Therefore, the Sauerbrey equation was utilized to quantify the amount of DAd adsorbed on the β-CD-modified surface, and a surface density of ~22.52 ng cm−2, which corresponds to 2.07 × 1017 molecules m−2, was obtained (see the Methods section for details).
Interaction energy of a single β-CD–DAd inclusion complex
The molecular interaction energy of a single β-CD–DAd inclusion complex was calculated from the interaction energy per area (from the SFA data) and the surface density of DAd on the β-CD-modified surface (from the QCM-D data) (Table 1). The calculated single molecular energy (~9.51 kBT) was comparable to previously reported values determined using molecular dynamics simulations53 as well as ITC and SPR measurements22. This result demonstrates that the measured interaction force could be converted into the single molecular interaction energy without complicated procedures, thus providing a reliable Wad value, comparable to those determined by other analytical measurements.
Table 1.
Analysis of the interaction of DAd with β-CD-modified surfaces and the corresponding single molecular interaction energy at CG = 0.05 mM.
| Normalized force (mN m−1) |
Adhesion energy (mJ m−2) |
Frequency shift (Hz) |
Surface density (ng cm−2) |
Single molecular interaction energy (10−21 J) |
|---|---|---|---|---|
| 38.02 ± 2.82 | 8.07 ± 0.60 | −1.30 ± 0.06 | 23.11 ± 1.12 |
38.88 ± 3.45 (9.51 ± 0.84 kBT) |
Discussion
In summary, we quantified the host–guest interaction forces between symmetric β-CD-modified surfaces and DAd under aqueous conditions using an SFA. The interaction force increased drastically with increasing DAd concentration until complete host–guest complexation and then decreased slightly. Notably, with excess DAd, the interaction force was maintained owing to the rearrangement of unbound adamantyl moieties in DAd to reach thermodynamic equilibrium. Furthermore, the binding affinity of DAd with CDs was shown to depend on the CD cavity size (α-, β-, and γ-CD). As expected, a strong adhesion force was only measured for β-CD owing to the compatibility of the cavity size with Ad, as also confirmed by NMR and QCM-D measurements. Finally, the single molecular interaction energy was calculated from the interaction energy per area and the surface density determined using SFA and QCM-D measurements, respectively. The calculated single molecular interaction energy was comparable to previously reported thermodynamic parameters based on ITC and SPR measurements. These results demonstrate that the experimental approach used in this study provides not only insights into the binding energies and kinetics of host–guest interactions but also reliable parameters on the single-molecule scale that could be applied to thermodynamic systems. Therefore, we anticipate that this approach will provide fundamental information for furthering the understanding and applications of supramolecular chemistry.
Methods
Materials
1-Adamantyl bromomethyl ketone, GPTMS, and 1-adamantylamine were purchased from Sigma-Aldrich (USA). α-, β-, and γ-CD were purchased from Tokyo Chemical Industry Co., Ltd. (Japan). 4,4′-Bipyridine was purchased from Alfa Aesar (USA). N,N′-Dimethylformamide (DMF), toluene, and diethyl ether were purchased from Samchun Pure Chemical (South Korea). Sodium hydroxide (NaOH) and methanol were purchased from Daejung Chemicals & Metals Co. Ltd. (South Korea). All reagents were of analytical grade and were used as received without further purification.
Characterization
NMR spectra (1H NMR, 13C NMR and 2D ROESY) were recorded on a 400 MHz FT-NMR spectrometer (AVANCE III HD, Bruker, USA) using DMSO-d6 or D2O as the solvent. The chemical shifts were referenced to the deuterated solvent. Fourier transform infrared (FTIR) spectra were recorded on a FTIR spectrometer (670-IR, Varian, USA) using an attenuated total reflection (ATR) mode. High resolution mass spectra (HRMS) analyses were performed using a direct analysis in real time (DART) method (AccuToF 4 G+ DART, JEOL, USA). The C, H, O, and N content of organic compound was determined using an elemental analyzer (Flash 2000, Thermo Scientific, USA). The surface topography was analyzed using AFM (Multimode V AFM, Veeco, USA). WCA measurements were performed using a goniometer (DSA100, KRÜSS, Germany). Droplets of ~5 µL were placed on the modified surfaces, and the WCA value was obtained as an average of at least five measurements.
Synthesis of DAd
1-Adamantyl bromomethyl ketone (1.0 g, 3.89 mmol) and 4,4′-bipyridine (0.270 g, 1.73 mmol) were dissolved in DMF (10 mL) and allowed to react at 100 °C for 3 h under an argon atmosphere. After cooling the reaction mixture to room temperature, the yellow precipitate was collected by centrifugation at 300 g for 3 min. The crude product was washed with diethyl ether and recrystallized from methanol for 12 h. Subsequent drying under vacuum at 40 °C for 12 h gave the final product in 66% yield. 1H NMR (400 MHz, DMSO-d6): δ 9.15 (d, 4H), 8.84 (d, 4H), 6.07 (s, 4H), 2.09 (s, 6H), 1.95 (s, 12H), 1.74 (q, 12H); 13C NMR (100 MHz, DMSO-d6): δ 205.5, 149.1, 146.9, 126.4, 65.1, 45.3, 37.2, 35.9, 27.2; IR (ATR): 3047 cm−1, 2913 cm−1, 2854 cm−1, 1712 cm−1, 1637 cm−1, 1199 cm−1; HRMS (m/z, H2O, DART−): [C34H42N2O2Br2-H]− calcd. for C34H41N2O2Br2, 669.1520; found, 669.1523; Elemental analysis (calcd., found for C34H42N2O2Br2 ∙ 2(H2O): C (57.80, 58.37), H (6.56, 6.56), N (3.96, 4.15), O (9.06, 8.96). The characterizations of the DAd were shown in Supplementary Fig. 2.
Preparation of CD-grafted surfaces
Muscovite mica (Grade #1, S&J Trading, USA) was used as a substrate for the CD-grafted surfaces. To prepare a mica surface, freshly cleaved back-silvered mica was glued onto a cylindrical glass disc (R = 2 cm) using an optical adhesive (NOA 81, Norland Products Inc., USA).
The grafting process involved (i) GPTMS functionalization of the bare mica surface and (ii) CD–GPTMS grafting. For GPTMS functionalization, the glued mica surface was activated using air plasma for 3 min at 100 W under 20 Pa and a GPTMS solution (1 vol% in toluene) was added dropwise onto the activated mica surface for 30 min. Then, the mica surface was rinsed with toluene and dried with N2. For GPTMS–CD grafting, the GPTMS-functionalized surface was immersed in a CD solution (0.075 mg mL−1 α-, β-, or γ-CD in 0.1 mM NaOH) for 10 min. Subsequently, the surface was rinsed with DI water to remove unbound residues and dried with N2. All reactions were performed in a humidity-controlled environment (RH = 10%, T = 23 °C).
Interaction force measurements using an SFA
An SFA (SFA 2000, SurForce LLC, USA) was used to measure the interaction energies and the absolute distances between the CD-grafted surfaces54. The CD-grafted surfaces were arranged in the SFA chamber with a cross-cylindrical geometry, and 40 µL of guest solution was injected between the surfaces. The interaction forces between the host and guest molecules were measured as a function of the guest molecule concentration (0, 0.0001, 0.001, 0.01, 0.05, 0.1, 0.5, and 1 mM DAd in DI water). In addition, to determine the effect of the guest molecule type (monotopic or ditopic) on the interaction energy, measurements were performed with 0.1 mM MAd. For each measurement, the SFA chamber was sealed with a water-soaked dust-free wiper to minimize evaporation during the experiment, and then the system was equilibrated for 1 h.
During the force measurements, the approach and retraction of two opposing surfaces were performed using a microscale motor at a constant speed (~5 nm s−1, otherwise mentioned). The interaction force (F) was measured by deflection of a double cantilever spring (k = 2451.7 N m−1) connected to the lower surface as a function of the absolute distance (D) between the opposing surfaces. The mica–mica distance (D) was confirmed using the fringes of equal chromatic order measured by multiple-beam interferometry55. The normalized adhesion force (Fad/R), which was determined using the absolute value of the minimum F/R, Fad/R = abs[min(F/R)], during separation, was converted to the adhesion energy per unit area (Wad = 2Fad/3πR) according to the Johnson–Kendall–Roberts model39,56, which is the conventional method for converting measured forces to adhesion energies in SFA experiments. All force–distance measurements were conducted at room temperature (T = 23 °C) and repeated more than 4 times under each condition to confirm reproducibility.
Static force-runs were performed by using extra-fine control of piezoelectric tube which supports the upper disk54. The constant voltage steps of 1.0 V was applied to the piezoelectric crystal to move the upper surface with regular shifts of distance (~5 nm). The surfaces were equilibrated at each distance (D) for ~40 s before moving to next D. Voltage-to-distance calibration of piezoelectric crystal was evaluated at separated distance (D > 400 nm), where no interaction forces were measured. All static force-runs were repeated 3 times for reproducibility.
Surface density measurements using QCM-D
The adsorption of guest molecules on the host-modified surfaces was investigated using QCM-D (QCM-D E4, Q-Sense, Sweden). The SiO2-coated quartz sensor chip (QSX 303, Q-Sense, Sweden) was cleaned by UV–ozone treatment for 10 min, rinsed twice with ethanol, and dried with N2. Then, the surface of the sensor chip was modified with β-CD using the same surface modification procedure as for the mica surface. All experiments were performed at a constant flow rate of 50 µL min−1 and a constant temperature of 25 °C. The monitored shifts in ∆f and ∆D were analyzed using the QTools software (Q-Sense, Sweden). The baseline drift was subtracted from the data. The surface density (∆m) was calculated from the negative shift in ∆f using the Sauerbrey equation57:
| 3 |
where ρq is the density of quartz (2.648 g cm−3), µq is the shear modulus of quartz for an AT-cut crystal (29.47 GPa), f0 is the resonant frequency of the fundamental mode (5 MHz), C is the mass sensitivity constant (17.7 ng cm−2 Hz−1), and n is the overtone number58. In this work, the seventh overtone was adopted for this calculation.
Supplementary information
Acknowledgements
This work was supported by the Basic Science Research Program (2019R1A2C2005854) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science of Korea. We thank Prof. Sebyung Kang at UNIST for providing QCM-D equipment.
Source data
Author contributions
J.-T.P. designed and performed the syntheses and the surface characterization. J.-W.P., J.L. performed SFA measurements. C.L. and D.W.L. supervised this project. J.-T.P., J.-W.P., C.L., and D.W.L. wrote the paper. All authors read and approved the final paper.
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files. Additional data related to this study can be requested from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jintae Park, Jinwoo Park.
Contributor Information
Chanoong Lim, Email: c.lim@unist.ac.kr.
Dong Woog Lee, Email: dongwoog.lee@unist.ac.kr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-27659-w.
References
- 1.Leckband D, Israelachvili J. Intermolecular forces in biology. Q. Rev. Biophys. 2001;34:105–267. doi: 10.1017/s0033583501003687. [DOI] [PubMed] [Google Scholar]
- 2.Lehn J. Supramolecular chemistry. Science. 1993;260:1762–1763. doi: 10.1126/science.8511582. [DOI] [PubMed] [Google Scholar]
- 3.Webber MJ, Appel EA, Meijer EW, Langer R. Supramolecular biomaterials. Nat. Mater. 2016;15:13–26. doi: 10.1038/nmat4474. [DOI] [PubMed] [Google Scholar]
- 4.Surin M. From nucleobase to DNA templates for precision supramolecular assemblies and synthetic polymers. Polym. Chem. 2016;7:4137–4150. [Google Scholar]
- 5.McLaughlin CK, Hamblin GD, Sleiman HF. Supramolecular DNA assembly. Chem. Soc. Rev. 2011;40:5647–5656. doi: 10.1039/c1cs15253j. [DOI] [PubMed] [Google Scholar]
- 6.Hannon MJ. Supramolecular DNA recognition. Chem. Soc. Rev. 2007;36:280–295. doi: 10.1039/b606046n. [DOI] [PubMed] [Google Scholar]
- 7.Dang DT, Nguyen HD, Merkx M, Brunsveld L. Supramolecular Control of Enzyme Activity through Cucurbit[8]uril-Mediated Dimerization. Angew. Chem. Int. Ed. 2013;52:2915–2919. doi: 10.1002/anie.201208239. [DOI] [PubMed] [Google Scholar]
- 8.Cram DJ, Cram JM. Host-Guest Chemistry. Science. 1974;183:803–809. doi: 10.1126/science.183.4127.803. [DOI] [PubMed] [Google Scholar]
- 9.Steed, J. W. & Atwood, J. L. Supramolecular chemistry (John Wiley & Sons, Chichester, 2009).
- 10.Nakahata M, Takashima Y, Yamaguchi H, Harada A. Redox-responsive self-healing materials formed from host–guest polymers. Nat. Commun. 2011;2:511. doi: 10.1038/ncomms1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kakuta T, et al. Preorganized hydrogel: self-healing properties of supramolecular hydrogels formed by polymerization of host–guest-monomers that contain cyclodextrins and hydrophobic guest groups. Adv. Mater. 2013;25:2849–2853. doi: 10.1002/adma.201205321. [DOI] [PubMed] [Google Scholar]
- 12.Feng Q, et al. Mechanically resilient, injectable, and bioadhesive supramolecular gelatin hydrogels crosslinked by weak host-guest interactions assist cell infiltration and in situ tissue regeneration. Biomaterials. 2016;101:217–228. doi: 10.1016/j.biomaterials.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 13.Hoang Thi TT, et al. Supramolecular cyclodextrin supplements to improve the tissue adhesion strength of gelatin bioglues. ACS Macro Lett. 2017;6:83–88. doi: 10.1021/acsmacrolett.6b00841. [DOI] [PubMed] [Google Scholar]
- 14.Hwang I, et al. Noncovalent Immobilization of Proteins on a Solid Surface by Cucurbit[7]uril-Ferrocenemethylammonium Pair, a Potential Replacement of Biotin−Avidin Pair. J. Am. Chem. Soc. 2007;129:4170–4171. doi: 10.1021/ja071130b. [DOI] [PubMed] [Google Scholar]
- 15.Xie S, Zhang J, Yuan Y, Chai Y, Yuan R. An electrochemical peptide cleavage-based biosensor for prostate specific antigen detection via host–guest interaction between ferrocene and β-cyclodextrin. Chem. Commun. 2015;51:3387–3390. doi: 10.1039/c4cc10363g. [DOI] [PubMed] [Google Scholar]
- 16.Duan Q, et al. pH-Responsive Supramolecular Vesicles Based on Water-Soluble Pillar[6]arene and Ferrocene Derivative for Drug Delivery. J. Am. Chem. Soc. 2013;135:10542–10549. doi: 10.1021/ja405014r. [DOI] [PubMed] [Google Scholar]
- 17.Guo Y, et al. Cyclodextrin functionalized graphene nanosheets with high supramolecular recognition capability: synthesis and host−guest inclusion for enhanced electrochemical performance. ACS Nano. 2010;4:4001–4010. doi: 10.1021/nn100939n. [DOI] [PubMed] [Google Scholar]
- 18.Herbst F, Döhler D, Michael P, Binder WH. Self-healing polymers via supramolecular forces. Macromol. Rapid Commun. 2013;34:203–220. doi: 10.1002/marc.201200675. [DOI] [PubMed] [Google Scholar]
- 19.Ji X, et al. Adhesive supramolecular polymeric materials constructed from macrocycle-based host–guest interactions. Chem. Soc. Rev. 2019;48:2682–2697. doi: 10.1039/c8cs00955d. [DOI] [PubMed] [Google Scholar]
- 20.Rüdiger V, et al. Conformational, calorimetric and NMR spectroscopic studies on inclusion complexes of cyclodextrins with substituted phenyl and adamantane derivatives. J. Chem. Soc., Perkin Trans. 2. 1996;21:2119–2123. [Google Scholar]
- 21.Schibilla F, et al. Host–guest complexes of cyclodextrins and nanodiamonds as a strong non-covalent binding motif for self-assembled nanomaterials. Chem. Eur. J. 2017;23:16059–16065. doi: 10.1002/chem.201703392. [DOI] [PubMed] [Google Scholar]
- 22.de Jong MR, Huskens J, Reinhoudt DN. Influencing the binding selectivity of self-assembled cyclodextrin monolayers on gold through their architecture. Chem. Eur. J. 2001;7:4164–4170. doi: 10.1002/1521-3765(20011001)7:19<4164::aid-chem4164>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 23.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Florin E, Moy V, Gaub H. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 25.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor–ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 26.Kado S, Kimura K. Single Complexation Force of 18-Crown-6 with Ammonium Ion Evaluated by Atomic Force Microscopy. J. Am. Chem. Soc. 2003;125:4560–4564. doi: 10.1021/ja029397s. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Yu Y, Gao J, Wang Z, Zhang X. Water-soluble supramolecular polymerization driven by multiple host-stabilized charge-transfer interactions. Angew. Chem. Int. Ed. 2010;49:6576–6579. doi: 10.1002/anie.201002415. [DOI] [PubMed] [Google Scholar]
- 28.Pandey S, et al. Chaperone-assisted host–guest interactions revealed by single-molecule force spectroscopy. J. Am. Chem. Soc. 2019;141:18385–18389. doi: 10.1021/jacs.9b09019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schönherr H, et al. Individual supramolecular host−guest interactions studied by dynamic single molecule force spectroscopy. J. Am. Chem. Soc. 2000;122:4963–4967. [Google Scholar]
- 30.Zapotoczny S, et al. Chain length and concentration dependence of β-cyclodextrin−ferrocene host−guest complex rupture forces probed by dynamic force spectroscopy. Langmuir. 2002;18:6988–6994. [Google Scholar]
- 31.Auletta T, et al. β-cyclodextrin host−guest complexes probed under thermodynamic equilibrium: thermodynamics and AFM force spectroscopy. J. Am. Chem. Soc. 2004;126:1577–1584. doi: 10.1021/ja0383569. [DOI] [PubMed] [Google Scholar]
- 32.Blass J, Albrecht M, Bozna BL, Wenz G, Bennewitz R. Dynamic effects in friction and adhesion through cooperative rupture and formation of supramolecular bonds. Nanoscale. 2015;7:7674–7681. doi: 10.1039/c5nr00329f. [DOI] [PubMed] [Google Scholar]
- 33.Blass J, Albrecht M, Wenz G, Zang YN, Bennewitz R. Single-molecule force spectroscopy of fast reversible bonds. Phys. Chem. Chem. Phys. 2017;19:5239–5245. doi: 10.1039/c6cp07532k. [DOI] [PubMed] [Google Scholar]
- 34.Blass J, Bozna B, Albrecht M, Wenz G, Bennewitz R. Molecular kinetics and cooperative effects in friction and adhesion of fast reversible bonds. Phys. Chem. Chem. Phys. 2019;21:17170–17175. doi: 10.1039/c9cp03350e. [DOI] [PubMed] [Google Scholar]
- 35.Leckband DE, Schmitt FJ, Israelachvili JN, Knoll W. Direct force measurements of specific and nonspecific protein interactions. Biochemistry. 1994;33:4611–4624. doi: 10.1021/bi00181a023. [DOI] [PubMed] [Google Scholar]
- 36.Raman S, Utzig T, Baimpos T, Ratna Shrestha B, Valtiner M. Deciphering the scaling of single-molecule interactions using Jarzynski’s equality. Nat. Commun. 2014;5:5539. doi: 10.1038/ncomms6539. [DOI] [PubMed] [Google Scholar]
- 37.Banquy X, Lee DW, Kristiansen K, Gebbie MA, Israelachvili JN. Interaction forces between supported lipid bilayers in the presence of PEGylated Polymers. Biomacromolecules. 2016;17:88–97. doi: 10.1021/acs.biomac.5b01216. [DOI] [PubMed] [Google Scholar]
- 38.Lim C, et al. Probing nanomechanical interaction at the interface between biological membrane and potentially toxic chemical. J. Hazard. Mater. 2018;353:271–279. doi: 10.1016/j.jhazmat.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 39.Israelachvili, J. N. Intermolecular and Surface Forces (Academic, San Diego, 2011).
- 40.Ohga K, et al. Preparation of supramolecular polymers from a cyclodextrin dimer and ditopic guest molecules: control of structure by linker flexibility. Macromolecules. 2005;38:5897–5904. [Google Scholar]
- 41.Tsukruk VV, Luzinov I, Julthongpiput D. Sticky molecular surfaces: epoxysilane self-assembled monolayers. Langmuir. 1999;15:3029–3032. [Google Scholar]
- 42.Yu Z, et al. Preparation of a novel anti-fouling β-cyclodextrin–PVDF membrane. RSC Adv. 2015;5:51364–51370. [Google Scholar]
- 43.Nietzold C, Dietrich PM, Lippitz A, Panne U, Unger WES. Cyclodextrin—ferrocene host–guest complexes on silicon oxide surfaces. Surf. Interface Anal. 2016;48:606–610. [Google Scholar]
- 44.Su Y-C, Chen W-C, Chang F-C. Preparation and characterization of polyseudorotaxanes based on adamantane-modified polybenzoxazines and β-cyclodextrin. Polymer. 2005;46:1617–1623. [Google Scholar]
- 45.Donaldson SH, et al. Developing a general interaction potential for hydrophobic and hydrophilic interactions. Langmuir. 2015;31:2051–2064. doi: 10.1021/la502115g. [DOI] [PubMed] [Google Scholar]
- 46.Lee DW, Lim C, Israelachvili JN, Hwang DS. Strong adhesion and cohesion of chitosan in aqueous solutions. Langmuir. 2013;29:14222–14229. doi: 10.1021/la403124u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim C, et al. Probing molecular mechanisms of M13 bacteriophage adhesion. Commun. Chem. 2019;2:96. [Google Scholar]
- 48.Deepankumar K, et al. Supramolecular β-Sheet Suckerin–Based Underwater Adhesives. Adv. Funct. Mater. 2020;30:1907534. [Google Scholar]
- 49.Li L, Guo X, Fu L, Prud’homme RK, Lincoln SF. Complexation behavior of α-, β-, and γ-cyclodextrin in modulating and constructing polymer Networks. Langmuir. 2008;24:8290–8296. doi: 10.1021/la800859w. [DOI] [PubMed] [Google Scholar]
- 50.Enright GD, Udachin KA, Ripmeester JA. Complex packing motifs templated by a pseudo-spherical guest: the structure of β-cyclodextrin–adamantane inclusion compounds. CrystEngComm. 2010;12:1450–1453. [Google Scholar]
- 51.Jelínková K, et al. Binding study on 1-adamantylalkyl(benz)imidazolium salts to cyclodextrins and cucurbit[n]urils. N. J. Chem. 2020;44:7071–7079. [Google Scholar]
- 52.Moy VT, Jiao Y, Hillmann T, Lehmann H, Sano T. Adhesion energy of receptor-mediated interaction measured by elastic deformation. Biophys. J. 1999;76:1632–1638. doi: 10.1016/S0006-3495(99)77322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cova TFGG, Milne BF, Nunes SCC, Pais AACC. Drastic stabilization of junction nodes in supramolecular structures based on host–guest complexes. Macromolecules. 2018;51:2732–2741. [Google Scholar]
- 54.Israelachvili J, et al. Recent advances in the surface forces apparatus (SFA) technique. Rep. Prog. Phys. 2010;73:036601. [Google Scholar]
- 55.Israelachvili JN. Thin film studies using multiple-beam interferometry. J. Colloid Interface Sci. 1973;44:259–272. [Google Scholar]
- 56.Johnson KL, Kendall K, Roberts AD, Tabor D. Surface energy and the contact of elastic solids. Proc. R. Soc. Lond., Ser. A. 1971;324:301–313. [Google Scholar]
- 57.Sauerbrey G. Verwendung von Schwingquarzen zur Wägung dünner Schichten und zur Mikrowägung. Z. Phys. 1959;155:206–222. [Google Scholar]
- 58.Guo H, et al. Real-Time QCM-D Monitoring of Deposition of Gold Nanorods on a Supported Lipid Bilayer as a Model Cell Membrane. ACS Omega. 2019;4:6059–6067. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files. Additional data related to this study can be requested from the corresponding author upon request. Source data are provided with this paper.