Abstract
Background
A major goal of spermiation is to degrade the apical ectoplasmic specialization (ES) junction between Sertoli cells and elongating spermatids in preparation for the eventual disengagement of spermatids into the lumen. E3 ubiquitin ligases mediate the process of ubiquitination and the subsequent proteasomal degradation, but their specific role during spermiation remains largely unexplored.
Methods
Ankyrin repeat and SOCS box protein 17 (Asb17)-knockout mice were generated via a CRISPR/Cas9 approach. Epididymal sperm parameters were assessed by a computer-assisted sperm analysis (CASA) system, and morphological analysis of testicular tissues were performed based on histological and immunostaining staining, and transmission electron microscopy (TEM). The interactions between ASB17 and Espin (ESPN) were predicted by HawkDock server and validated through protein pull-down and immunoprecipitation assays.
Results
We report that ASB17, an E3 ligase, is required for the completion of spermiation and that mice lacking Asb17 are oligozoospermic owing to spermiation failure. ASB17-deficient mice are fertile; however, spermatids exhibit a disorganized ES junction, resulting in retention within the seminiferous epithelium. Mechanistically, ASB17 deficiency leads to excess accumulation of ESPN, an actin-binding essential structural component of the ES. We determined that ASB17 regulates the removal of the ES through ubiquitin mediated protein degradation of ESPN.
Conclusions
In summary, our study describes a role for ASB17 in the regulation of cell-cell junctions between germ cells and somatic cells in the testis. These findings establish a novel mechanism for the regulatory role of E3 ligases during spermatogenesis.
Keywords: Spermiation, ectoplasmic specialization (ES), E3 ligase, ASB17, ESPN
Introduction
Spermatogenesis is a well-organized process that encompasses the proliferation of spermatogonia, meiosis of spermatocytes, and differentiation of spermatids to generate mature spermatozoa (1). Ubiquitination, a major posttranslational modification in eukaryotes that determines protein stability and activity, controls cell remodeling and protein turnover during multiple stages of spermatogenesis, from the gonocyte to the differentiated spermatid stage (2). The outcome of a recent study provides evidence for the extensive involvement of ubiquitin ligases (E3) in mouse spermatogenesis: at least 29 different E3 enzymes are expressed in the testis and execute a diverse range of functions at different stages of spermatogenesis (3). In addition, definitive roles of E3 enzymes during spermatogenesis have also been shown using mouse knock-out (KO) studies. For instance, disruption of E3 ubiquitin-protein ligase HUWE1 depleted spermatogonial pools and caused a Sertoli cell-only histopathology (4); loss of ring finger protein 216 (RNF216) or ubiquitin protein ligase E3 component n-recognin 2 (UBR2) led to meiotic arrest and spermatocyte apoptosis (5,6); and deficiency of PHD finger protein 7 (PHF7) or ring finger protein 8 (RNF8) caused aberrant nuclear condensation in elongated spermatids (7-9). In our previous study, we found that E3 ubiquitin-protein ligase makorin-2 (MKRN2)-deficient mice exhibit spermiation failure and oligozoospermia (10), raising the possibility that spermiation also depends on ubiquitin-based protein degradation as well.
Spermiation, also known as sperm release, is a complex process by which mature spermatids detach from the adluminal compartment and are released into the tubule lumen (11). During spermatogenesis, germ cells are transported by Sertoli cells, a somatic cell type found in spermatogenic tubules, across the seminiferous epithelium from the basal to the adluminal compartment, so that mature spermatozoa can be released during spermiation and enter the epididymis for further maturation (12). Yet, during this process the developing spermatids must remain attached to the epithelium via a specialized Sertoli-germ cell adherens junction (AJ) named the apical ectoplasmic specialization (ES) (13). The ES is an actin-based anchoring junction found uniquely in the testis at two distinct sites, termed the basal ES and the apical ES. The basal ES describes the interface between Sertoli cells at the blood-testis barrier, whereas the interface between Sertoli cells and elongating spermatids is designated the apical ES (14). Prior to spermiation, the apical ES, as characterized by hexagonally-packed actin bundles sandwiched between the Sertoli cell membrane and the cisternae of endoplasmic reticulum, surrounds the acrosome of an elongating spermatid and mechanically grasps the nuclei of the spermatid (15,16). As spermiation proceeds, the apical ES structure is disassembled and replaced by tubulobulbar complexes (TBCs) (17). These changes are facilitated by the endocytosis of the ES proteins so that the target proteins can be degraded or recycled to assemble new ES (18,19). However, the molecular regulators that mediate the dynamic changes of apical ES during spermiation remain elusive.
ASB17 is one of 18 members of the ankyrin repeat and suppressor of cytokine signaling (SOCS) box containing protein (ASB) family, for which all of the ASB proteins contain two functional domains, a C-terminal SOCS box and an N-terminal ankyrin repeat (20). The SOCS box recruits elongin B/C, Cullin-2/5, and RING-box proteins Rbx1/2 to form the Elongin–Cul–SOCS (ECS) E3 ubiquitin ligase complex, promoting the target proteins for polyubiquitination and subsequent proteasomal degradation, while the ankyrin repeat directs substrate recognition (21). Expression profiling of ASB17 in 16 human tissues and 14 mouse tissues by Northern blot has shown that ASB17 is expressed exclusively in the testis, suggesting a potential role of ASB17 in spermatogenesis (22,23). Recently, using a gene KO approach, Yang et al., have shown that ASB17-deficient mice undergo normal testicular development and fertility (24). However, ASB17 deficiency leads to a significant reduction of apoptosis in germ cells and prevents spermatogonial apoptosis as induced by etoposide in vivo. Mechanistically, the authors found that ASB17 mediated the ubiquitylation and degradation of two anti-apoptotic factors, BCLW and MCL1 in vitro (24). Taken together, the above data suggests that ASB17 plays a minor but not crucial role during spermatogenesis.
Herein, we report that ASB17 acts as a molecular switch to regulate the ES junction assembly and spermiation. Specifically, we demonstrate that ASB17 interacts with the ES protein Espin (ESPN) and directs its ubiquitination in vivo. Our study proposes a novel mechanism for the regulatory role of E3 ubiquitin ligases during spermiation. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/tau-21-789).
Methods
Mice
Mice were maintained in a specific-pathogen-free environment at Nanjing Medical University. All animal experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and approved by the Animal Ethical and Welfare Committee of Nanjing Medical University (No. IACUC-2004020). To generate Asb17-KO mice, two sgRNAs were designed based on exon 1 of Asb17. The target sgRNA sequences were 5'-GTGCGAATACCATTCTGTACTGG-3' and 5'-GAAAGGTAATCCTGACTTCGTGG-3', respectively. Cas9 mRNA and sgRNAs were co- microinjected into zygotes of CD-1 mice and transferred into pseudo-pregnant recipients according to our previously reported procedure (25,26). The genotype of Asb17 was identified by PCR amplification and further Sanger sequencing using the following primers (forward 5'-CTGTGACTGAGGAATGGCAGCAT-3' and reverse 5'-GTATGGCGTAAGGCAAGGTCTGT-3').
Histological analysis
For light microscope assessment, samples were fixed in modified Davidson’s fluid, dehydrated, and paraffin embedded. Sections (5 μm-thick) were cut, mounted on glass slides, deparaffinized with xylene, and stained with hematoxylin-eosin (H&E). After dehydration through a series of graded ethanol baths and clearing in xylene, samples were mounted and viewed under a bright-field microscope (Axioskop 2 plus, Carl Zeiss, Oberkochen, Germany). For ultrastructural examination, 4% (vol/vol) glutaraldehyde-fixed testes were postfixed with 2% (wt/vol) OsO4 and embedded in Araldite. Sections (80 nm-thick) were analyzed using a transmission electron microscope (JEM-1410, JEOL, Tokyo, Japan).
Computer-assisted sperm analysis (CASA)
Sperm were collected from the cauda epididymis of adult mice and then incubated with a warm human tubal fluid culture medium containing 10% bovine serum albumin (BSA) for 5 min. Parameters including sperm concentration, motility, and progressive motility were analyzed using a Hamilton Thorne’s sperm analysis system (Beverly, MA, USA). Sperm morphology evaluation was carried out as described previously (27).
Fertility test
Adult males of Asb17 wild-type (WT, +/+), heterozygous (+/−), and KO (−/−) were continuously mated with WT females at a ratio of 1:2 for 3 months. The presence of vaginal plugs was recorded and the number of pups born were counted for subsequent statistical analyses.
Immunofluorescence
Immunofluorescence staining was performed as described previously (28,29). Testicular sections were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked by incubation in 3% hydrogen peroxide for 15 min at 37 °C. After antigen retrieval in sodium citrate buffer, sections were blocked and incubated overnight at 4 °C with primary antibodies (Table S1). After three washes with phosphate-buffered saline (PBS), sections were incubated with Alexa Fluor series secondary antibodies (Thermo Scientific, Waltham, USA) and images were captured on a Zeiss laser confocal microscope (LSM800, Carl Zeiss).
RNA isolation and Real-time quantitative PCR
Total RNAs from testicular tissues were extracted using an RNeasy Plus Micro Kit (74034, Qiagen, Duesseldorf, Germany) following the manufacturer’s protocol. cDNA was reverse transcribed using a PrimeScript RT reagent Kit (Vazyme, Nanjing, China) and then processed through a SYBR Green-based StepOne-Plus real-time PCR system (Applied Biosystems, Foster City, CA, USA) with 18S rRNA as a normalization control. Primers targeting Espn and 18S rRNA have been previously listed (26).
Terminal deoxynucleotidyl transferase-dUTP nick-end labeling (TUNEL)
Apoptotic cells were detected according to our previously described procedure (30). In brief, paraffin testicular sections were deparaffinized, rehydrated, and treated with proteinase K (10 µg/mL) for 10 min. Subsequently, the slides were reacted with BrightRed Labeling Buffer (Vazyme, Nanjing, China) for 1 h at 37 °C. After washing with PBS thrice, sections were stained with 4’,6-diamidino-2-phenylindole (DAPI) before being mounted. Images were then analyzed using a LSM800 confocal laser microscope (Carl Zeiss).
Protein pull-down and immunoprecipitation
The full length of mouse ASB17 protein was expressed as a 6*His fusion protein in E. coli using the pET-28a vector and affinity purified with a Ni-NTA Resin (TransGene, Beijing, China). The resin-ASB17 protein complex was incubated with testicular lysates on a roller for 30 min at 4 °C. Then the resin-protein complex was washed with 1X Wash Buffer (50 mM sodium phosphate, 300 mM NaCl, 0.01% Tween-20) and eluted with 1% SDS buffer for 15 min at 98 °C. Samples were stored at −20 °C before use for western blot analysis. For immunoprecipitation (IP) assays, testicular tissues were lysed with RIPA buffer (Beyotime, Nantong, China) and precleared with the protein A-Sepharose (Invitrogen, Carlsbad, CA, USA), then incubated with the anti-ESPN antibody overnight. The lysates were incubated with the protein A-Sepharose for 2 h at 4 °C, and the precipitants were washed with PBS, eluted with 1% SDS buffer, and resolved by SDS-PAGE.
Western blot
Testicular lysates and immunoprecipitated proteins were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane as previously reported (31,32). The membrane was blocked and incubated overnight at 4 °C with the indicated primary antibodies (Table S1). The membrane was washed and incubated at room temperature for 2 h with secondary antibodies. The signals of the detected proteins were visualized on the SuperSignal West Femto Chemiluminescent Substrate detection system (Thermo Scientific).
Statistical analysis
All experiments were independently repeated for three times. Quantitative data are presented as mean ± standard deviation (SD). Statistical analysis was examined using Student’s t-test or one-way ANOVA with P<0.05 (*).
Results
ASB17 is distributed at the apical ES junction of the mouse testis
We first analyzed the Asb17 mRNA transcript profile in multiple murine tissues. Bioinformatic analysis showed that Asb17 was predominantly expressed in the testis (Figure S1A), which was further validated using reverse transcription PCR (RT-PCR) analysis (Figure S1B). The above findings were highly consistent with previous work (22,23).
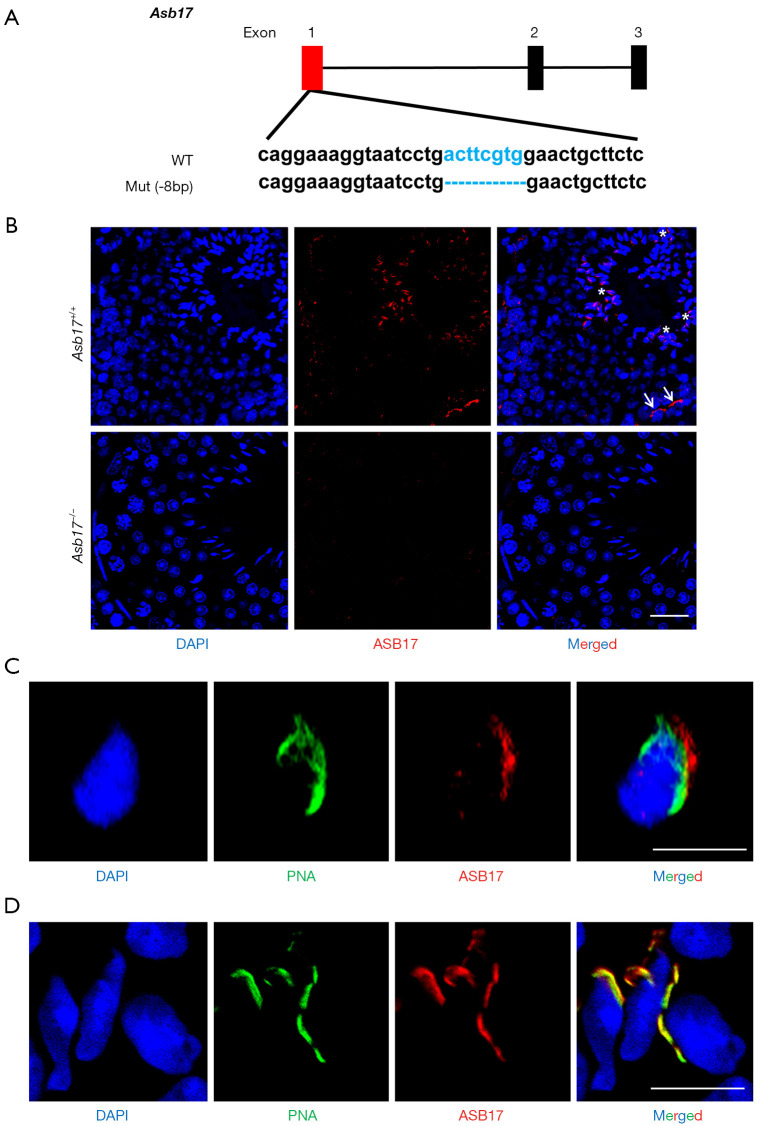
To better understand the role of Asb17 in testes, we generated mice with ubiquitous knockout of Asb17 (Asb17-KO) by using CRISPR/Cas9-based gene editing to cause an 8-bp deletion in exon 1 of Asb17 (Figure 1A). Asb17-KO mice were confirmed by PCR amplification and further Sanger sequencing (Figure S2). Moreover, immunofluorescence staining analysis of testes in wild-type (WT) showed an obvious distribution of ASB17 protein around spermatid heads, while in KO mice, the fluorescence signal was undetectable, suggesting an absence of ASB17 protein in KO testes (Figure 1B). To uncover the precise location of ASB17 relative to spermatid heads, we performed co-immunostaining of ASB17 and PNA (acrosome marker) (Figure 1C) or F-actin (ES junction maker) (Figure 1D) in elongated spermatids. The results revealed that ASB17 was located outside of the acrosome, but co-located with F-actin, indicating that ASB17 was distributed in the apical ES junction.
Figure 1.
Generation of Asb17−/− mice. (A) Schematic diagram of CRISPR/Cas9-mediated Asb17 gene editing. (B) Immunostaining of ASB17 in Asb17+/+ and Asb17−/− adult testes. In Asb17+/+ testes, ASB17 is mainly distributed around spermatid heads (asterisks). Meanwhile, in several spermatogonia, ASB17 can also be labeled (arrows). Scale bar, 20 µm. (C) Co-immunostaining of ASB17 and PNA in elongating spermatids. Scale bar, 10 µm. (D) Co-immunostaining of ASB17 and F-actin in elongating spermatids. Scale bar, 10 µm. ASB17, Ankyrin repeat and SOCS box protein 17; PNA, Peanut agglutinin.
Asb17−/− mice are fertile
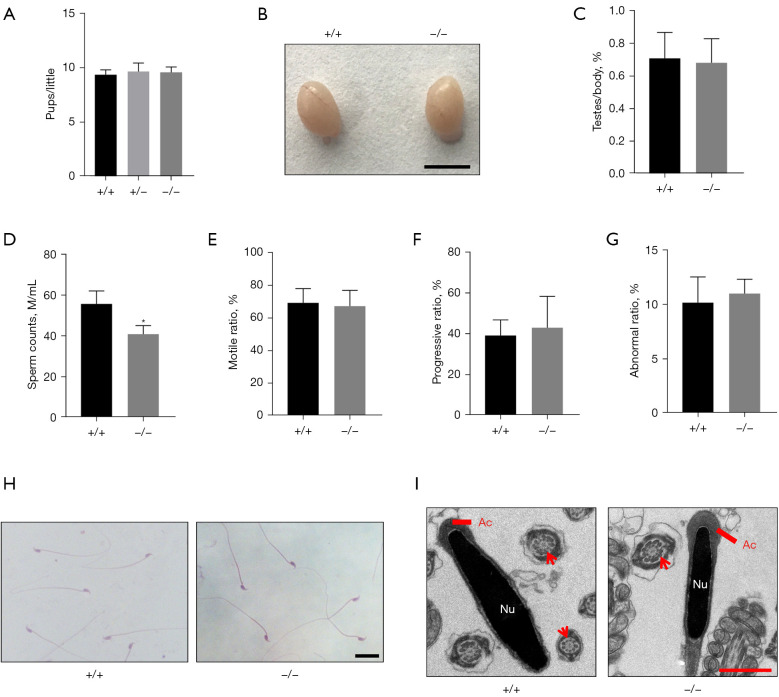
In line with previous findings (24), Asb17-KO mice were viable, and fertility tests revealed normal fertility in Asb17-KO adult males, compared with WT or heterozygous (+/−) mice (Figure 2A). The testes weight was comparable between Asb17-KO and WT mice (Figure 2B,2C). However, compared with WT, epididymal sperm analysis based on CASA revealed a significant decrease in sperm concentration in Asb17-KO (Figure 2D), whereas sperm motility was not altered (Figure 2E,2F). In addition, sperm morphology evaluation was also comparable between the two groups, as demonstrated by optical microscopy (Figure 2G,2H) or transmission electron microscopy (TEM) (Figure 2I). We therefore concluded that deficiency of ASB17 caused oligozoospermia in mice, although this had no effect on male fertility.
Figure 2.
Asb17−/− mice are fertile. (A) Fertility test of Asb17+/+, Asb17+/− and Asb17−/− males. n=3. P>0.05. (B) Gross morphology of representative testes from adult Asb17+/+ and Asb17−/− males. Scale bar: 5 mm. (C) Testis/body weight. n=4. P>0.05. (D) Sperm concentration in Asb17+/+ and Asb17−/− mice. n=3. *, P<0.05. (E) Sperm motility in Asb17+/+ and Asb17−/− mice. n=3. P>0.05. (F) Progressive ratio in Asb17+/+ and Asb17−/− mice. n=3. P>0.05. (G) Sperm abnormality in Asb17+/+ and Asb17−/− mice. n=3. P>0.05. (H) H&E staining of cauda epididymal sperm in Asb17+/+ and Asb17−/− mice. Scale bar, 20 µm. (I) TEM analysis of cauda epididymal sperm in Asb17+/+ and Asb17−/− mice. Axoneme with typical “9+2” microtubule are indicated with red arrows. Scale bar, 1 µm. Ac, acrosome; Nu, nucleus; ASB17, Ankyrin repeat and SOCS box protein 17.
Complete spermatogenesis, but spermiation failure in Asb17−/− mice
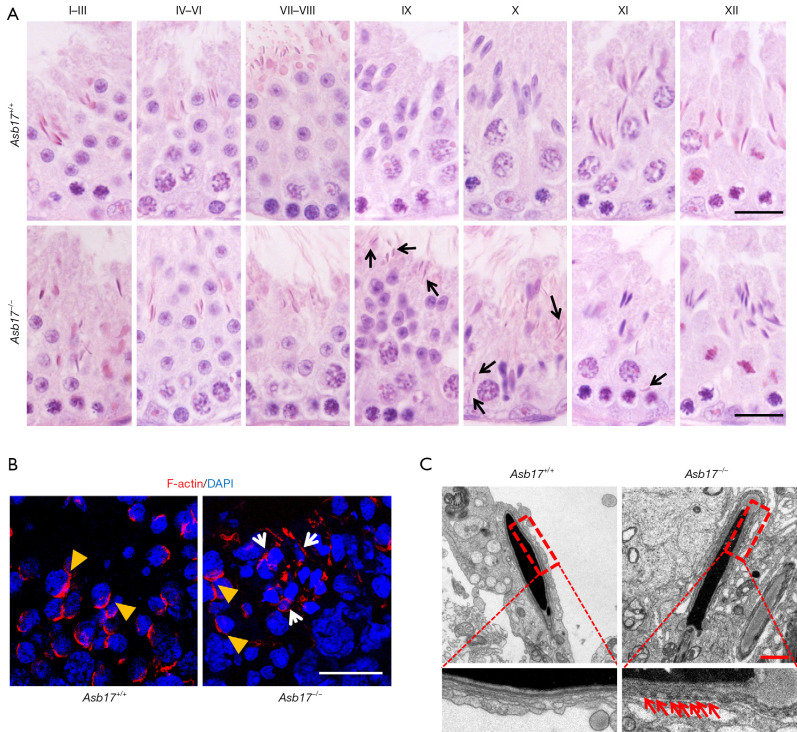
We next assessed germ cell development within the seminiferous epithelium using H&E staining. Similar to WT, Asb17-KO testes exhibited complete spermatogenesis, displaying all stages of the spermatogenetic cycle (Figure 3A). However, at stage IX of the seminiferous epithelium, when all mature spermatozoa have been released from the lumen in WT, many mature spermatozoa were still retained in the seminiferous epithelium of ASB17-deficient mice. Moreover, the retained spermatozoa persisted until stage XI (Figure 3A and Figure S3). Spermiation failure always results from a disorganized apical ES; we therefore performed F-actin staining in Asb17-KO and WT testes. As shown in Figure 3B, at stage IX, the apical ES first appeared as a highly ordered stack of filamentous (F)-actin-containing hoops around the apical region of the elongating spermatid nuclei of step 9; these were easily observed in both Asb17-KO and WT (Figure 3B). However, the F-actin bundles in Asb17-KO testes were unable to disassemble and were still surrounded by the retained mature spermatozoa (Figure 3B). Taken together, our data revealed that ASB17 is not required for the formation of the apical ES, but is required for its removal.
Figure 3.
Spermiation failure in Asb17−/− mice. (A) H&E staining of testis sections in Asb17+/+ and Asb17−/− mice. The epithelial cycle can be divided into 12 stages according to the morphological changes of the germ cells. At stages IX to XI, the retained mature spermatozoa were found in Asb17−/− testes as indicated with black arrows. Scale bar, 20 µm. (B) Phalloidin labeled F-actin staining of testis sections in Asb17+/+ and Asb17−/− mice. Elongating spermatids of step 9 are indicated by yellow arrowheads, while the retained mature spermatozoa in Asb17−/− testes are indicated with white arrows. Scale bar, 20 µm. (C) TEM analysis of mature spermatozoa in Asb17+/+ and Asb17−/− testes. The retained actin bundles in Asb17−/− mice are indicated with red arrows. Scale bar, 1 µm. ASB17, Ankyrin repeat and SOCS box protein 17.
To further determine the above findings, we examined the ultrastructure of the testes using TEM. In WT mature spermatozoa, the F-actin bundles were completely removed from the acrosome region, whereas in KO mature spermatozoa, the apical ES synchronously stretched along with the acrosome, as characterized by the presence of the F-actin bundles (Figure 3C). These data suggest that the aberrant F-actin organization in apical ES may be a major cause of the spermiation failure observed in Asb17-KO testes.
ASB17 interacts with ESPN
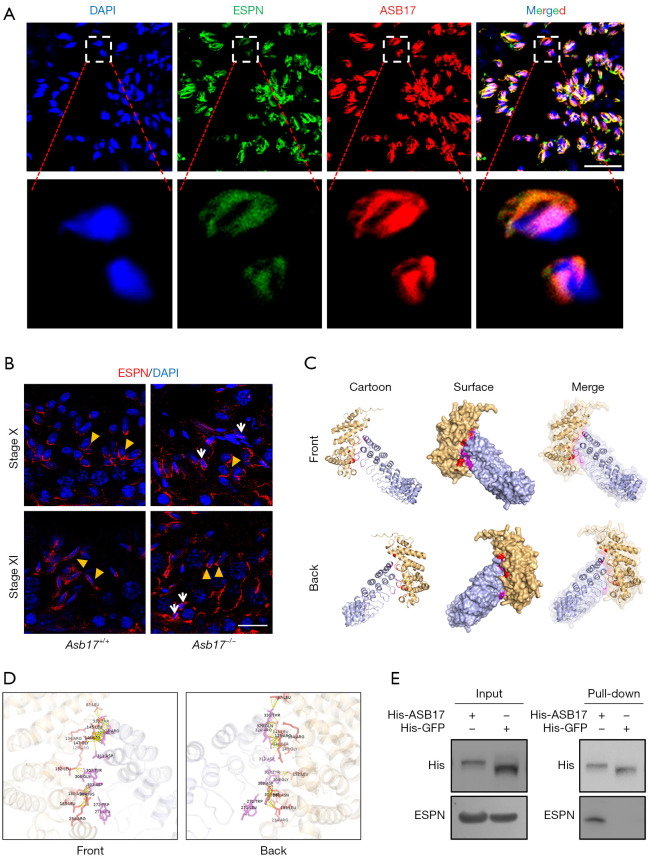
ESPN, or Espin, is an actin-binding protein that is responsible for the dynamic remodeling of actin bundles (33). ESPN-1, the largest isoform, was first identified in the Sertoli cell-spermatid ES junction, while other shorter spliced isoforms (ESPN-2B, ESPN-3A, and ESPN-4) were shown to be expressed in other actin bundle structures, such as hair cells and Purkinje cells (34,35). By co-immunostaining of ESPN and ASB17 in testes, we observed that ASB17 co-localized with ESPN at the apical ES (Figure 4A). Moreover, in Asb17-KO testes, we observed marked accumulation of ESPN in the retained mature spermatozoa (Figure 4B), suggesting that ASB17 was required for ESPN degradation during spermiation. To further elucidate the interactions between ASB17 and ESPN, we employed the HawkDock server (36) to predict ASB17-ESPN interactions. As shown in Figure 4C, the 3D structures of ASB17-ESPN complexes were predicted by HawkDock and visualized by PyMOL (version 2.1.0). The binding interface was formed by the residues 87-234 of ASB17 and bound to the residues 271-330 of ESPN (Figure 4D); the key residues for protein-protein interactions (PPIs) are provided in Table S2. According to protein sequence information on UniProt (https://www.uniprot.org/), the binding interface of ASB17 contained the ankyrin repeat that was responsible for substrate recognition, while the binding interface of ESPN also encompassed the ankyrin repeats that were reported for protein binding (37,38). To ascertain the bioinformatic results, we performed ASB17 protein pull-down assay. Indeed, the results revealed that ASB17 interacted with ESPN (Figure 4E). We therefore speculated that ESPN was a direct substrate of the E3 ligase ASB17.
Figure 4.
ASB17 interacts with ESPN. (A) Co-immunostaining of ASB17 and ESPN in testes. Scale bar, 20 µm. (B) Immunostaining of ESPN in Asb17+/+ and Asb17−/− testes. Elongating spermatids are indicated by yellow arrowheads, while the retained mature spermatozoa in Asb17−/− testes are indicated with white arrows. Scale bar, 20 µm. (C) Predicted 3D structures of ASB17–ESPN complexes. The cartoon mode represents the backbone and secondary structures. The surface mode represents the solvent accessible surface area. ESPN and ASB17 are labeled as orange and blue, respectively. The binding residues between ESPN and ASB17 are labeled as red and purple, respectively. (D) Magnification of the binding interface. (E) Protein pull-down assay. His-tagged ASB17 and its negative control (his-tagged GFP) were purified from E. coli with a Ni-NTA Resin and incubated with testicular lysates. The protein complexes were resolved by SDS-PAGE and detected by western blot analysis. ASB17, Ankyrin repeat and SOCS box protein 17; ESPN: Espin.
ASB17 ubiquitinates ESPN in vivo
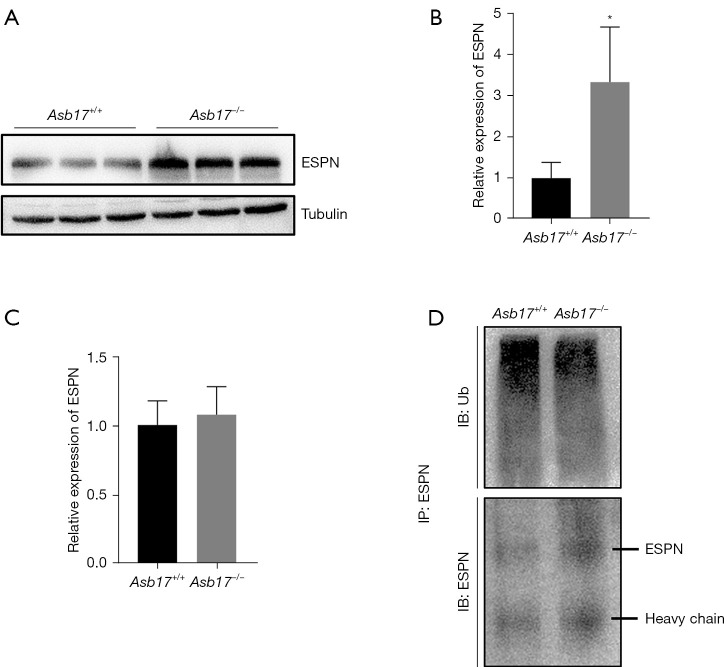
Western blot analysis confirmed markedly increased protein levels of ESPN in Asb17-KO testes (Figure 5A,5B), whereas ESPN mRNA levels in Asb17-KO testes levels were similar to the WT, according to q-PCR analysis (Figure 5C), suggesting that ASB17 destabilized RAI14 protein. Finally, we assessed ESPN ubiquitination in the testis. ESPN-IP followed by western blot analysis for total ubiquitin revealed significantly lower levels of poly-ubiquitinated ESPN in Asb17-KO testis compared with the WT (Figure 5D). Taken together, the results indicate that ASB17 ubiquitinates ESPN protein for its subsequent degradation.
Figure 5.
ASB17 poly-ubiquitinates ESPN. (A) Western blot analysis of ESPN in Asb17+/+ and Asb17−/− testes. (B) Quantification of (A). n=3. *, P<0.05. (C) Q-PCR analysis of Espn in Asb17+/+ and Asb17−/− testes. n=3. P>0.05. (D) Poly-ubiquitination analysis of ESPN in Asb17+/+ and Asb17−/− testes. Testicular lysates were subjected to IP with anti-ESPN antibody. The immunoprecipitates were resolved by SDS-PAGE and detected using anti-ESPN and anti-ubiquitin antibody, respectively. ASB17, ankyrin repeat and SOCS box protein 17; ESPN, Espin; Ub, ubiquitin.
Discussion
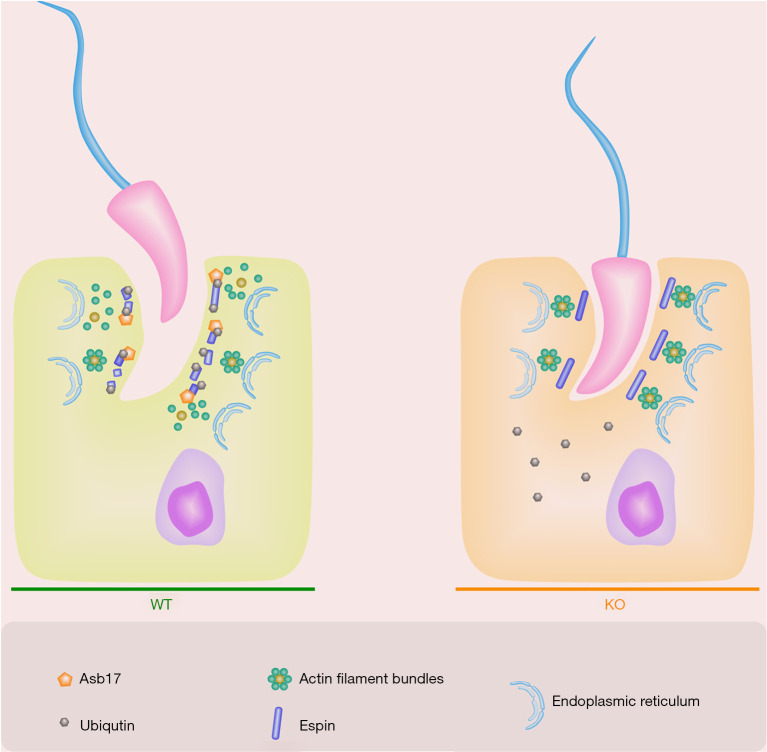
In addition to the known roles of ASB17 in promoting spermatogenic cell apoptosis (24), in the present study, we uncovered a novel mechanism in spermiation involving ASB17-mediated ubiquitination and degradation of the ES associated protein, ESPN. Consistent with the previous work (24), we found that Asb17-KO males were fertile and showed complete spermatogenesis. However, we also showed that Asb17-KO males exhibited oligozoospermia, which was considered to be a major result of spermiation failure found in Asb17-KO. Specifically, ASB17 supports the removal of the ES by adding degradation-linked poly-ubiquitin chains to the actin-binding protein ESPN. This ensures the dynamic remodeling of the ES during spermiation. ASB17 is therefore required to maintain normal sperm release, and its depletion results in spermiation failure owing to the aberrant accumulation of ESPN (Figure 6).
Figure 6.
Schematic illustration of ASB17 during spermiation. At the beginning of spermiation, ESPN is degraded through the ASB17-mediated ubiquitin proteasome system (UPS), thereby leading to apical ES disassembly and mature spermatozoa disengagement. The absence of ASB17 stabilizes ESPN and finally causes spermiation failure. ASB17, ankyrin repeat and SOCS box protein 17.
Our findings provide novel insight into the role of E3 ligases during spermiation. In eukaryotes, autophagy and ubiquitination are thought to be the two main pathways for protein degradation; therefore these pathways may contribute to the degradation of the ES (18,19,39). For instance, PDZ and LIM domain protein 1 (PDLIM1), a negative cytoskeleton organization regulator, can be degraded through the autophagy protein 5 (ATG5) or the ATG7-mediated autophagy-lysosome pathway. In Atg5-KO or Atg7-KO mouse testes, PDLIM1 was found to be significantly accumulated in the apical ES region, thus finally causing disruption of apical ES assembly and spermiation failure (40). Besides autophagy, growing evidence from in vitro and in vivo experiments exists, indicating that ubiquitination also directs dynamic remodeling of cell junctions (41). Studies by Liu and Lee show that ubiquitination regulates the bioavailability of targeted junction proteins at the site of cell-cell contact, resulting in the opening and closing of cell junctions (42). Fujita et al. report that Hakai, an E3 ubiquitin ligase, targets E-cadherin and thus triggers ubiquitination and degradation of the E-cadherin complex in the adherens junction (43). In addition, an E2 ubiquitin-conjugating enzyme (UBE2J1) KO mouse line was generated that appeared to have defective spermiation (44), suggesting that sperm release depends on the ubiquitin proteasome system (UPS). In our previous study, we found that the E3 ligase MKRN2 was located in the apical ES, and KO of Mkrn2 in mice led to the disarrangement of the apical ES and spermiation failure, and finally caused oligoasthenozoospermia (10). Although the study delineates a direct role of E3 ligase on the regulation of the ES and spermiation in vivo, the definitive substrate of MKRN2 involved in UPS remains unknown. To the best of our knowledge, ASB17 is the second E3 ligase required for the dynamic remodeling of the apical ES in the testis. Our study identified a novel mechanism for the physiological role of E3 ligases during spermiation.
Transcript level analysis of Asb17 at different stages of postnatal testicular development reveals that Asb17 is expressed from the third week and reaches the peak at the fourth week onwards (23,24), which corresponds to the stage of spermatid development. In line with this expression pattern, in the current study, using immunostaining, we found that ASB17 was mainly distributed in the apical ES of the testes. Meanwhile, ASB17 fluorescence signals were undetectable in Asb17-KO mice, further validating the specificity of the antibody against ASB17. Unfortunately, the commercial antibody against ASB17 was not suitable for western blot or IP analysis, so we carried out ASB17 protein pull-down experiments to identify its interacting proteins. Alternatively, transcript level analysis of Asb17 also reveals that it is not expressed at the first or second weeks of postnatal testicular development (22-24), suggesting that Asb17 is not required for spermatogonia or spermatocyte development. In addition, few location of ASB17 was observed in spermatogonia or spermatocytes based on immunostaining in this study. However, Yang et al. found that KO of ASB17 significantly prevented spermatogonial apoptosis induced by etoposide in mice (24), thus demonstrating a role for ASB17 in promoting spermatogonial apoptosis. Taken together, these findings suggest that etoposide-induced apoptosis in spermatogonia may trigger expression of the pro-apoptotic factor ASB17.
Conclusions
In summary, this study presented the loss-of-function analysis of the role of E3 ligase ASB17 during spermiation. We uncovered a novel mechanism in spermiation involving ASB17-mediated ES protein degradation and provided a potential therapeutic candidate for the treatment of spermiation failure.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81901532 to BZ and 81901533 to CS), Natural Science Foundation of Jiangsu Province (BK20190188 to BZ), Open Fund of State Key Laboratory of Reproductive Medicine of Nanjing Medical University (SKLRM-K202010 to BZ), and Gusu Health Talent Program of Suzhou (GSWS2020068 to BZ).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and approved by the Animal Ethical and Welfare Committee of Nanjing Medical University (No. IACUC-2004020).
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-789
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tau-21-789
Peer Review File: Available at https://dx.doi.org/10.21037/tau-21-789
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-789). The authors have no conflicts of interest to declare.
References
- 1.Wu S, Yan M, Ge R, et al. Crosstalk between Sertoli and Germ Cells in Male Fertility. Trends Mol Med 2020;26:215-31. 10.1016/j.molmed.2019.09.006 [DOI] [PubMed] [Google Scholar]
- 2.Bose R, Manku G, Culty M, et al. Ubiquitin-proteasome system in spermatogenesis. Adv Exp Med Biol 2014;759:181-213. 10.1007/978-1-4939-0817-2_9 [DOI] [PubMed] [Google Scholar]
- 3.Hou X, Zhang W, Xiao Z, et al. Mining and characterization of ubiquitin E3 ligases expressed in the mouse testis. BMC Genomics 2012;13:495. 10.1186/1471-2164-13-495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fok KL, Bose R, Sheng K, et al. Huwe1 Regulates the Establishment and Maintenance of Spermatogonia by Suppressing DNA Damage Response. Endocrinology 2017;158:4000-16. 10.1210/en.2017-00396 [DOI] [PubMed] [Google Scholar]
- 5.Li D, Li F, Meng L, et al. RNF216 regulates meiosis and PKA stability in the testes. FASEB J 2021;35:e21460. 10.1096/fj.202002294RR [DOI] [PubMed] [Google Scholar]
- 6.Kwon YT, Xia Z, An JY, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol 2003;23:8255-71. 10.1128/MCB.23.22.8255-8271.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim CR, Noda T, Kim H, et al. PHF7 Modulates BRDT Stability and Histone-to-Protamine Exchange during Spermiogenesis. Cell Rep 2020;32:107950. 10.1016/j.celrep.2020.107950 [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Kang JY, Wei L, et al. PHF7 is a novel histone H2A E3 ligase prior to histone-to-protamine exchange during spermiogenesis. Development 2019;146:dev175547. 10.1242/dev.175547 [DOI] [PubMed] [Google Scholar]
- 9.Lu LY, Wu J, Ye L, et al. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 2010;18:371-84. 10.1016/j.devcel.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian X, Wang L, Zheng B, et al. Deficiency of Mkrn2 causes abnormal spermiogenesis and spermiation, and impairs male fertility. Sci Rep 2016;6:39318. 10.1038/srep39318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng CY, Mruk DD. A local autocrine axis in the testes that regulates spermatogenesis. Nat Rev Endocrinol 2010;6:380-95. 10.1038/nrendo.2010.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian X, Mruk DD, Cheng YH, et al. Actin binding proteins, spermatid transport and spermiation. Semin Cell Dev Biol 2014;30:75-85. 10.1016/j.semcdb.2014.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee NP, Cheng CY. Ectoplasmic specialization, a testis-specific cell-cell actin-based adherens junction type: is this a potential target for male contraceptive development? Hum Reprod Update 2004;10:349-69. 10.1093/humupd/dmh026 [DOI] [PubMed] [Google Scholar]
- 14.Bu T, Wang L, Wu X, et al. A laminin-based local regulatory network in the testis that supports spermatogenesis. Semin Cell Dev Biol 2021. [Epub ahead of print]. 10.1016/j.semcdb.2021.03.025 [DOI] [PubMed] [Google Scholar]
- 15.Wong EW, Mruk DD, Cheng CY. Biology and regulation of ectoplasmic specialization, an atypical adherens junction type, in the testis. Biochim Biophys Acta 2008;1778:692-708. 10.1016/j.bbamem.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mruk DD, Cheng CY. Cell-cell interactions at the ectoplasmic specialization in the testis. Trends Endocrinol Metab 2004;15:439-47. 10.1016/j.tem.2004.09.009 [DOI] [PubMed] [Google Scholar]
- 17.Upadhyay RD, Kumar AV, Ganeshan M, et al. Tubulobulbar complex: cytoskeletal remodeling to release spermatozoa. Reprod Biol Endocrinol 2012;10:27. 10.1186/1477-7827-10-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng CY, Mruk DD. Regulation of spermiogenesis, spermiation and blood-testis barrier dynamics: novel insights from studies on Eps8 and Arp3. Biochem J 2011;435:553-62. 10.1042/BJ20102121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lui WY, Cheng CY. Regulation of cell junction dynamics by cytokines in the testis: a molecular and biochemical perspective. Cytokine Growth Factor Rev 2007;18:299-311. 10.1016/j.cytogfr.2007.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kile BT, Schulman BA, Alexander WS, et al. The SOCS box: a tale of destruction and degradation. Trends Biochem Sci 2002;27:235-41. 10.1016/S0968-0004(02)02085-6 [DOI] [PubMed] [Google Scholar]
- 21.Liu P, Verhaar AP, Peppelenbosch MP. Signaling Size: Ankyrin and SOCS Box-Containing ASB E3 Ligases in Action. Trends Biochem Sci 2019;44:64-74. 10.1016/j.tibs.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 22.Kim KS, Kim MS, Kim SK, et al. Murine Asb-17 expression during mouse testis development and spermatogenesis. Zygote 2004;12:151-6. 10.1017/S0967199404002722 [DOI] [PubMed] [Google Scholar]
- 23.Guo JH, Saiyin H, Wei YH, et al. Expression of testis specific ankyrin repeat and SOCS box-containing 17 gene. Arch Androl 2004;50:155-61. 10.1080/01485010490425485 [DOI] [PubMed] [Google Scholar]
- 24.Yang G, Wan P, Xiang Q, et al. E3 Ubiquitin Ligase ASB17 Promotes Apoptosis by Ubiquitylating and Degrading BCLW and MCL1. Biology (Basel) 2021;10:234. 10.3390/biology10030234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin M, Lv J, Zhao D, et al. MRNIP is essential for meiotic progression and spermatogenesis in mice. Biochem Biophys Res Commun 2021;550:127-33. 10.1016/j.bbrc.2021.02.143 [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Wang T, Zhao Z, et al. Retinoic Acid Induced Protein 14 (Rai14) is dispensable for mouse spermatogenesis. PeerJ 2021;9:e10847. 10.7717/peerj.10847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng T, Zhou S, Shi X, et al. Eef2k is not required for fertility in male mice. Transl Androl Urol 2021;10:1988-99. 10.21037/tau-21-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao T, Lin M, Shao B, et al. BMI1 promotes steroidogenesis through maintaining redox homeostasis in mouse MLTC-1 and primary Leydig cells. Cell Cycle 2020;19:1884-98. 10.1080/15384101.2020.1779471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng B, Yu J, Guo Y, et al. Cellular nucleic acid-binding protein is vital to testis development and spermatogenesis in mice. Reproduction 2018;156:59-69. 10.1530/REP-17-0666 [DOI] [PubMed] [Google Scholar]
- 30.Gao T, Lin M, Wu Y, et al. Transferrin receptor (TFRC) is essential for meiotic progression during mouse spermatogenesis. Zygote 2021;29:169-75. 10.1017/S0967199420000659 [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Wu Y, Li H, et al. BMI1 Drives Steroidogenesis Through Epigenetically Repressing the p38 MAPK Pathway. Front Cell Dev Biol 2021;9:665089. 10.3389/fcell.2021.665089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao D, Shen C, Gao T, et al. Myotubularin related protein 7 is essential for the spermatogonial stem cell homeostasis via PI3K/AKT signaling. Cell Cycle 2019;18:2800-13. 10.1080/15384101.2019.1661174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanagishita T, Yajima I, Kumasaka M, et al. Actin-binding protein, Espin: a novel metastatic regulator for melanoma. Mol Cancer Res 2014;12:440-6. 10.1158/1541-7786.MCR-13-0468-T [DOI] [PubMed] [Google Scholar]
- 34.Bartles JR, Zheng L, Li A, et al. Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J Cell Biol 1998;143:107-19. 10.1083/jcb.143.1.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekerková G, Loomis PA, Changyaleket B, et al. Novel espin actin-bundling proteins are localized to Purkinje cell dendritic spines and bind the Src homology 3 adapter protein insulin receptor substrate p53. J Neurosci 2003;23:1310-9. 10.1523/JNEUROSCI.23-04-01310.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weng G, Wang E, Wang Z, et al. HawkDock: a web server to predict and analyze the protein-protein complex based on computational docking and MM/GBSA. Nucleic Acids Res 2019;47:W322-30. 10.1093/nar/gkz397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merritt RC, Manor U, Salles FT, et al. Myosin IIIB uses an actin-binding motif in its espin-1 cargo to reach the tips of actin protrusions. Curr Biol 2012;22:320-5. 10.1016/j.cub.2011.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salles FT, Merritt RC, Jr, Manor U, et al. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat Cell Biol 2009;11:443-50. 10.1038/ncb1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin J, Ni B, Tian ZQ, et al. Regulatory effects of autophagy on spermatogenesis. Biol Reprod 2017;96:525-30. 10.1095/biolreprod.116.144063 [DOI] [PubMed] [Google Scholar]
- 40.Liu C, Wang H, Shang Y, et al. Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy 2016;12:814-32. 10.1080/15548627.2016.1159377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lui WY, Lee WM. Regulation of junction dynamics in the testis--transcriptional and post-translational regulations of cell junction proteins. Mol Cell Endocrinol 2006;250:25-35. 10.1016/j.mce.2005.12.021 [DOI] [PubMed] [Google Scholar]
- 42.Lui WY, Lee WM. cAMP perturbs inter-Sertoli tight junction permeability barrier in vitro via its effect on proteasome-sensitive ubiquitination of occludin. J Cell Physiol 2005;203:564-72. 10.1002/jcp.20254 [DOI] [PubMed] [Google Scholar]
- 43.Fujita Y, Krause G, Scheffner M, et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol 2002;4:222-31. 10.1038/ncb758 [DOI] [PubMed] [Google Scholar]
- 44.Koenig PA, Nicholls PK, Schmidt FI, et al. The E2 ubiquitin-conjugating enzyme UBE2J1 is required for spermiogenesis in mice. J Biol Chem 2014;289:34490-502. 10.1074/jbc.M114.604132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as