Abstract
The D-type cyclins (CCND1, CCND2, and CCND3) in association with CDK4/6 are known drivers of cell cycle progression. We reported previously that inactivation of FOXO1 confers growth arrest and apoptosis in B-ALL, partially mediated by subsequent depletion of CCND3. Given that previously the canonical MYC target CCND2 has been considered to play the major role in B-ALL proliferation, further investigation of the role of FOXO1 in CCND3 transcription and the role of CCND3 in B-ALL is warranted. In this study, we demonstrated that CCND3 is essential for the proliferation and survival of B-ALL, independent of the mutational background. Respectively, its expression at mRNA level exceeds that of CCND1 and CCND2. Furthermore, we identified FOXO1 as a CCND3-activating transcription factor in B-ALL. By comparing the effects of CCND3 depletion and CDK4/6 inhibition by palbociclib on B-ALL cells harboring different driver mutations, we found that the anti-apoptotic effect of CCND3 is independent of the kinase activity of the CCND3-CDK4/6 complex. Moreover, we found that CCND3 contributes to CDK8 transcription, which in part might explain the anti-apoptotic effect of CCND3. Finally, we found that increased CCND3 expression is associated with the development of resistance to palbociclib. We conclude that CCND3 plays an essential role in the maintenance of B-ALL, regardless of the underlying driver mutation. Moreover, downregulation of CCND3 expression might be superior to inhibition of CDK4/6 kinase activity in terms of B-ALL treatment.
Subject terms: Leukaemia, Cell division
Introduction
B-cell lymphoblastic leukemia (B-ALL) is the most common pediatric neoplasia [1]. Pediatric B-ALL is associated with long-term survival of over 90%, whereas adult B-ALL shows dismal outcomes with cure rates below 40% [2]. Therapy in both children and adults typically consists of combinatory administration of cytostatic agents [3], which in many cases induce life treating toxicity and complications. Therefore, the search for novel, less toxic targeted therapies is warranted [4].
We reported previously that the tightly regulated expression of the transcription factor forkhead box protein O1 (FOXO1) is essential for the maintenance of B-ALL [5]. Given that FOXO1 had been identified as a canonical tumor suppressor, our finding was paradoxical [6]. However, our data were later corroborated in a BCR-ABL1-transformed B-ALL mouse model with inducible Foxo1 deletion [7]. Most importantly, we demonstrated that genetic and pharmacological inhibition of FOXO1 downregulates Cyclin D3 (CCND3) expression and the cytotoxic effects of FOXO1 depletion could be ameliorated by CCND3 overexpression [5].
The three D-cyclins CCND1, CCND2, and CCND3 work in a holoenzyme complex with the cyclin-dependent kinases CDK4 and CDK6. Upon mitogenic stimulation, the complex phosphorylates retinoblastoma protein 1 (RB1), which represses the transcriptional activity of E2Fs [8]. Following phosphorylation, RB1 is released, thereby permitting the E2F mediated initiation of the G1-S transcriptional program.
Throughout B-cell development, expression levels of CCND2 and CCND3 vary, while expression of CCND1 is stopped after differentiation from hematopoietic stem cells [9]. CCND3 plays a key role during the development of B-cell precursor cells and cannot be substituted by another D-type cyclin [9]. Additionally, at later stages of B-cell development, CCND3 is essential for the expansion of germinal center B-cells [10].
The role of CCND3 was extensively investigated in T-cell acute lymphoblastic leukemia (T-ALL). It was shown that Ccnd3−/− mice are less susceptible to the development of T-ALL [11], and the ablation of Ccnd3 in Notch1-driven T-ALL induces apoptosis [12]. In B-ALL however, the role of CCND3 has been neglected since in BCR-ABL1+ B-ALL it has been demonstrated that the proto-oncogene MYC canonically induces transcription of CCND2 [13], suggesting its role as the supreme D-cyclin in B-ALL [14].
Since our previous work on FOXO1-depletion indicated a major role of CCND3 in B-ALL, we investigated the transcriptional regulation of CCND3 by FOXO1 and the effects of acute loss of CCND3 in B-ALL cells of different genetic backgrounds. In addition, we addressed the CDK4/6-independent role of CCND3.
Results
CCND3 is the highest expressed D-Cyclin in all subtypes of B-ALL
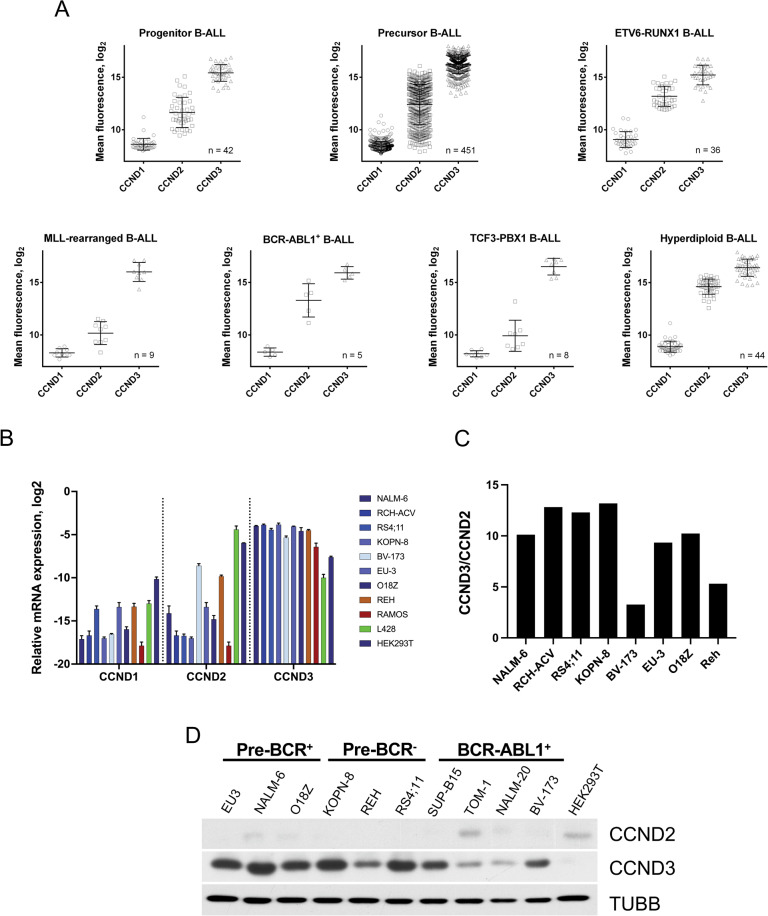
To clarify the role of D-type cyclins in B-ALL, we compared their expression at mRNA level in the most common B-ALL subtypes (Fig. 1A). Compared to all other indexed types of cancer, CCND3 showed the highest expression levels in B-ALL (Supplementary Fig. 1). Furthermore, CCND3 was expressed at much higher levels than CCND1 and CCND2 in all B-ALL subtypes. CCND1 appeared to be the least expressed D-cyclin in all genetic groups of B-ALL. To clarify whether the observed differences in the expression levels of D-type cyclins indicate the exceptional functional significance of CCND3, we used publicly available data of genome-wide CRISPR/Cas9 loss-of-function viability screenings (https://depmap.org/portal/). For all indexed B-ALL cell lines (SEM, SEMK2, 697, RCH-ACV, REH, JM-1, NALM-6, HB1119, and P300HK), estimated dependency on CCND3 was much higher than for CCND1 and CCND2 (Supplementary Fig. 2A, B).
Fig. 1. CCND3 exceeds expression of CCND1 and CCND2 at mRNA and protein levels in B-ALL.
A The mRNA expression data of CCND1, CCND2, and CCND3 in different B-ALL subtypes were mined from publicly available databases using the GENEVESTIGATOR software (http://www.genevestigator.com/). The experiment IDs are listed in Supplementary Materials and Methods. Data shown as mean ± SD. B The mRNA expression levels of D-type cyclins in B–ALL (blue) and control cell lines was measured by qRT-PCR. The control cell lines included Burkitt lymphoma (Ramos), cHL (L428), and HEK293T which express high levels of CCND3, CCND2, and CCND1, respectively. Data shown as mean ± SD, n = 3. C Ratio of mRNA levels CCND3/CCND2 measured by qRT-PCR in B-ALL cell lines. D Expression of CCND2 and CCND3 proteins in B-ALL cell lines was measured by immunoblot. TUBB is used as loading control. Image representative of n = 3.
Next, we asked whether the high CCND3 expression is maintained in human B-ALL cell lines. To this end, we measured mRNA and protein expression levels of the D-cyclins in B-ALL cell lines harboring different mutations (Fig. 1B,D). The ratio of CCND3/CCND2 mRNA expression in all cell lines was similar to the primary B-ALL samples (Fig. 1C). CCND2 expression varies throughout genotypes with higher expression in BCR-ABL1+ cell lines. CCND1 was not detectable on protein level in any B-ALL cell line (Supplementary Fig. 3).
This data indicates that CCND3 is by far the highest expressed D-type cyclin in B-ALL patient samples and cell lines, independent of the underlying driver mutation.
CCND3 transcription is directly regulated by FOXO1
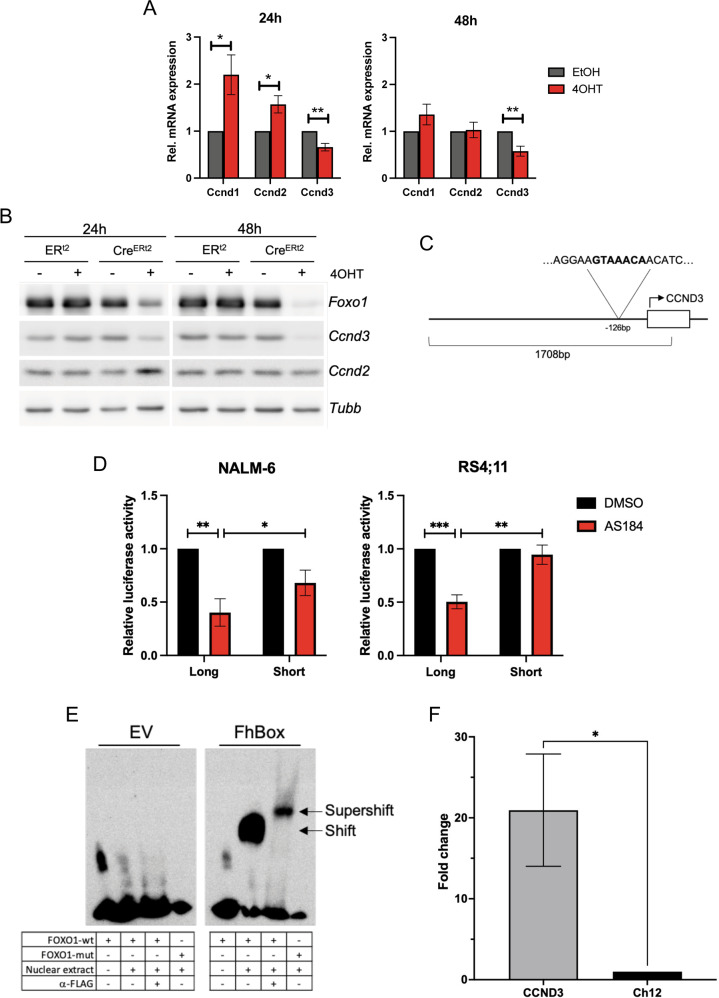
FOXO1 was shown to repress the transcription of CCND1 and CCND2 [15]. At the same time, FOXO1 activates Ccnd3 transcription in murine pancreatic cells [16]. Moreover, we have shown that FOXO1 inactivation decreases CCND3 expression in human B-ALL cell lines [5]. To further investigate the dependency of CCND3 transcription on FOXO1, we induced depletion of Foxo1 in a BCR-ABL1+ Foxo1fl/fl murine B-ALL model (Fig. 2A). This exclusively decreased Ccnd3 but not Ccnd1 or Ccnd2 at mRNA and protein levels (Fig. 2B). In line with the known role of FOXO1 as a negative regulator of Ccnd1 and Ccnd2 transcription, both Cyclins were significantly upregulated on mRNA level after 24 h. Furthermore, 24 h after Foxo1 deletion, CCND2 was upregulated on protein level. However, this upregulation of CCND2 could not substitute CCND3 in the maintenance of cell cycle progression (Supplementary Fig. 4).
Fig. 2. FOXO1 regulates CCND3 transcription by DNA binding.
A Deletion of Foxo1 in BCR-ABL1+ murine B-ALL cells decreases mRNA and B protein expression of Ccnd3 but increases Ccnd1 and Ccnd2 mRNA expression. Ccnd1 was not detectable on protein level. Data shown as mean ± SD, n = 3. Statistical analysis was performed with students t-test. *=p < 0.05 **=p < 0.01. C CCND3 promoter region −1389 to +259 bp from the CCND3 TSS (NC_000006.12, https://www.ncbi.nlm.nih.gov, 22 May 2020) used for luciferase assay containing the FOXO binding motif GTAAACA −126 bp from the transcriptional start site. D NALM-6 and RS4;11 cells were co-transfected with either pGL4.22-CCND3-promoter (long) or a truncated version containing the core promoter without the FOXO binding motif (short), together with Ubi-Renilla luciferase-expressing vectors and treated for 24 h with the FOXO1 inhibitor AS1842856 at a concentration of 80 nM. The luminescence intensities of the experimental vectors were normalized to Ubi-Renilla. Data shown as mean ± SD, n = 3. Statistical analysis was performed with students t-test. *=p < 0.05 **=p < 0.01. ***=p < 0.005 E Binding of FOXO1 to the CCND3 promoter (NC_000006.12, https://www.ncbi.nlm.nih.gov, 22 May 2020) shown by EMSA. Nuclear extracts from HEK293T cells transfected with either pFLAG-CMV2-Empty Vector (EV) or pFLAG-CMV2-FhBox (FhBox) were co-incubated with DNA probes containing FOXO binding motif −126 to −101 bp from the CCND3 TSS (FOXO1-wt) or scrambled FOXO binding motif (FOXO1-mut). Image representative of n = 2. F Chromatin immunoprecipitation. NALM-6 cells transfected with humanized biotin ligase BirA and either a constitutively active variant of FOXO1 with N-terminal biotinylation signal or empty control vector. Pulldown was performed with magnetic streptavidin particles. Enrichment of the DNA fragment containing the FOXO binding motif (CCND3) was analyzed by qRT-PCR with two primers flanking the binding motif. Ct values were first normalized to the respective input and then calculated relative to the signal obtained by amplification with primers targeting the Ch12 gene desert (Ch12). Data are shown as mean ± SD, n = 3. Statistical analysis was performed with students t-test. *=p < 0.05.
Next, we addressed the mechanism underlying the decrease of Ccnd3 transcription induced by Foxo1 depletion. Previously we reported on the decrease of MYC transcription and RB1 phosphorylation by FOXO1 depletion in human B-ALL cell lines [5]. Given that both MYC as well as the RB1 repression target E2F1 might activate CCND3 transcription [8, 17, 18], we investigated their role in the observed Ccnd3 downregulation.
We found that Foxo1 deletion decreased Myc transcription in the BCR-ABL1-transformed murine pre-B-cells (Supplementary Fig. 5A). Given that BCR-ABL1 activates Myc transcription and increases the inactivating phosphorylation of FOXO1 in this B-ALL model [7], we treated the cells with 1 µM of the BCR-ABL1 inhibitor imatinib to abolish Myc expression. Imatinib strongly decreased MYC expression at mRNA and protein levels (Supplementary Fig. 5B,C), but simultaneously increased FOXO1 protein level and reduced the FOXO1-inactivating S256 phosphorylation. Ccnd3 mRNA expression remained stable. These data indicated that Ccnd3 transcription does not depend on Myc. To address the role of E2Fs in Ccnd3 transcription [18], we treated the cells with the CDK4/6 inhibitor palbociclib at an effective concentration of 1 µM, which completely arrested cell cycle progression in human B-ALL cells. Palbociclib increased FOXO1 protein expression (Supplementary Fig. 6A) and concomitantly increased Ccnd3 expression at mRNA (Supplementary Fig. 6B) and protein levels (Supplementary Fig. 6C) suggesting that also E2Fs are not essential for Ccnd3 transcription in B-ALL.
We further investigated the effect of FOXO1 on the activity of the CCND3 promoter using a luciferase-reporter assay. To this end we transfected NALM-6 and RS4;11 B-ALL cells with a reporter construct driven by the CCND3 promoter spanning from −1389 to +259 bp from the transcriptional start site (TSS) and harboring the FOXO binding motif GTAAACA −126 bp from the TSS (“long”) [19] (Fig. 2C). As a control, we used the core promoter region of CCND3, which lacks the FOXO binding motif (“short”). Treatment with the FOXO1 inhibitor AS1842856 at a concentration of 80 nM significantly decreased the activity of the “long” but not the “short” reporter construct (Fig. 2D).
To further elaborate on the transcriptional regulation of CCND3 by FOXO1, we analyzed the binding of the forkhead DNA-binding domain (FhBox) to the FOXO-binding site located −126 bp from the CCND3 TSS. To this end, we performed electrophoretic mobility shift assay (EMSA). Nuclear extracts of HEK293T cells transfected with either FhBox-FLAG or empty vector (EV) were co-incubated with probes corresponding to −142 to −117 bp from the TSS and containing the FOXO binding motif. The construct containing the FOXO-binding motif was only shifted by the nuclear extracts expressing FhBox, but not by the control extracts (Fig. 2E and Supplementary Fig. 7A). Mutation of the FOXO binding motif completely abolished the FhBox binding. The specificity of the binding of FhBox to the FOXO binding site was confirmed by super-shift assay.
In order to confirm the direct binding of FOXO1 at the CCND3 promoter, we first mined available ChIP-seq data on binding of FOXO1 to transcription regulatory elements in pre-leukemic stem cells and mature B-lymphocytes (Supplementary Fig. 8). Both sample tracks showed significant binding events at the CCND3 promoter in the proximity of the FOXO binding site which we identified by EMSA. Next, we experimentally corroborated FOXO1 binding to the CCND3 promoter with help of ChIP-qRT-PCR. To this end we used a constitutively active variant of FOXO1, containing an N-terminal biotinylation signal, as we described previously (Supplementary Fig. 7B) [20].
Using this approach, we demonstrated direct binding of FOXO1 at the CCND3 promoter (Fig. 2F).
CCND3 is essential for the growth and survival of B-ALL cells
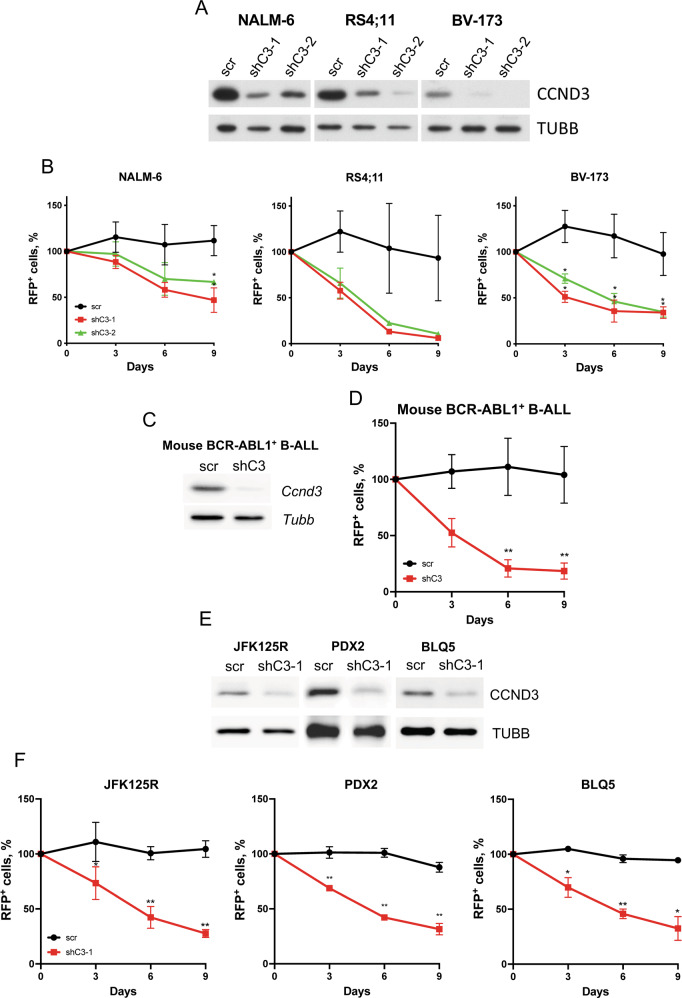
Previously we have shown that CCND3 knockdown induces growth arrest and apoptosis in the B-ALL cell lines NALM-6 and RS4;11, which harbor ETV6-PDGFR [21] and MLL-AF4 [22] translocations, respectively. Here we set to investigate the role of CCND3 in the oncogenic program of other B-ALL subtypes, including BCR-ABL1+ B-ALL cell lines, which are distinguished by the highest CCND2 expression compared to other genotypes. To deplete CCND3, we used two shRNAs. Both shRNAs efficiently downregulated CCND3 (Fig. 3A) and conferred a significant growth disadvantage compared to control cells transduced with a non-targeting construct in a competitive growth assay in all cell lines (Fig. 3B and Supplementary Fig. 9A). This effect of shRNA-dependent knockdown was corroborated by CRISPR/Cas9-editing (Supplementary Fig. 9B, C).
Fig. 3. Downregulation of CCND3 is toxic for B-ALL cells.
A Efficiency of the shRNA-dependent CCND3 knockdown was assessed by immunoblot. Image is representative of n = 3. B B-ALL cell lines were transduced with plasmids expressing RFP and either a non-targeting control, or CCND3-targeting shRNAs. Data shown as mean ± SD, n = 3. Statistical analysis was performed with student’s t-test. *=p < 0.05. C Immunoblot shows downregulation of CCND3 expression by specific shRNA in BCR-ABL1 transformed mouse pre-B-cells. Image representative of n = 3. D shRNA-dependent knockdown of Ccnd3 in BCR-ABL1 transformed murine pre-B-cells significantly inhibits their fitness in a competitive growth assay. Data shown as mean ± SD, n = 3. Statistical analysis was performed by student’s t-test. **=p < 0.01. E B-ALL PDXs JFK125R, PDX2, and BLQ5 were transduced with vectors expressing a shRNA targeting CCND3. CCND3 downregulation was analyzed by immunoblot. Image representative of n = 3. F The cytotoxic effect of CCND3 depletion was analyzed by competitive growth test as described in legend to Fig. 3B. Data shown as mean ± SD, n = 3. Statistical analysis was performed by student’s t-test. *=p < 0.05 **=p < 0.01.
The pro-apoptotic and growth-inhibitory effect of CCND3 knockdown was also observed in an ex vivo BCR-ABL1+ murine B-ALL model with a shRNA construct targeting Ccnd3 (Fig. 3C, D). Additionally, we corroborated our data on the essential role of CCND3 by including three B-ALL patient-derived xenografts (PDX). JFK125R, PDX2, and BLQ5. JFK125R represents refractory, KRASG12D-driven B-ALL, PDX2 harbors the BCR-ABL1 translocation and IKZF1-deletion, and BLQ5 was derived from a relapsed BCR-ABL1+ B-ALL patient after treatment with imatinib [23]. In all three PDXs, CCND3 knockdown was detrimental to survival.
We conclude that CCND3 is essential for the survival of B-ALL independently of the type of driver mutation.
The pro-apoptotic effect of CCND3 downregulation is independent of CDK4/6 kinase activity
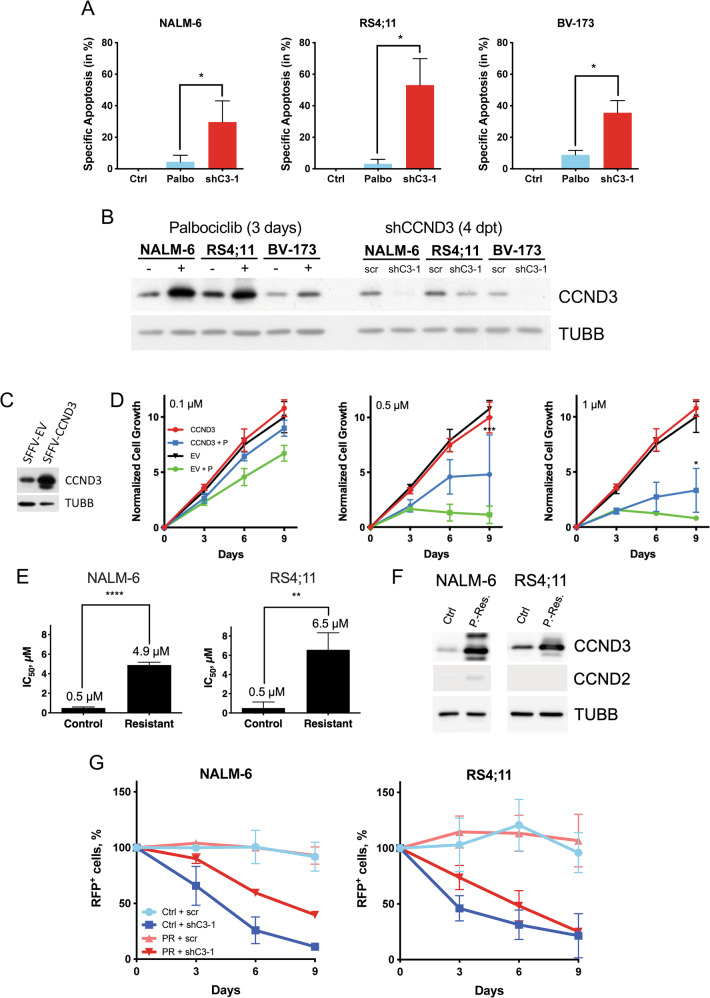
In T-ALL cell lines, the anti-apoptotic effect of CCND3 depends on the kinase activity of the CCND3-CDK6 complex [24]. To clarify if the same holds true in B-ALL, we treated the B-ALL cell lines NALM-6, RS4;11 and BV-173 with the highly specific CDK4/6 inhibitor palbociclib at a concentration of 1 µM, which has been shown to induce apoptosis in other B-and T-cell malignancies [25], and was approximately two-fold higher than the maximally achievable plasma concentration in patients [26]. Both palbociclib and CCND3 knockdown significantly decreased the number of cells in S-phase in NALM-6, RS4;11, and BV-173 cell lines (Supplementary Fig. 10). However, only CCND3 knockdown but not treatment with palbociclib induced strong apoptosis (Fig. 4A). Induction of apoptosis by Ccnd3 knockdown was also confirmed in an ex vivo BCRABL1+ mouse B-ALL model (Supplementary Fig. 11).
Fig. 4. CCND3 protects B-ALL from apoptosis in a kinase-independent manner and confers resistance to palbociclib.
A B-ALL cell lines were treated with 1 µM palbociclib for 3 days or transduced with either a scrambled (control) or a CCND3-shRNA containing vector, harboring a fluorescent marker (RFP). The RFP+ cells were sorted by FACS 4 days post transduction and cultured for 3 more days. Apoptosis was analyzed by PI/annexin V staining, with help of flow cytometry. Data shown as mean ± SD, n = 3. Statistical analysis was performed by student’s t-test. *=p < 0.05 B CCND3 protein expression level was analyzed by immunoblot 3 days after treatment with 1 µM palbociclib and 4 days after transduction with either scrambled control or CCND3 targeting shRNA. Image is representative of n = 3. C NALM-6 cells were transduced with SFFV-CCND3 vector expressing wtCCND3 and a fluorescent marker (EGFP) or by control vector (EV). CCND3 overexpression was controlled by immunoblot. D Cell growth of CCND3 overexpressing NALM-6 cells and EV control expressing NALM-6 cells was determined by calculating GFP+ cells relative to the absolute cell number of living cells. Data shown as mean ± SD, n = 3. Statistical analysis was performed by Student’s t-test. *=p < 0.05, ***=p < 0.005. E, F Cells were cultured in the presence of increasing (from 0.01 to 1.7 µM) concentrations of palbociclib for 3 months. After reaching 1.7 µM concentration with cells growing at a speed comparable to the vehicle-treated control cells, their sensitivity to palbociclib was measured using MTT Assay (E) and CCND3 and CCND2 expression at the same time point was measured by immunoblot (F). Image is representative for n = 3. Statistical analysis was performed by student’s t-test. **=p < 0.01, ****=p < 0.001. G Control (Ctrl) and palbociclib-resistant (PR) NALM-6 and RS4;11 cells were transduced with either a scrambled shRNA (scr) or shRNA against CCND3 (shC3), co-expressing RFP. The proportion of RFP+ cells was monitored throughout 9 days, starting 4 days post transduction (Day 0). Data shown as mean ± SD, n = 3.
We concluded that other mechanisms than inhibition of CDK4/6 kinase activity are responsible for the pro-apoptotic effect of CCND3 depletion in B-ALL.
Increase of CCND3 expression contributes to the resistance to palbociclib
We noticed that short treatment with palbociclib strongly increased the expression of CCND3 protein in all cell lines (Fig. 4B). CCND1 protein remained undetectable in all cell lines, whereas CCND2 was slightly upregulated in NALM-6, not detectable in RS4;11 and increased in BV-173 (Supplementary Fig. 12A). In order to assess if the upregulation of CCND3 represents an adaptation mechanism, we overexpressed CCND3 in NALM-6 cells (Fig. 4C) and incubated them in presence of increasing concentrations of palbociclib (Fig. 4D). Ectopic CCND3 expression did not affect the proliferation parameters of control cells, but significantly decreased sensitivity of NALM-6 to the growth-inhibitory effects of palbociclib.
In order to interrogate whether CCND3 upregulation poses a mechanism for acquired resistance to palbociclib, NALM-6 and RS4;11 were cultured in the presence of gradually increased concentrations of palbociclib. In due course of the selection, the IC50 of palbociclib increased from 0.5 to 4.9 µM in NALM-6 and from 0.5 to 6.9 µM in RS4;11, respectively (Fig. 4E). The palbociclib-resistant NALM-6 and RS4;11 cells showed highly elevated expression levels of CCND3 and FOXO1, much higher than after short-term treatment (Fig. 4F and Supplementary Fig. 12B). CCND2 protein was slightly increased in resistant NALM-6 but not detectable in RS4;11 (Fig. 4F). Importantly, B-ALL cells resistant to palbociclib were still sensitive to the growth-inhibitory effect of CCND3 knockdown (Fig. 4G).
Thus, upregulation of CCND3 contributes to the acquisition of resistance to palbociclib in B-ALL.
CDK8 repression contributes to the cytotoxic effects of CCND3 knockdown
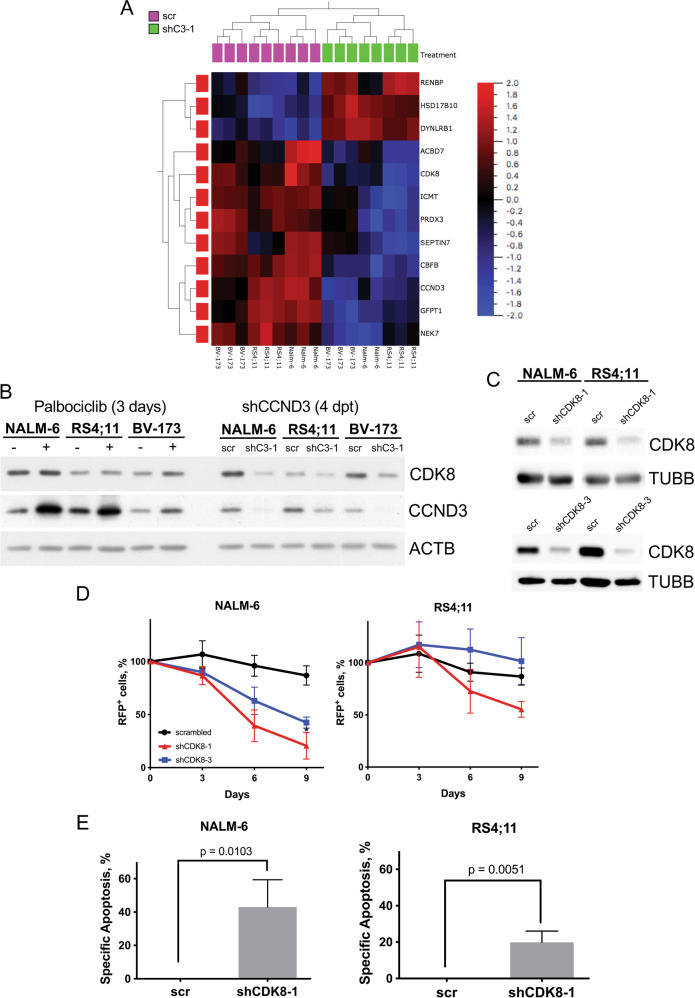
To identify mechanisms responsible for the CDK4/6 kinase-independent anti-apoptotic function of CCND3, we compared the effects of palbociclib and CCND3 knockdown at transcriptome level in three B-ALL cell lines of different subtypes using RNA-sequencing (Supplementary Table S1). Using hierarchical clustering and Venn analysis, we identified genes differentially regulated by CCND3 knockdown and palbociclib treatment throughout all three cell lines (Fig. 5A and Supplementary Fig.13). Among the genes repressed only by CCND3 knockdown were genes regulating metabolism (ABCD7 [27], RENBP [28], SEPTIN7 [29]), oxidative stress (PRDX3 [30], HSD17B10 [31]) and genes involved in oncogenic processes (NEK7 [32], DYNLRB1 [33], ICMT [34], CBFB [35], GFPT1 [36], CDK8 [37]). Given that the non-canonical kinase CDK8 was recently identified as an essential transcriptional regulator in BCR-ABL1+ B-ALL [37], we focused on this gene. Since the role of CDK8 was investigated in detail only in the oncogenic program of BCR-ABL1+ B-ALL, we addressed the function of CDK8 in other B-ALL subtypes. Using immunoblot, we corroborated CDK8 protein downregulation by CCND3 knockdown but not by treatment with palbociclib in all three cell lines (Fig. 5B).
Fig. 5. CDK8 downregulation contributes to the cytotoxicity induced by CCND3-knockdown.
A Heatmap generated from RNA-Seq data performed for the three B-ALL cell lines NALM-6, RS4;11 & BV-173, which were either treated with 1 µM palbociclib for 3 days or sorted after 4 days post lentiviral transduction with either a shRNA targeting CCND3 (shC3-1) or control vector (scr). The heatmap represents the result of unsupervised hierarchical clustering of the 12 genes that were exclusively regulated only by CCND3-knockdown in all cell lines. (Supplementary Fig. 13). q = 0.1, p < 0.05. B CDK8 protein levels are downregulated by CCND3-knockdown, not by palbociclib treatment. CDK8 levels were analyzed by immunoblot either after three days of 1 µM palbociclib treatment or four days post transduction with a CCND3-targeting shRNA. n = 3, image representative. C ShRNA-dependent CDK8 knockdown was controlled by immunoblot, image representative of n = 3. D Both CDK8-targeting shRNAs reduce cell fitness in a competitive growth assay (see legend to Fig. 3B). Data shown as mean ± SD, n = 3. E CDK8-knockdown by shRNA induces significant apoptosis in B-ALL cell lines. Cells were transduced as described in the legend of Fig. 5D and sorted four days after transduction for 100% RFP+ cells, then cultured for two more days. Specific apoptosis was determined by staining with PI and for Annexin-V, measured via flow cytometry. Data shown as mean ± SD, n = 3 Statistical analysis was performed by Student’s t-test.
Since it has been shown that loss of CDK8 protein but not inhibition of its enzymatic activity is cytotoxic for BCRABL1+ B-ALL [37], we assessed the sensitivity of NALM-6 and RS4;11 cells to the CDK8 inhibitor SEL120. Both cell lines did not differ in sensitivity to SEL120 compared to the non-sensitive BCR-ABL1+ cell line BV-173 (Supplementary Fig. 14) [37].
To investigate whether loss of CDK8 protein is detrimental to B-ALL cells regardless of the underlying genotype, we repressed CDK8 in NALM-6 and RS4;11 by using two shRNAs (Fig. 5C). Depletion of CDK8 significantly decreased the fitness of cells in a competitive growth assay (Fig. 5D). Importantly, CDK8 knockdown induced apoptosis in both cell lines (Fig. 5E).
We concluded that CCND3 regulates CDK8 transcription and that downregulation of CDK8 might contribute to the anti-apoptotic effect of CCND3 in B-ALL.
Discussion
We demonstrated that CCND3 is indispensable for the maintenance of B-ALL, regardless of the underlying oncogenic genotype. The anti-apoptotic effect of CCND3 did not depend on the catalytic activity of the CCND3-CDK4/6 complex. Furthermore, we identified a role of CCND3 in the transcription of the anti-apoptotic kinase CDK8 and revealed the role of FOXO1 in the regulation of CCND3 transcription.
We found that CCND3 is the highest expressed D-cyclin throughout all major subtypes of B-ALL. Moreover, the data of genome wide loss-of-function screening indicate the superior role of CCND3 specifically in oncogenic programs of B-ALL subtypes. This is surprising, since so far, CCND2 has widely been considered to be the most important D-cyclin during B-cell development and in B-ALL. CCND2 is a direct transcriptional target of the proto-oncogene MYC, a key driver of B-lymphoblast expansion and oncogenic signaling in B-ALL [38]. CCND2 is abundant throughout all stages of early B-cell development [9]. In non-neoplastic pro- and pre-B-cells, survival signals converge in the activation of MYC transcription and its stabilization.
In B-ALL, the essential role of MYC was especially well studied in BCR-ABL1+ cases [14]. The BCR-ABL1 oncogene activates transcription only of Ccnd2 via MYC, but not Ccnd3 as it has been shown in murine pre-B-cells [7]. Indeed, our analysis has demonstrated that BCR-ABL1+ B-ALL expressed higher levels of CCND2 than other subtypes.
CCND1 has been shown to be expressed in very low levels in pro- and pre-B-cells [9]. Indeed, our analysis demonstrated that this carries over to their malignant counterparts, since no B-ALL cell line showed detectable CCND1 protein expression. Overall, even in BCR-ABL1+ cases, CCND3 was always the highest expressed D-cyclin.
Importantly, by using loss-of-function experiments we demonstrated an essential role of CCND3 in cell lines and PDXs representing different subtypes of B-ALL including BCR-ABL1+ ones. The observed cytotoxic effect of CCND3 depletion indicates a unique role of CCND3, which depletion cannot be compensated by the relatively high CCND2 levels of BCR-ABL1+ B-ALL.
We demonstrated that apoptosis in B-ALL cell lines can only be efficiently induced by genetic depletion of CCND3 but not by inhibition of the CCND3-associated kinases CDK4/6 by palbociclib. This finding indicates principal differences of B-ALL to T-ALL, where palbociclib induces apoptosis by repressing the pentose phosphate pathway, thereby inducing oxidative stress [24]. Our transcriptome analysis did not reveal enrichment of the oxidative stress signature neither after treatment with palbociclib nor after CCND3 knockdown. Interestingly, although it was documented that palbociclib is not able to induce apoptosis in B-ALL, genetic depletion, or a PROTAC-dependent degradation of CDK6, the predominantly expressed CDK that forms a holoenzyme with CCND3, induces cell death in BCRABL1+ B-ALL. This, together with our findings, indicates kinase-independent effects of the components of the CCND-CDK holoenzyme [39].
We conclude that CCND3 indeed possesses unique, non-canonical, essential functions in B-ALL that cannot be substituted by another D-type cyclin, as previously suggested by others [40, 41].
Comparing the transcriptome changes induced by CCND3 knockdown versus palbociclib, we identified CDK8 among the genes exclusively repressed by CCND3 knockdown and demonstrated that knockdown of CDK8 induces apoptosis in B-ALL cell lines harboring ETV6-PDGFRB and MLL-AF4 translocations. It has been demonstrated that the pro-survival effects of CDK8 include facilitation of glycolysis, activation of STAT1 and STAT5 oncogenic signaling, and activation of mTORC1 [37]. The sensitivity of BCR-ABL1+ B-ALL to genetic and PROTAC-mediated CDK8 depletion has been already shown to particularly depend on subsequent inactivation of mTORC1 activity [37]. Our data indicate a role of CCND3 in the regulation of CDK8 transcription and show that the anti-apoptotic effect of CDK8 is not limited to the BCR-ABL1+ subtype.
Although CCND3 is mostly known as a canonical component of the CCND-CDK4/6 holoenzyme complex, which inactivates RB1 and thereby activates E2F-dependent transcription, this mechanism most possibly is not involved in the regulation of CDK8 transcription. Even relatively high concentrations of palbociclib which we used did not influence CDK8 expression. At the same time, there is much evidence on the involvement of CCND3 in the regulation of transcriptional activity independently of E2Fs. In particular, CCND3 binds to the nuclear matrix, restricting access to multiple genes including the Vk locus [41, 42]. CCND3 can also regulate the activity of transcription factors such as Androgen Receptor (AR) or RUNX1 by direct binding to them, thereby modulating their activity [43, 44].
Thus, direct transcriptional regulation might be instrumental in the anti-apoptotic effect of CCND3 in B-ALL.
We identified increased CCND3 expression as an adaptation mechanism facilitating the development of resistance to palbociclib treatment. The increase of CCND3 protein expression after treatment with palbociclib might in part be explained by a paradoxical stabilization of CCND3-CDK4/6 complexes [45]. However, we observed an increase of CCND3 expression not only at protein, but also at mRNA levels, indicating involvement of transcriptional mechanisms.
Interestingly, in a KRAS-driven non-small cell lung cancer mouse model, ERK-induced concomitant upregulation of Ccnd1, as well as Ccnd3, was observed, rendering the tumor resistant to palbociclib [46]. Taking this into account, we cannot exclude that the observed increase of CCND2 protein in BV-173 after treatment with palbociclib represents an additional adaptation mechanism. In T-ALL, upregulation of CCND3 after treatment with palbociclib was also observed, albeit not further explained [24].
We identified FOXO1 as a direct transcriptional activator of CCND3. This finding is not absolutely unexpected, since FOXO1 has been shown to directly bind to the Ccnd3 promoter and activate its transcription in murine pancreatic cells [16]. In early B-cells, PAX5 has been considered as a main activator of CCND3 transcription [47]. Interestingly, FOXO1 is a negative regulator of PAX5 in healthy B-cell precursors, inhibiting proliferation and inducing differentiation [48].
On the contrary, the direct FOXO1 transcription target EBF1, which, together with FOXO1 comprises a positive feedback loop, also activates CCND3 transcription [49]. This is in line with ours and others data, showing the essential role of FOXO1 for the maintenance and proliferation of B-ALL [5, 7], and in pro-B-cells where FOXO1 knockdown induces growth arrest and apoptosis [50].
Apart from PAX5 and EBF1, MYC and E2F might also activate CCND3 transcription [18, 51]. However, our functional analysis showed that inhibition of neither MYC with imatinib nor E2F with palbociclib negatively impacted CCND3 transcription, as long as FOXO1 is present, which was upregulated by both treatments.
Of note, unstimulated germinal center B-cells express high levels of FOXO1 as well as CCND3, but both are downregulated upon stimulation of the BCR, simultaneously with MYC upregulation. Moreover, forced activation of MYC and thereby activation of CCND2 transcription was not able to overcome the growth arrest after CCND3 deletion in germinal center B-cells [52].
Taken together, we conclude that although transcriptional regulation of CCND3 expression is complex and requires cooperative interaction of multiple factors, involvement of FOXO1 is essential for complete CCND3 transcription, particularly in B-ALL.
Conclusively, we demonstrated an essential role of CCND3 in the maintenance of B-ALL of major genetic subtypes using different in vitro and ex vivo models. Moreover, we revealed the mechanistic basis of CCND3 downregulation after FOXO1 depletion by showing direct binding of FOXO1 to the CCND3 promoter, activating its transcription. Furthermore, we revealed that the anti-apoptotic effect of CCND3 does not depend on kinase activity. Lastly, we identified CDK8 as an anti-apoptotic, transcriptionally regulated target of CCND3, providing further incentive for therapeutic intervention by CDK8 degradation or transcriptional inhibition.
Materials and methods
Additional and detailed information on all experimental procedures and reagents is provided in Supplementary Materials and Methods.
Data mining
Publicly available RNA-expression data were mined by using GENEVESTIGATOR software. Experiment IDs are listed in Supplementary Table 1.
Cell culture
B-ALL cell lines NALM-6, RS4;11, BV-173, REH, TOM-1, NALM-20, SUP-B15, KOPN-8, and EU-3/697, the classical Hodgkin lymphoma (cHL) cell line L428 and Burkitt lymphoma cell line RAMOS, HEK293T, and LentiX cells were purchased from DSMZ (Braunschweig, Germany). The B-ALL cell line 018Z was provided by Meyer L-H., (Ulm University, Germany). Details for the human cell lines used are listed in Supplementary Table 2. Cre-ERT2 and ERT2 murine pre-B-cells homozygous for loxP flanked Foxo1 (Foxo1fl/fl) transformed with BCR-ABL1 were a gift of Jumaa H. (Ulm University, Germany) [7]. The B-ALL patient-derived xenografts (PDX) JFK125R, PDX2, and BLQ5 were a kind gift of Müschen M. (Yale School of Medicine, USA).
Immunoblot and qRT-PCR
Immunoblot and qRT-PCR were done as described previously [5]. Primers used for qRT-PCR are listed in Supplementary Table 3. Antibodies used for immunoblot are listed in Supplementary Table 4.
Luciferase assay, EMSA, and ChIP
Primers used for cloning of the luciferase reporter constructs are listed in Supplementary Table 5. Oligos used for EMSA are listed in Supplementary Table 6. Primers for qRT-PCR analysis of ChIP are listed in Supplementary Table 7.
Vectors and lentiviral transduction
Lentiviral transduction was done as described previously [5]. Sequences used for cloning of the shRNA constructs are listed in Supplementary Table 8. CRISPR/Cas9 gRNA sequences are listed in Supplementary Table 9.
Flow cytometry, cell sorting, cell cycle, and apoptosis measurement
Growth dynamics, cell sorting, cell cycle analysis, and apoptosis measurement of lentivirally transduced and treated cells were performed as described previously [5].
IC50 determination
The sensitivities of the cell lines to the CDK4/6 inhibitor palbociclib and the CDK8 inhibitor SEL120 were assessed by MTT assay as we described previously [5].
Data analysis
Data were analyzed by two-tailed Student’s t-test analysis and by one-way ANOVA using GraphPad PRISM (GraphPad Software, San Diego, CA).
Supplementary information
Acknowledgements
The authors thank Anita Kick for excellent technical assistance and Franziska Gehringer (Institute of Physiological Chemistry, Ulm University) for fruitful discussions. Further, the authors want to acknowledge Nina Ushmorova (Institute of Pathology, Ulm University) for conducting STR analysis of the cell lines used in this study.
Author contributions
Performed research and analyzed data: FK. Designed research and wrote the manuscript: FK, AU, and TW. Provided murine B-ALL cells and helped with the experimental design: HA and HJ. Provided experimental support and helped with the experimental design: MV. Provided PDX cells, helped with experimental design: MM. Conducted STR analysis, quality control of RNA-seq samples and helped with experimental design: RM. All authors reviewed and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The RNA-sequencing datasets generated and analyzed in this study are available at NCBI GEO database (https://www.ncbi.nlm.nih.gov/), accession number GSE178660.
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Thomas Wirth, Email: thomas.wirth@uni-ulm.de.
Alexey Ushmorov, Email: alexey.ushmorov@uni-ulm.de.
Supplementary information
The online version contains supplementary material available at 10.1038/s41389-021-00377-0.
References
- 1.Bethesda M. SEER Cancer Statistics Review, 1973–2013 SEER Cancer Statistics. National Cancer Institute. 2016. https://seer.cancer.gov/archive/csr/1975_2011/results_single/sect_28_table.03.pdf%0A, https://seer.cancer.gov/archive/csr/1975_2013/%0A, http://seer.cancer.gov/csr/1973_1996/.
- 2.Rowe JM. Prognostic factors in adult acute lymphoblastic leukaemia. Br J Haematol. 2010;150:389–405. doi: 10.1111/j.1365-2141.2010.08246.x. [DOI] [PubMed] [Google Scholar]
- 3.Larson RA, Dodge RK, Burns CP, Lee EJ, Stone RM, Schulman P, et al. A five-drug remission induction regimen with intensive consolidation for adults with acute lymphoblastic leukemia: Cancer and leukemia group B study 8811. Blood. 1995;85:2025–37. [PubMed] [Google Scholar]
- 4.Mohseni M, Uludag H, Brandwein JM. Advances in biology of acute lymphoblastic leukemia (ALL) and therapeutic implications. Am J Blood Res. 2018;8:29–56. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F, Demir S, Gehringer F, Osswald CD, Seyfried F, Enzenmüller S, et al. Tight regulation of FOXO1 is essential for maintenance of B-cell precursor acute lymphoblastic leukemia. Blood. 2018;131:2929–42. doi: 10.1182/blood-2017-10-813576. [DOI] [PubMed] [Google Scholar]
- 6.Köhrer S, Havranek O, Seyfried F, Hurtz C, Coffey GP, Kim E, et al. Pre-BCR signaling in precursor B-cell acute lymphoblastic leukemia regulates PI3K/AKT, FOXO1 and MYC, and can be targeted by SYK inhibition. Leukemia. 2016;30:1246–54. doi: 10.1038/leu.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelrasoul H, Vadakumchery A, Werner M, Lenk L, Khadour A, Young M, et al. Synergism between IL7R and CXCR4 drives BCR-ABL induced transformation in Philadelphia chromosome-positive acute lymphoblastic leukemia. Nat Commun. 2020;11:3194.. doi: 10.1038/s41467-020-16927-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimova DK, Dyson NJ. The E2F transcriptional network: Old acquaintances with new faces. Oncogene. 2005;24:2810–26. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 9.Cooper AB, Sawai CM, Sicinska E, Powers SE, Sicinski P, Clark MR, et al. A unique function for cyclin D3 in early B cell development. Nat Immunol. 2006;7:489–97. doi: 10.1038/ni1324. [DOI] [PubMed] [Google Scholar]
- 10.Cato MH, Chintalapati SK, Yau IW, Omori SA, Rickert RC. Cyclin D3 is selectively required for proliferative expansion of germinal center B cells. Mol Cell Biol. 2011;31:127–37. doi: 10.1128/MCB.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sicinska E, Aifantis I, Le Cam L, Swat W, Borowski C, Yu Q, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–61. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 12.Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–51. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–33. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng H, Hurtz C, Lenz KB, Chen Z, Baumjohann D, Thompson S, et al. Self-enforcing feedback activation between BCL6 and Pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell. 2015;27:409–25. doi: 10.1016/j.ccell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kode A, Mosialou I, Silva BC, Rached MT, Zhou B, Wang J, et al. FOXO1 orchestrates the bone-suppressing function of gut-derived serotonin. J Clin Invest. 2012;122:3490–503. doi: 10.1172/JCI64906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T, Kim DH, Xiao X, Lee S, Gong Z, Muzumdar R, et al. Foxo1 plays an important role in regulating β-cell compensation for insulin resistance in male mice. Endocrinology. 2016;157:1055–70. doi: 10.1210/en.2015-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coller HA, Forman JJ, Legesse-Miller A. “Myc’ed messages”: Myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet Public Libr Sci. 2007;3:1319–24. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y, Yuan J, Huang M, Jove R, Cress WD. Regulation of the cyclin D3 promoter by E2F1. J Biol Chem. 2003;278:16770–6. doi: 10.1074/jbc.M212702200. [DOI] [PubMed] [Google Scholar]
- 19.Shin DJ, Joshi P, Hong SH, Mosure K, Shin DG, Osborne TF. Genome-wide analysis of FoxO1 binding in hepatic chromatin: Potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic Acids Res. 2012;40:11499–509. doi: 10.1093/nar/gks932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogel MJ, Xie L, Guan H, Tooze RM, Maier T, Kostezka U, et al. FOXO1 repression contributes to block of plasma cell differentiation in classical Hodgkin lymphoma. Blood. 2014;124:3118–29. doi: 10.1182/blood-2014-07-590570. [DOI] [PubMed] [Google Scholar]
- 21.Drexler HG, Macleod RA. History of leukemia-lymphoma cell lines. Hum Cell. 2010;23:75–82. doi: 10.1111/j.1749-0774.2010.00087.x. [DOI] [PubMed] [Google Scholar]
- 22.Stong RC, Korsmeyer SJ, Parkin JL, Arthur DC, Kersey JH. Human acute leukemia cell line with the t(4;11) chromosomal rearrangement exhibits B lineage and monocytic characteristics. Blood. 1985;65:21–31. [PubMed] [Google Scholar]
- 23.Chan LN, Murakami MA, Robinson ME, Caeser R, Sadras T, Lee J, et al. Signalling input from divergent pathways subverts B cell transformation. Nature. 2020;583:845–51. doi: 10.1038/s41586-020-2513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Nicolay BN, Chick JM, Gao X, Geng Y, Ren H, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature. 2017;546:426–30. doi: 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–20. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Mukai H, Naito Y, Yonemori K, Kodaira M, Tanabe Y, et al. Phase I study of palbociclib, a cyclin-dependent kinase 4/6 inhibitor, in Japanese patients. Cancer Sci. 2016;107:755–63. doi: 10.1111/cas.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanfray D, Caron A, Roy MC, Laplante M, Morin F, Leprince J, et al. Involvement of the Acyl-CoA binding domain containing 7 in the control of food intake and energy expenditure in mice. Elife. 2016;5:e11742.. doi: 10.7554/eLife.11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly TN, Li C, Hixson JE, Gu D, Rao DC, Huang J, et al. Resequencing study identifies rare renin-angiotensin-aldosterone system variants associated with blood pressure salt-sensitivity: The gensalt study. Am J Hypertens. 2017;30:495–501. doi: 10.1093/ajh/hpx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wasik AA, Polianskyte-Prause Z, Dong M-Q, Shaw AS, Yates JR, Farquhar MG, et al. Septin 7 forms a complex with CD2AP and nephrin and regulates glucose transporter trafficking. Mol Biol Cell. 2012;23:3370–9. doi: 10.1091/mbc.E11-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wonsey DR, Zeller KI, Dang CV. The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc Natl Acad Sci USA. 2002;99:6649–54. doi: 10.1073/pnas.102523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Chen S, Yu M, Ge C, Ren M, Liu B, et al. Deacetylation of HSD17B10 by SIRT3 regulates cell growth and cell resistance under oxidative and starvation stresses. Cell Death Dis. 2020;11:1–14. doi: 10.1038/s41419-020-02763-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L, Wang Z, Xu X, Wan Y, Qu K, Fan H, et al. Nek7 is overexpressed in hepatocellular carcinoma and promotes hepatocellular carcinoma cell proliferation in vitro and in vivo. Oncotarget. 2016;7:18620–30. doi: 10.18632/oncotarget.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raza A, Pandey MS, Jin Q, Mulder KM. km23-1/DYNLRB1 regulation of MEK/ERK signaling and R-Ras in invasive human colorectal cancer cells. Cell Biol Int. 2020;44:155–65. doi: 10.1002/cbin.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Zhu Y, Wang F, Zhou Y, Xia G, Xu W. ICMT contributes to hepatocellular carcinoma growth, survival, migration and chemoresistance via multiple oncogenic pathways. Biochem Biophys Res Commun. 2019;518:584–9. doi: 10.1016/j.bbrc.2019.08.094. [DOI] [PubMed] [Google Scholar]
- 35.Faber ZJ, Chen X, Gedman AL, Boggs K, Cheng J, Ma J, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48:1551–6. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, Lian H, Xie L, Yin N, Cui Y. LncRNA ELFN1-AS1 promotes esophageal cancer progression by up-regulating GFPT1 via sponging miR-183-3p. Biol Chem. 2020;401:1053–61. doi: 10.1515/hsz-2019-0430. [DOI] [PubMed] [Google Scholar]
- 37.Menzl I, Zhang T, Berger-Becvar A, Grausenburger R, Heller G, Prchal-Murphy M, et al. A kinase-independent role for CDK8 in BCR-ABL1+ leukemia. Nat Commun. 2019;10:4741.. doi: 10.1038/s41467-019-12656-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchner M, Müschen M. Targeting the B-cell receptor signaling pathway in B lymphoid malignancies. Curr Opin Hematol. 2014;21:341–9. doi: 10.1097/MOH.0000000000000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Dominici M, Porazzi P, Xiao Y, Chao A, Tang HY, Kumar G, et al. Selective inhibition of Ph-positive ALL cell growth through kinase-dependent and -independent effects by CDK6-specific PROTACs. Blood. 2020;135:1560–73. doi: 10.1182/blood.2019003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–92. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powers SE, Mandal M, Matsuda S, Miletic AV, Cato MH, Tanaka A, et al. Subnuclear cyclin D3 compartments and the coordinated regulation of proliferation and immunoglobulin variable gene repression. J Exp Med. 2012;209:2199–213. doi: 10.1084/jem.20120800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karki S, Kennedy DE, Mclean K, Grzybowski AT, Maienschein-Cline M, Banerjee S, et al. Regulated capture of Vκ gene topologically associating domains by transcription factories. Cell Rep. 2018;24:2443–56. doi: 10.1016/j.celrep.2018.07.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olshavsky NA, Groh EM, Comstock CES, Morey LM, Wang Y, Revelo MP, et al. Cyclin D3 action in androgen receptor regulation and prostate cancer. Oncogene. 2008;27:3111–21. doi: 10.1038/sj.onc.1210981. [DOI] [PubMed] [Google Scholar]
- 44.Peterson LF, Boyapati A, Ranganathan V, Iwama A, Tenen DG, Tsai S, et al. The hematopoietic transcription factor AML1(RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol Cell Biol. 2005;25:10205–19. doi: 10.1128/MCB.25.23.10205-10219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paternot S, Colleoni B, Bisteau X, Roger PP. The CDK4/CDK6 inhibitor PD0332991 paradoxically stabilizes activated cyclin D3-CDK4/6 complexes. Cell Cycle. 2014;13:2879–88. doi: 10.4161/15384101.2014.946841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haines E, Chen T, Kommajosyula N, Chen Z, Herter-Sprie GS, Cornell L, et al. Palbociclib resistance confers dependence on an FGFR-MAP kinase-mTOR-driven pathway in KRAS-mutant non-small cell lung cancer. Oncotarget. 2018;9:31572–89. doi: 10.18632/oncotarget.25803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, et al. Identification of Pax5 target genes in early B cell differentiation. J Immunol. 2008;180:1719–28. doi: 10.4049/jimmunol.180.3.1719. [DOI] [PubMed] [Google Scholar]
- 48.Abdelrasoul H, Werner M, Setz CS, Okkenhaug K, Jumaa H. PI3K induces B-cell development and regulates B cell identity. Sci Rep. 2018;8:1327. doi: 10.1038/s41598-018-19460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Györy I, Boller S, Nechanitzky R, Mandel E, Pott S, Liu E, et al. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 2012;26:668. doi: 10.1101/gad.187328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–98. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 2010;30:4149–58. doi: 10.1128/MCB.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramezani-Rad P, Chen C, Zhu Z, Rickert RC. Cyclin D3 governs clonal expansion of dark zone germinal center B cells. Cell Rep. 2020;33:108403.. doi: 10.1016/j.celrep.2020.108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing datasets generated and analyzed in this study are available at NCBI GEO database (https://www.ncbi.nlm.nih.gov/), accession number GSE178660.