Abstract
Increased stiffness of the extracellular matrix is an important hallmark of melanoma development and progression, but its regulatory role and related mechanisms remain unclear. We adapted polydimethylsiloxane (PDMS)-micropillar-based matrix platform and investigated the effect of matrix stiffness on the proliferation, epithelial-mesenchymal transition (EMT), and immune escape of melanoma cells. We observed a stiff matrix enhanced cell proliferation, EMT, and immune escape of A375 cells. Furthermore, the expression of SNF5 on the stiffer matrix was higher than that on the softer matrix. Next, we investigated whether SNF5 is an important transducer in response to matrix stiffness. Our results revealed that knockdown of SNF5 significantly decreased stiff matrix-induced activation of cell proliferation, EMT and immune escape. Meanwhile, the overexpression of SNF5 showed its ability to increase cell proliferation, invasion and immune escape by activating the STAT-3 pathway in vitro. Furthermore, SNF5 deficiency elevated the level of tumor-infiltrating CD8+T cells and decreased the number of PD-L1 positive cells in vivo. Together, our findings suggested that stiffer substrate enhanced melanoma development by upregulating SNF5 expression, and SNF5 is a key mediator of stiffer matrix-induced immune evasion of melanoma cancer cells.
Keywords: PDMS, Polydimethylsiloxane; EMT, Epithelial-mesenchymal transition; ECM, Extracellular matrix; hPSCs, Human pluripotent stem cells; FBS, Fatal bovine serum; cDNA, Complementary DNA; PVDF, Polyvinylidene fluoride; IHC, Immunohistochemistry; AFM, Atomic force microscope; SNF5, Sucrose Non-Fermenting gene number 5; PD-L1, Programmed cell death-ligand 1; PD-L2, Programmed cell death-ligand 2
Keywords: SNF5, Matrix stiffness, Melanoma, Immune escape
Introduction
Melanoma is the most common primary malignancy of the skin, and its incidence rate increases continuously [1,2]. Although the development of targeted therapy and immunotherapy have improved the prognosis of patients with melanoma, primary or secondary therapy evasion still limits the therapeutic effectiveness [3]. Therefore, it is important to explore the mechanism of tumor development and find new therapeutic targets.
The mechanical microenvironment, especially the extracellular matrix (ECM) microenvironment, is a promoter of oncogenesis, progression, and treatment resistance. The process of ECM remodeling is accompanied by the expression changes in collagen, fibronectin, actin, and tubulin content, which gradually increases or decreases matrix stiffness. Recent studies have shown that ECM stiffness influences tumor growth, immunotherapy, treatment responsiveness and relapse, migration, and invasion [4], [5], [6], [7]. Immune escape is an important way for tumors to resist clearance by immune cells. There is emerging evidence that matrix stiffness enhances the immune escape potential of cancer cells. Azadi et al. [8] demonstrated that matrix stiffness activated the expression of PD-L1 and EMT-related transcription factors in breast cancer cells. This conclusion was also verified in lung cancer cells. Miyazawa et al. [9] used polyacrylamide hydrogels with different stiffness (25 and 2 kPa) to evaluate the effects of matrix stiffness on PD-L1 expression. They showed that the expression of PD-L1 was higher on stiffer matrixes than on softer ones, accompanied by increased stress fiber formation and cell growth.
SWI/SNF chromatin remodeling complexes include catalytic ATPase subunits (BRM or BRG1), core subunits (SNF5, BAF155 and BAF170), and variant subunits (e.g., BAF250a, BAF250b, BAF180, BAF200, BAF60a/b/c, BAF57, BAF53a/b, and BAF45a/b/c/d). They usually utilize the energy of ATP hydrolysis to modulate transcription and remodel nucleosomes [10]. The subunits of SWI/SNF have abundant functions in tumor regulation. On one hand, there is growing evidence that patients with aberrant expression of SWI/SNF genes have a more powerful tumor-specific immune response. For example, patients with aberrant expression of PBRM1 show better responsiveness to immune-checkpoint inhibition by facilitating immune-related signaling pathways [11]. Furthermore, SWI/SNF mutations alter the tumor immune-microenvironment by increasing cytotoxic T cell infiltration and PD-L1 expression to enhance the sensitivity to immune-checkpoint therapy [12]. On the other hand, we previously reported that BAF57 responded to a cyclic stretch and acted as a regulator in the splicing process of cyclin D1 [13]. A recent study has also shown that ARID1A is a mechanoregulated inhibitor of YAP/TAZ. Chang et al. [14] divided the mechanical signals into two cases. In low mechanical signals, ARID1A bound with YAP and reduced the expression of TEAD, one of the YAP/TAZ direct target genes. Conversely, at high mechanical stress, ARID1A bound with F-actin and recovered the association of YAP/TAZ with TEAD. Another subunit of the SWI/SNF complex, BRG1, was also confirmed to regulate the ECM-related genes [15]. Above all, the SWI/SNF complex may serve as a bridge between mechanical signals and immune evasion, but its suppressor or oncogene role and the underlying mechanisms in melanoma are still unclear.
As one of the core subunits of the SWI/SNF complex, SNF5 (also called SMARCB1, INI-1, or BAF47) has widespread roles in stem cell self-renewal and differentiation, cellular senescence, tumor suppression and tumor-specific immune response. For example, SNF5 is often aberrantly expressed in nearly all malignant rhabdoid tumors of children [16]. Additional functions of SNF5 have been discovered in recent years. In normal immortalized human cell lines, loss of SNF5 induced cell cycle arrest and elevated the expression level of IL-6 [17]. In human pluripotent stem cells (hPSCs), SNF5 regulated cell differentiation and the interactions with the ECM via the Wnt pathway [18]. Interestingly, contrary to other studies that considered SNF5 to be a tumor suppressor, Hong et al. [2] proved that SNF5 is an oncogene in liver cancer. Loss of SMARCB1 reduced cell proliferation, wound healing capacity in vitro, and tumor growth in vivo.
In this study, we investigated the roles of SNF5 in melanoma and found that SNF5 responded to different levels of stiffness of matrix topography and affected melanoma cell proliferation and EMT. More importantly, SNF5 affected the immune escape of melanoma by activating the phosphorylating STAT3. Furthermore, SNF5 deficiency elevated the level of tumor-infiltrated CD8+ T cells in vivo.
Materials and methods
Polydimethylsiloxane (PDMS) topological matrixes
The Polydimethylsiloxane (PDMS) topological matrixes were performed as previously described [19]. The spacing between the micropillars was 4 µm; the diameter of the micropillars was 4 µm; the height of the micropillars was also 4 µm. PDMS (Sylgard 184, Dow-Corning, Midland, MI) consisting of an oligomeric base and a curing agent was thoroughly mixed at two ratios (oligomeric base/Sylgard184 = 10:1 and 30:1). The mixture was cast onto the silicon mold, degassed under vacuum for about 20 min and then cross-linked at 80°C for 12 h. Finally, the PDMS matrixes were peeled off from the silicon mold.
Cell culture
Human melanoma cell line A375 was kindly given by Prof. Zhong Li from Chongqing University. Mouse melanoma cell line B16-F10 was kindly given by Prof. Ye Lilin from the Third Military Medical University. Human embryonic kidney cell line 293T was purchased from the cell bank of Chinese Academy of Sciences. A375 cells and B16-F10 cells were cultured in a DMEM medium (Gibco, 12800017) containing 10% fatal bovine serum (FBS) (HyClone, SH30088.03HI), 100 units of penicillin and 100 units of streptomycin (Sigma, P4458), and cultured in 25 cm2 flasks (Corning, 3815). HEK293T cells were cultured in a DMEM medium (Gibco, 12800017) containing 10% FBS, 100 units of penicillin and 100 units of streptomycin, and cultured in 10 cm dishes. The cells were incubated in a humidified cell culture incubator with 5% CO2 at 37 °C. A375 cells were plated on a PDMS matrix, which was preprocessed by 10 µg/mL fibronectin for 12 h at 4 °C.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted by using the RNAiso Plus reagent (Takara, 9109, China) in accordance with the manufacturer's instructions. Complementary DNA (cDNA) was synthesized by using the Takara reverse-transcription system kit (Takara, 6110A, China). PCR reactions were performed using TB GreenTM Premix Ex Taq TMII (Takara, RR820A, China).
The following RT-PCR primer sequences were used:
| Gene | Forward (5′- 3′) | Reverse (5′- 3′) |
|---|---|---|
| β-actin | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| E-cadherin | ATTTTTCCCTCGACACCCGAT | TCCCAGGCGTAGACCAAGA |
| N-cadherin | AGCCAACCTTAACTGAGGAGT | GGCAAGTTGATTGGAGGGATG |
| Vimentin | TGCCGTTGAAGCTGCTAACTA | CCAGAGGGAGTGAATCCAGATTA |
| SNF5 | GACGACGGCGAGTTCTAC | TCCTCTTGGCCTTCTGTT |
| PD-L2 | ACCCTGGAATGCAACTTTGAC | AAGTGGCTCTTTCACGGTGTG |
| FAS | TCTGGTTCTTACGTCTGTTGC | CTGTGCAGTCCCTAGCTTTCC |
| IDO1 | TCTCATTTCGTGATGGAGACTGC | GTGTCCCGTTCTTGCATTTGC |
| PD-L1 | TGGCATTTGCTGAACGCATTT | TGCAGCCAGGTCTAATTGTTTT |
| p16 | GGGTTTTCGTGGTTCACATCC | CTAGACGCTGGCTCCTCAGTA |
| pRB | TTGGATCACAGCGATACAAACTT | AGCGCACGCCAATAAAGACAT |
Western blot
Total protein lysates were obtained by lysing the cells with RIPA buffer supplemented with a phosphatase inhibitor cocktail (BioTeke, China). The protein content was measured by using the Bradford assay (Beyotime, China). Then, 20–40 μg of each sample was mixed with loading buffer and boiled for 5 min. All the samples were electrophoresed in a polyacrylamide gel and electro-blotted onto a polyvinylidene fluoride (PVDF) membrane. All the primary antibodies were used in accordance with to the manufacturer's instructions (anti-GAPDH, Proteintech, 10,494–1-AP; anti-SNF5, Proteintech, 20,654–1-AP; anti-BAF60c, Proteintech, 12,838–1-AP; anti-vimentin, Santa, sc-373,717; anti-E-cadherin, CST, #3195; anti-N-cadherin, CST, #13,116 anti-PD-L1, CST, #13,684; anti-Phospho-Stat3 (Tyr705), CST, #9131; anti-Stat3, CST, #9139; anti-PD-L2, ABCAM, ab187662; anti- IDO1, ABCAM, ab211017; anti- FAS, Proteintech, 13,098–1-AP; anti- SNAI1, Zen-bio, 340,942; anti-ZEB1, Zen-bio, 220,860; anti-β-Actin, Proteintech, 20,536–1-AP). The western blots were visualized under chemiluminescence conditions (Millipore, USA) using the ChemiDoc XRS machine (Bio-rad, USA). The bands were analyzed using the Image J Analysis software (Bio-rad, USA).
Lentivirus induction and infection
To induce the lentiviruses, HEK293T cells were co-transfected with pLenti-CMV plasmids or pLKO.1 plasmids containing the target gene fragments (10 μg), pCMV-dR8.2dvpr (7.5 μg), and pCMV–VSVG (2 μg) using Effectene transfection reagent (QIAGEN, Cat#301425) in a 10 cm dish. Before transfection, the HEK293T cells were replaced with a fresh DMEM culture medium without antibiotics. The viruses were collected and filtered through a 0.22 μm filter after 72 h.
For infection, A375 cells were cultured in a 6-well plate. On the following day, the cells were changed replaced with 1 mL fresh culture medium, and 1 mL of virus supernatant was added. Then, 72 h later, a DMEM medium containing 1 μg/mL puromycin was used to select the infected cells. The selection lasted for one week. Then, the RT-PCR and western blot were performed to confirm the recombinant expression. The A375 cells stably expressing SNF5 and knockdown SNF5 cells were used for further research.
Migration and invasion assay
Transwell chambers (8 µm polycarbonate membrane) were used for cell migration and invasion assay. The matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was added to the upper chamber for cell invasion assay. A375 cells were cultured in the upper chamber with 1% FBS DMEM medium, while 10% FBS DMEM medium was added to the lower chamber. Then the transwell chambers were incubated in a humidified cell culture incubator with 5% CO2 at 37 °C for 24 h. After that, the migrated or invasive cells were fixed with methanol and stained with crystal violet. The cells on the inner layer were gently removed with a cotton swab and three randomly selected views were counted. Finally, the average number of cells per view was calculated.
Animal experiments
The animal protocols used for the study were approved by the Committee on the Use and Care of Animals (Chongqing University Cancer Hospital, Chongqing, China), and the research was performed in accordance with the institution's guidelines. Mice were housed at an ambient temperature of 22 ℃, with a humidity of 30–70%, and a light cycle of 12 h on/12 h off set from 8 am to 8 pm. A375-shNC and A375-shSNF5 tumor xenografts were established by subcutaneously inoculating 5 × 106 cells into flanks of six-week-old female BALB/c nude mice. Before injecting the tumor cells, eight-week-old female C57BL/6 J mice were shaved at the flank; then, 5 × 106 B16-shNC and B16-shSNF5 tumor cells were injected into the shaved flank subcutaneously.
We depleted CD8+ T cells by using 100 ug/mL of intraperitoneally injected anti- CD8α antibody (2.43, Bio X Cell) on days −3, 0, 3, and 6. Tumor size was measured using a caliper and calculated using the following formula: volume = (length)(width)2/2.
Immunohistochemistry (IHC)
Mouse tumor tissues were fixed in 4% paraformaldehyde, washed by tap water, dehydrated using graded ethanol series, vitrificated by dimethylbenzene, embedded with paraffin, and sectioned longitudinally at 6 µm. Then, the tissue sections were dewaxed and rehydrated; antigens were retrieved with sodium citrate; endogenous peroxidase activity was blocked with 0.3% H2O2, and then nonspecific binding was blocked with goat serum. The tissue sections were stained for CD8α, and PD-L1. Human tissue sections were stained for SNF5 and PD-L1. Images were captured by Aperio Imagescope (Leica Biosystems) software.
Human samples
Human melanoma specimens were purchased from Iwill Biotech, Wuhan, comprising specimens from 37 melanoma patients and five melanocytic nevi patients. This study was approved by the Institutional Review Board of Chongqing University Cancer Hospital.
Statistical analysis
Two-tailed Student's t-tests with unequal variances were used for statistical analysis. All the results shown in Figures had at least three independent repeats. Survival analysis was performed using the log-rank test. A P < 0.05 was considered to indicate a statistically significant result.
Results
The stiffness of the micropillar-arrayed matrix affects the proliferation, EMT, and immune escape of A375 cells
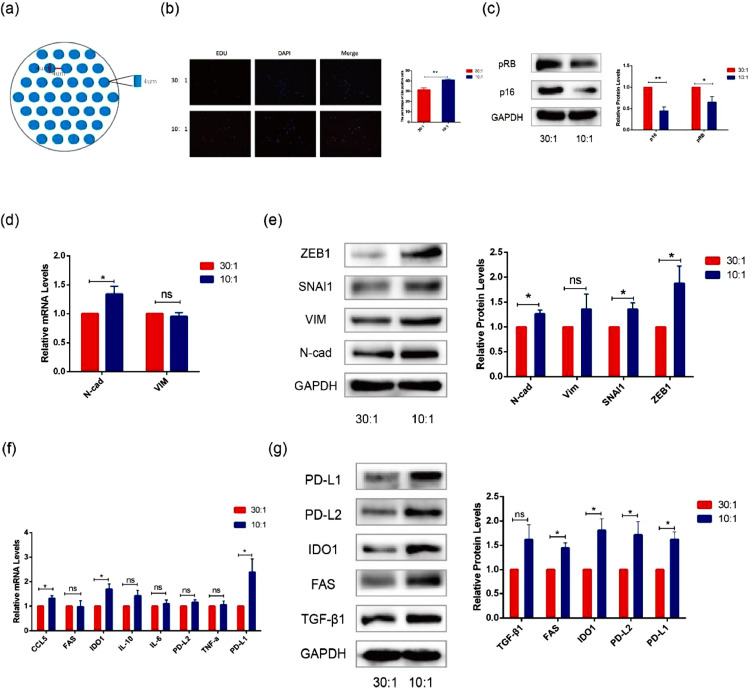
We used PDMS micropillars patterned with different degrees of stiffness to simulate the mechanical microenvironment of cell growth. The size of the micropillar arrays was 4 µm in height, 4 µm in diameter, 4 µm spacing allowed the cells to be supported by micropillars (Fig. 1a). The PDMS consisting of an oligomeric base and a curing agent was thoroughly mixed at two ratios (oligomeric base/Sylgard184 = 10:1 and 30:1). Based on our previous atomic force microscope (AFM) results, the 10:1 matrix had a higher Young's modulus than the 30:1 matrix [19].
Fig. 1.
Regulation of cellular proliferation, EMT, and immune evasion by different degrees of stiffness of micropillar-arrayed matrixes. (a) The schematic shows the structure of the micropillar-arrayed matrixes. (b) Representative immunofluorescence images of Edu and DAPI in A375 cells cultured on the matrixes with different degrees of stiffness(left). Bar graphs show the percentage of Edu positive cells (right). (c) The protein expression levels of p16 and pRB were detected by western blot in A375 cells cultured on the matrixes with different degrees of stiffness. (d) The mRNA expression levels of N-Cad and VIM were detected by RT-PCR in A375 cells cultured on the matrixes with different degrees of stiffness. (e) The protein expression levels of ZEB1, SNAI1, VIM and N-Cad were detected by western blot in A375 cells cultured on the matrixes with different degrees of stiffness. (f) The mRNA expression levels of CCL5, FAS, IDO1, IL-10, IL-6, PD-L2, TNF-α and PD-L1 were detected by RT-PCR in A375 cells cultured on the matrixes with different degrees of stiffness. (g) The protein expression levels of PD-L1, PD-L2, IDO1, FAS and TGF-β1 were detected by western blott in A375 cells cultured on the matrixes with different degrees of stiffness. (Values are shown as the mean ± standard deviation [SD.]. n = 3, a representative experiment is shown; ‘ns’ means P > 0.05, ‘*’means P < 0.05)
Mechanical factors play an important role in the development of melanoma. Extracellular matrix stiffness has a great effect on cell growth, migration and invasion [20]. In addition, in vivo, most tumors are mechanically stiffer than the surrounding tissue [21]. To confirm the influence of the mechanical microenvironment on A375 cells, we detected the proliferation, EMT, and immune escape with different degrees of matrix stiffness. The EdU assay showed that the cells cultured on the 10:1 matrix had higher viability (Fig. 1b). The phenomenon may be induced by the cell cycle regulation, as reflected by the reduced expression of p16 and pRB (Fig. 1c). We also observed the expression of EMT-related genes (N-cad, VIM, SNAI1 and ZEB1) and found that their mRNA and protein expression levels were upregulated in the cells cultured on the 10:1 matrix compared with the cells cultured on the 30:1 matrix (Fig. 1d, e). Interestingly, we found that the stiffness of the micropillar-arrayed matrix also had effects on the immune escape of A375 cells. As shown in Fig. 1f and g, the expression of immune escape-related genes TGF-β1, FAS, IDO1, PD-L2, and PD-L1 was upregulated at mRNA and protein levels in the cells cultured on the 10:1 matrix compared with those cultured on the 30:1 matrix.
These data demonstrate that the increase in stiffness has a great influence on the development of melanoma. Specifically, we observed that the mechanical change affected the expression of immune escape-related factors.
The SWI/SNF complex responds to different degrees of matrix stiffness
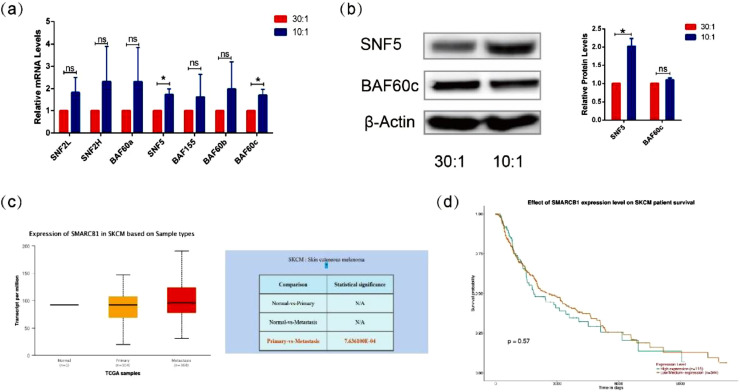
A previous study has shown that SWI/SNF is the dominant player in mechanical signaling to affect cell plasticity and tumorigenesis [14]. To investigate the influence of matrix structures with different degrees of stiffness on SWI/SNF subunits, we examined the expression of different SWI/SNF subunits at the gene level and the protein level. Our results showed that the mRNA expression of SNF5 was significantly upregulated in A375 cells cultured on stiffer micropillar patterned matrix, while the expression of other subunits of SWI/SNF was not dependent on the different degrees of stiffness of the micropillar patterned matrix (Fig. 2a). We further used western blot to confirm the upregulation of SNF5 in the cells cultured on the stiffer matrix (Fig. 2b). These results implied that the matrix stiffness might alter the expression of SNF5.
Fig. 2.
Response of SNF5 to the micropillar-arrayed matrixes. (a) The mRNA expression levels of SNF2L, SNF2H, BAF60a, SNF5, BAF155, BAF60b and BAF60c were detected by RT-PCR in A375 cells cultured on the matrixes with different degrees of stiffness. (b) Protein expression levels of SNF5 and BAF60c were detected by western blot in A375 cells cultured on the matrixes with different degrees of stiffness. (c) The relative mRNA expression of SNF5 in melanoma based on sample types. (d) The effect of SNF5 expression level on survival of melanoma patients. (Significant differences were determined using a one-way analysis of variance; values are shown as the mean ± standard deviation [SD.]. n = 3, a representative experiment is shown; ‘ns’ means P > 0.05, ‘*’ means P < 0.05).
To further confirm the role of SNF5 in melanoma, we analyzed the expression of SNF5 mRNA in normal melanoma tissue samples, primary melanoma tissue samples and metastasis tissue samples by TCGA database and Ualcan database. We found that the expression of SNF5 was higher in metastases than that in primary tumors (Fig. 2c). Furthermore, the survival time was prolonged in patients with low or medium expression levels of SNF5 (Fig. 2d). Taken together, SNF5 affects tumor metastasis and prognosis in response to mechanical stimulation.
SNF5 is a key factor that regulates tumor development in response to different degrees of matrix stiffness
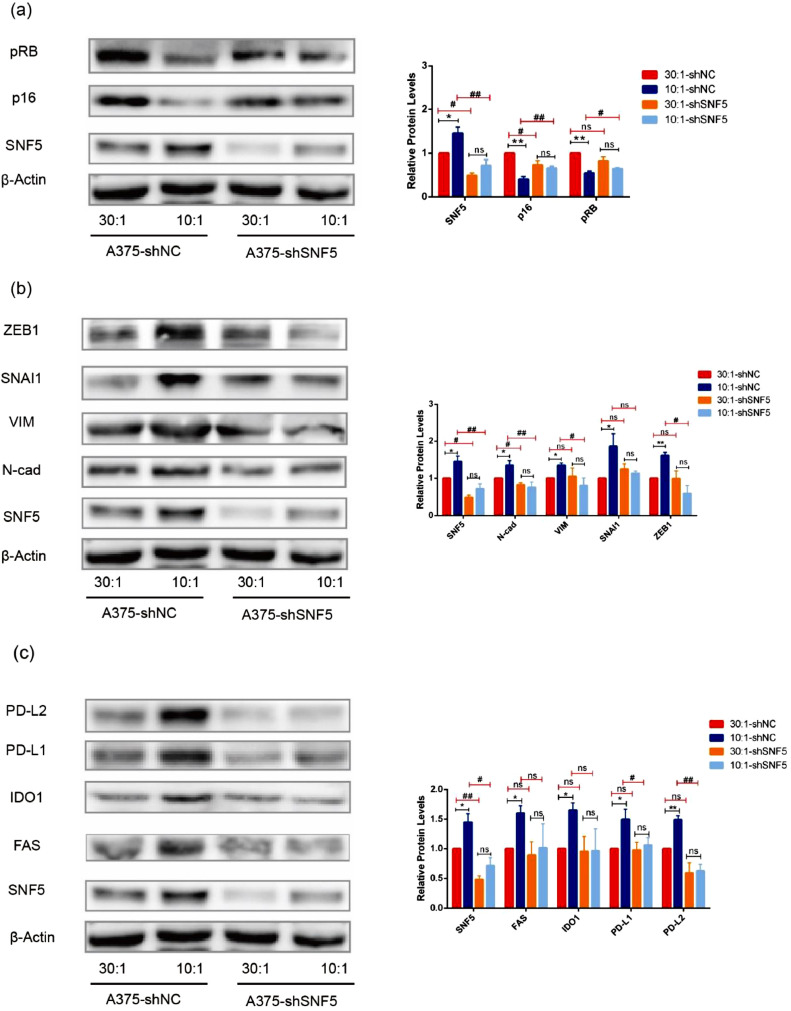
In our previous experiment, we found that the subunits of the SWI/SNF complex were possibly responsible for the mechanical stress and regulation of mechanotransduction-mediated alternative splicing [13]. To explore whether SNF5 affected tumor development in response to mechanical signals, we used shRNA to knock down the expression of SNF5 in A375 cells. Our data showed that the levels of p16 and pRB were lower in the cells on the 10:1 matrix compared with those on the 30:1 matrix with shNC treatment. However, after knocking down SNF5 with shSNF5 in A375 cells, the expression levels of p16 and pRB of the cells cultured on the 10:1 matrix became the same as those of the cells cultured on the 30:1 matrix (Fig. 3a). Similar results were obtained in the expression of EMT-related genes, but the expression of N-cad, VIM, SNAI1 and ZEB1 in response to the stiffer matrix was not up-regulated after the knockdown of SNF5 with shRNA (Fig. 3b). In addition, we further demonstrated that in the SNF5 knockdown group, the effects of matrix stiffness on the expression of immune escape-related genes, such as FAS, IDO1, PD-L2, and PD-L1, were neutralized (Fig. 3c).
Fig. 3.
shSNF5 neutralizes the effects of matrix stiffness on tumor development. (a) Western blot shows the expression of p16 and RB at the protein level in A375 cells cultured on the matrixes with different degrees of stiffness with shNC or shSNF5 treatment. (b) Western blot shows the expression of ZEB1, SNAI1, VIM and N-Cad at the protein level in A375 cells cultured on the matrixes with different degrees of stiffness with shNC or shSNF5 treatment. (c) Western blot shows the expression of PD-L1, PD-L2, IDO1 and FAS at protein level in A375 cells cultured on the matrixes with different stiffness with shNC or shSNF5 treatment. (Values are shown as the mean ± standard deviation [SD.]. n = 3, a representative experiment is shown; ‘ns’ means P > 0.05, ‘*’means P < 0.05, ‘**’ means P < 0.01, ‘#’means P < 0.05, ‘##’ means P < 0.01).
SNF5 plays an important role in regulating tumor development
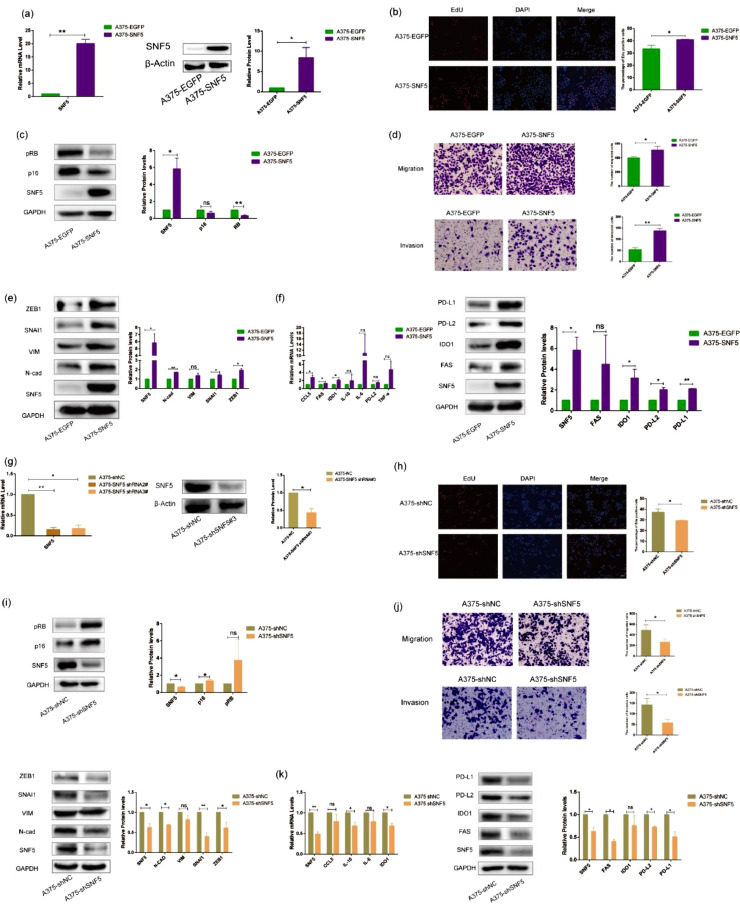
To explore the potential correlation between SNF5 and tumor development, we established EGFP or SNF5 stably transfected A375 cells by using lentiviral infection. RT-PCR and western blot results showed that all the target genes were successfully embedded in A375 cells (Fig. 4a). Then we used these infected cells (A375-EGFP and A375-SNF5) to assess the role of SNF5 in cell proliferation. Our data showed that the overexpression of SNF5 increased the number of EdU-positive cells (Fig. 4b). Furthermore, the expression levels of p16 and pRB were decreased in A375-SNF5 cells (Fig. 4c). We further investigated the effect of SNF5 on cell migration and invasion. Overexpression of SNF5 in A375 cells resulted in the increased numbers of migrating and invading cells as well as the increase in N-cad, VIM, SNAI1, and ZEB1 expression levels (Fig. 4d, e). Compared with the control cells, the expression levels of immune escape-related genes FAS, IDO1, PD-L2 and PD-L1 were higher in A375-SNF5 cells (Fig. 4f). Independently, we also stably knocked down SNF5 in A375 cells with lentiviruses expressing SNF5 shRNA. The RT-PCR and western blot results showed that SNF5 shRNA3# had a more powerful ability to silence SNF5 expression (Fig. 4g). In the later experiments, we used SNF5 shRNA3# to knockdown SNF5 expression. The number of EdU-positive cells was significantly lower in A375-shSNF5 cells (Fig. 4h). Similarly, the expression levels of p16 and pRB were significantly higher in A375-shSNF5 cells (Fig. 4i). Next, we observed that the loss of SNF5 expression reduced the numbers of migrating and invading cells, and it decreased N-cad, VIM, SNAI1 and ZEB1 expression levels (Fig. 4j). Compared with A375-shNC control cells, the expression levels of FAS, IDO1, PD-L2, and PD-L1 were reduced in A375-shSNF5 cells (Fig. 4k). Together, these results indicated that SNF5 might be involved in tumor development.
Fig. 4.
SNF5 plays an important role in regulating tumor development. (a) We constructed an SNF5 -overexpressing A375 cell line, and an A375-EGFP cell line served as a control. The overexpression of SNF5 was detected by RT-PCR (left) and western blot (right). (b) Representative images show EdU positive cells numbers after the overexpression of SNF5. (c) Representative images show p16 and pRB protein expression levels in A375 cells that recombinantly expressed SNF5 or EGFP. (d) Representative images show migrated cells and invasive cells in A375 cells that recombinantly expressed SNF5 or EGFP. (e) Western blotting results show the EMT-related protein expression in A375 cells that recombinantly expressed SNF5 or EGFP. (f) Representative images show immune-related factors expression at gene levels (left) and protein levels (right). (g) We constructed an SNF5 stable knockdown A375 cell line, with A375-shNC cell line served as a control. Knockdown of SNF5 was detected by RT-PCR (left) and western blot (right). (h) Representative images show Edu positive cells numbers after knockdown of SNF5. (i) Representative images show p16 and pRB protein expression levels in A375 cells that recombinantly expressed shSNF5 or shNC. (j) Representative images show migrated cells and invasive cells in A375 cells that recombinantly expressed shSNF5 or shNC (up). Western blotting results show the EMT-related protein expression levels in A375 cells that recombinantly expressed shSNF5 or shNC (down). (k) Representative images show immune-related factors expression at gene levels (left) and protein levels (right). (Values are shown as the mean ± standard deviation [SD.]. n = 3, a representative experiment is shown; ‘ns’ means P > 0.05, ‘*’means P < 0.05, ‘**’means P < 0.01).
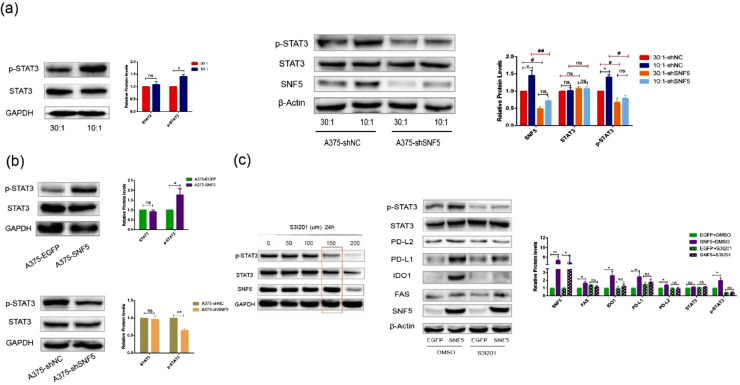
SNF5 regulates A375 cells immune escape via activating the STAT3/p-STAT3 signaling pathway
To investigate possible mechanism of SNF5 regulated immune escape in A375 cells, we first detected the expression levels of p-STAT3 and STAT3 in response to the different degrees of matrix stiffness. The results showed that the phosphorylation level of STAT3 was higher in A375 cells on the stiffer matrix. However, when we knocked down the expression of SNF5 in A375 cells cultured on the 10:1 matrix, the expression of p-STAT3 became the same as that in the cells cultured on the 30:1 matrix (Fig. 5a). Additionally, the phosphorylation level of STAT3 was higher in A375-SNF5 cells, while the phosphorylation level was decreased in A375-shSNF5 cells (Fig. 5b). As the above results showed, the STAT3 phosphorylation correlated with SNF5 expression. To further confirm whether p-STAT3 was involved in the regulation of immune evasion, we used a specific STAT3 inhibitor S3I-201 to inactivate STAT3 by blocking its phosphorylation and dimerization. S3I-201 was identified as an inhibitor of phosphorylation at Tyr-705 in the STAT3 transactivation domain and further inhibited the transcription of targe genes [22]. Based on our data, the optimum concentration S3I-201 was 100 µM with a processing time of and 24 h; this had no significant effect on the expression of STAT3 (Fig. 5c). The presented data confirmed that the expression levels of FAS, IDO1, PD-L2, and PD-L1 were increased in A375-SNF5 cells, while these increases were significantly reversed by S3I-201 administration, supporting the role of SNF5 in promoting immune evasion by activating the STST3 pathway in A375 cells (Fig. 5c).
Fig. 5.
SNF5 regulates immune escape through the STAT3/p-STAT3 signaling pathway in A375 cells. (a) Representative images show STAT3 and p-STAT3 protein expression levels in A375 cells cultured on the matrixes with different degrees of stiffness (left). Western blot shows the expression of STAT3 and p-STAT3 at the protein level in A375 cells cultured on the matrixes with different degrees of stiffness with shNC or shSNF5 treatment (right). (b) Representative images show STAT3 and p-STAT3 protein expression levels in A375 cells that recombinantly expressed SNF5 or EGFP (up). Representative images show STAT3 and p-STAT3 protein expression levels in A375 cells that recombinantly expressed shSNF5 or shNC (down). (c) Screening the optimum treatment duration and concentration of S3I201 (left). A375-EGFP and A375-SNF5 cells were treated with S3I201(150 µm) or DMSO for 24 h. Protein expression levels of p-STAT3, STAT3, PD-L2, PD-L1, IDO1 and FAS were detected by western blot (right). (Values are shown as the mean ± standard deviation [SD.]. n = 3, a representative experiment is shown; ‘ns’ means P > 0.05, ‘*’means P < 0.05, ‘**’means P < 0.01).
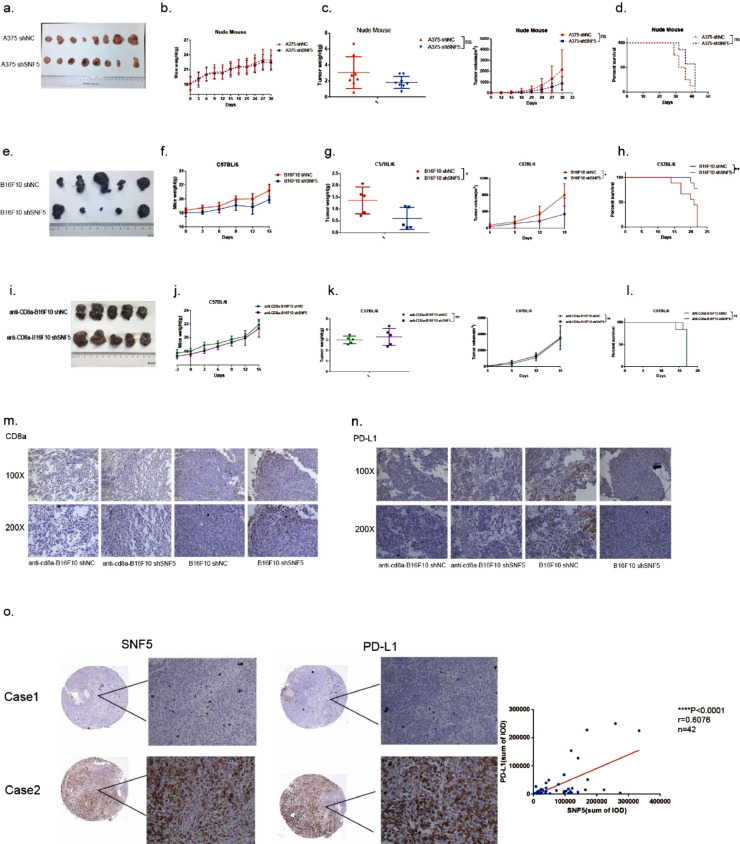
SNF5 disruption in melanoma enhances antitumor immunity through tumor-infiltrating T lymphocytes
To further confirm the antitumor immunity role of SNF5 in vivo, we constructed a subcutaneous tumor model in immunodeficient nude mice and immunocompetent C57BL/6 mice. Our results showed that tumor growth, tumor weight, and survival were not significantly different between the SNF5 knockdown group and the control group (Fig. 6a–d), indicating that the immune system is an indispensable part of the antitumor response. In an immunocompetent environment, we found that the knockdown of SNF5 significantly suppressed tumor growth, decreased tumor weight and prolonged survival in C57BL/6 mice (Fig. 6e–h). CD8+ cytotoxic T lymphocytes are major immune cells responsible for killing cancer cells. Usually, higher level of infiltrating CD8+T cells indicates a better prognosis. To confirm the unique function of infiltrating CD8+T cells, we used an anti-mouse CD8 antibody to deplete CD8 lymphocytes in the mice. Strikingly, tumor weight and volume grew faster after depleting CD8 lymphocytes. Furthermore, tumor growth, tumor weight, and survival were not significantly different in the SNF5 knockdown and control groups (Fig. 6i–l). Immunohistochemical staining revealed that the knockdown of SNF5 increased the number of infiltrating CD8+T cells and decreased the number of PD-L1 positive cells (Fig. 6m, n). Furthermore, we also analyzed the expression levels of SNF5 and PD-L1 in human normal tissue samples (n = 5) and human tumor tissue samples (n = 37). The results showed a significant relationship between SNF5 and PD-L1 (Fig. 6o). Taken together, SNF5 disruption in melanoma enhances antitumor immunity through tumor-infiltrating T lymphocytes.
Fig. 6.
SNF5 disruption in melanoma enhances antitumor immunity through tumor-infiltrating T lymphocytes. (a) SNF5-knockdown A375 cells and control cells were inoculated subcutaneously into nude mice and monitored for tumor formation. Mice body weight (b), tumor growth (c) and survival (d) were measured every three days, (n = 8). (e) SNF5-knockdown B16-F10 cells and control cells were inoculated subcutaneously into C57BL/6 mice and monitored for tumor formation. Mice body weight (f), tumor growth (g) and survival (h) were measured every three days, (n = 5). (i) SNF5-knockdown B16-F10 cells and control cells were inoculated subcutaneously into C57BL/6 mice after depleting the CD8 lymphocytes and were monitored for tumor formation. Mice body weight (j), tumor growth (k) and survival (l) were measured every three days, (n = 5). Immunohistochemical analysis of the expression of CD8a (m) and PD-L1 (n) in the tumors. (o) Immunohistochemical analysis of the expression of SNF5 and PD-L1 in human tissue samples. (Values are shown as the mean ± standard deviation [SD.]. n = 3, a representative experiment is shown; ‘ns’ means P > 0.05, ‘*’means P < 0.05, ‘**’means P < 0.01).
Discussion
The composition and physical characteristics of the ECM are altered during melanoma progression. Importantly, increased matrix stiffness has a great effect on tumor growth, metastasis, invasion, and infiltration by T- lymphocytes. For example, Reid et.al [5] demonstrated that the increased matrix stiffness facilitated melanoma cancer cell binding with blood vessels, a typical process of cancer cell migration. In addition, Long et.al [6] found that vemurafenib treatment-induced ECM remodeling was sufficient to alter cell differentiation and decrease treatment responses in melanoma. Furthermore, Miskolczi et.al [23] designed varying stiffness of collagen and found that increased collagen stiffness promoted 501 mel and WM266–4 cells proliferation and differentiation. In line with the above results, our results also confirmed that a 10:1 stiffer PDMS-micropillar matrix promoted A375 melanoma cancer cells proliferation and EMT (Fig. 1). Particularly, we found that SNF5 was involved in the stiffness-induced tumor progression (Fig. 2).
Notably, SNF5 is one of the core subunits of SWI/SNF. There is accumulating evidence that the expressions levels of many subunits of SWI/SNF are regulated by tumor matrix stiffness, which has profound effects on the development of cancer. Chang et al. [14] suggested that at high mechanical stress, ARID1A bound with F-actin rather than with YAP/TAZ and prompted the association of YAP/TAZ with TEAD, thereby inducing cell plasticity and tumorigenesis. In addition, BAF60a ameliorated the ECM degradation by inhibiting the upregulation of the proteolytic enzyme cysteine protease in smooth muscle cells [24]. Amelio et al. [25]found that SWI/SNF mediated the expression of ECM components and promoted tumor progression by binding with p53 mutant/HIF-1 complex in non-small cell lung cancer. It is worth noting that BRG1, the core subunit of the SWI/SNF chromatin remodeling complex, promoted liver fibrosis by activating hepatic stellate cells. Liver fibrosis is known as a promoter of liver cancer by providing a permissive environment for cancer progression [26]. We reported for the first time that SNF5 responds to matrix stiffness stimulation. Indeed, knocking down SNF5 inhibited the effects of matrix stiffness-induced cell growth, migration, invasion and EMT (Fig. 3). Hence, targeting the stiffness-induced changes in tumor cells, is a potential strategy to inhibit tumor growth and metastasis [5]. Therefore, we speculated that targeting SNF5 may be beneficial in reducing tumor growth, migration, and invasion.
However, our in vivo experiments showed that knocking down SNF5 did not inhibit tumor growth in immunodeficient nude mice, whereas the tumor growth suppression was obvious in immunocompetent C57BL/6 mice (Fig. 6). Thus, we speculated whether the immune environment in vivo was involved in the effect of SNF5 on tumor growth. Previous studies have shown that the increased matrix stiffness regulated the tumor immune escape. In breast cancer, the expression of PD-L1 in cells cultured on a stiff matrix was higher than that in cells cultured on semi-soft and soft matrixes [8]. Similar results also have been reported regarding in lung cancer cells, where the expression of PD-L1 on a stiffer matrix was higher than that on a softer matrix [9]. Extending this observation, our in vitro results showed that the increased matrix stiffness upregulated the expression of immune escape-related genes such as FAS, IDO1, PD-L2 and PD-L1. Furthermore, the knockdown of SNF5 neutralized the regulation of tumor immune escape by matrix stiffness, indicating that stiffer matrix promotes immune escape via SNF5 and facilitates tumor growth in melanoma. Moreover, the overexpression of SNF5 also confirmed that SNF5 upregulated the immune escape-related genes such as FAS, IDO1, PD-L2 and PD-L1 in vitro (Fig. 4). In vivo, SNF5 deficiency in immunocompetent mice decreased tumor weight and prolonged the survival time via increasing the tumor infiltration by CD8+ T cells (Fig. 6). In general, our findings underscore the important role of SNF5 in the melanoma immune escape. This is in agreement with a recent report that demonstrated that SNF5 binds to the IL6 promoter and inhibits IL6 transcription, reducing the immune response of the cell [17]. Unfortunately, there is a lack of studies on the mechanisms by which SNF5 would impact the tumor immune escape and cancer therapeutics. As the core subunit of SWI/SNF, SNF5 has potential benefits in antitumor immunity and clinical responses to immune- checkpoint blockade therapy. Therefore, it is necessary to reveal how SNF5 regulates the immune cell function in the complex tumor microenvironment. In addition, it is more important to utilize this regulatory mechanism of SNF5 to enrich the immune- checkpoint system and to exploit a combination treatment of SNF5 and immune- checkpoint inhibitors.
STAT3 plays a critical role in various cellular processes, including cell proliferation, apoptosis, angiogenesis, and metastasis. STAT3 is constitutively activated by Tyr705 and Ser727 phosphorylation in various cancers. Interestingly, phosphorylated STAT3 can dimerize and translocate to the nucleus, where it binds with the promoter of PD-L1 to increase PD-L1 transcription. This process finally induces immune escape [27]. In addition, p-STAT3 is also related to autophagy and creates an immune-depressed environment [28]. Bu et al. [29] confirmed that STAT3 signaling is essential for PD-1/PD-L1 regulation and the antitumor immune response of head and neck squamous cell carcinoma. Likewise, in melanoma, SNF5-regulated cellular immune escape also needs the involvement of p-STAT3 (Fig. 5a). We used S3I-201, an inhibitor of STAT3 phosphorylation, to disrupt the formation of the SNF5/p-STAT3 axis and found that SNF5 cannot mediate the expression of immune-related factors without the involvement of p-STAT3 (Fig. 5b). This result is in line with most of the previous research results. Zhang et al. [30] revealed that SMARCA2 promoted pancreatic cancer growth and chemoresistance by activating the STAT3 signaling.
In the process of carcinoma progression, the increased matrix stiffness promotes cancer cell proliferation. In addition, the tumor immune microenvironment blocks T cell infiltration and upregulates tumor cells’ surface expression of PD-L1, which can bind with PD-1 receptor on T cells to avoid the attack by CD8+ T cells. Our findings would be significant for melanoma therapy. We have considered the influence of the mechanical microenvironment on melanoma progression and focused on the effect of different 3D topology matrix stiffness on the progression of melanoma. In response to increasing matrix stiffness, human melanoma cell lines exhibited significantly increased cell proliferation, metastasis, and immune escape. Then, our study revealed that SNF5 acted as a mechanoregulated connector involved in cell proliferation, EMT, and immune escape. In line with the oncogene role of SNF5 in liver cancer [2], we found that SNF5 was upregulated in metastatic tumors and improved cell proliferation, EMT, and immune escape in vitro. In addition, we disclosed the role of the STAT3 signaling pathway in SNF5-mediated tumor immune escape. Finally, we explored the mechanism of SNF5-mediated tumor immune escape in vivo and found that SNF5 deficiency elevated the level of tumor-infiltrating CD8+ T cells. The relationship that we have identified between the mechanistic signal transduction and SNF5-mediated melanoma progression highlights the need to consider the role of the tumor mechanical microenvironment in cancer therapy.
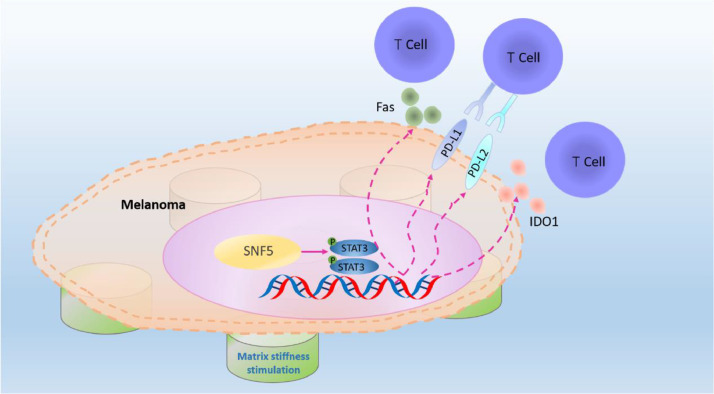
In general, our data provide new insights that it is of benefit for oncotherapy to combine of mechanistic signal transduction, immune checkpoint and cytotoxic T cell infiltration (Fig. 7). Therefore, it is worthwhile that SNF5 may be a promising candidate for the combination therapy with PD-1 monoclonal antibody.
Fig. 7.
Proposed model of the role of SNF5 in regulating immune escape upon mechanical stimulation in melanoma. The upregulated expression of SNF5 on the stiffer matrix activates the expression of immune escape genes by activating the phosphorylation of STAT3, thereby inhibiting T cells recognition and infiltration.
Institutional review board statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Animal Care Committee of Chongqing University Cancer Hospital.
Data availability statement
The data that support the findings of this research are available from the corresponding authors upon reasonable request.
Funding
The work was funded by the National Natural Science Foundation of Chongqing: cstc2021ycjh-bgzxm0169 and National Natural Science Foundation of China: NO.31670952.
CRediT authorship contribution statement
Ying Chen: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft. Meilian Zhao: Investigation. Lu Zhang: Validation, Methodology. Dongliang Shen: Investigation. Xichao Xu: Validation, Methodology. Qian Yi: Conceptualization, Supervision, Project administration, Writing – review & editing. Liling Tang: Conceptualization, Supervision, Project administration, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Dr. Li Zhong (Chongqing University, Chongqing, China) and Dr. Lilin Ye (Third Military Medical University) for sharing the A375 cell lines and B16-F10 cell lines, respectively.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101335.
Contributor Information
Qian Yi, Email: yiqian2010@yeah.net.
Liling Tang, Email: tangliling@cqu.edu.cn.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Hong S.H., Son K.H., Ha S.Y., Wee T.I., Choi S.K., Won J.E., Han H.D., Ro Y., Park Y.M., Eun J.W., Nam S.W., Han J.W., Kang K., You J.S. Nucleoporin 210 serves a key scaffold for smarcb1 in liver cancer. Cancer Res. 2021;81:356–370. doi: 10.1158/0008-5472.CAN-20-0568. [DOI] [PubMed] [Google Scholar]
- 3.Boumahdi S., de Sauvage F.J. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug. Discov. 2020;19:39–56. doi: 10.1038/s41573-019-0044-1. [DOI] [PubMed] [Google Scholar]
- 4.Pach E., Brinckmann J., Rübsam M., Kümper M., Mauch C., Zigrino P. Fibroblast MMP14-dependent collagen processing is necessary for melanoma growth. Cancers (Basel) 2021;13:1–15. doi: 10.3390/cancers13081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid S.E., Kay E.J., Neilson L.J., Henze A.T., Serneels J., McGhee E.J., Dhayade S., Nixon C., Mackey J.B., Santi A., Swaminathan K., Athineos D., Papalazarou V., Patella F., Román-Fernández Á., ElMaghloob Y., Hernandez-Fernaud J.R., Adams R.H., Ismail S., Bryant D.M., Salmeron-Sanchez M., Machesky L.M., Carlin L.M., Blyth K., Mazzone M., Zanivan S. Tumor matrix stiffness promotes metastatic cancer cell interaction with the endothelium. EMBO J. 2017;36:2373–2389. doi: 10.15252/embj.201694912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long J.E., Wongchenko M.J., Nickles D., Chung W.J., Wang B.E., Riegler J., Li J., Li Q., Sandoval W., Eastham-Anderson J., Modrusan Z., Junttila T., Carano R., Foreman O., Yan Y., Junttila M.R. Therapeutic resistance and susceptibility is shaped by cooperative multi-compartment tumor adaptation. Cell Death Differ. 2019;26:2416–2429. doi: 10.1038/s41418-019-0310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muñoz N.M., Williams M., Dixon K., Dupuis C., McWatters A., Avritscher R., Manrique S.Z., McHugh K., Murthy R., Tam A., Naing A., Patel S.P., Leach D., Hartgerink J.D., Young S., Prakash P., Hwu P., Sheth R.A. Influence of injection technique, drug formulation and tumor microenvironment on intratumoral immunotherapy delivery and efficacy. J. Immunother. Cancer. 2021;9:1–9. doi: 10.1136/jitc-2020-001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azadi S., Aboulkheyr E.H., Razavi B.S., Thiery J.P., Asadnia M., Ebrahimi W.M. Upregulation of PD-L1 expression in breast cancer cells through the formation of 3D multicellular cancer aggregates under different chemical and mechanical conditions. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866 doi: 10.1016/j.bbamcr.2019.118526. [DOI] [PubMed] [Google Scholar]
- 9.Miyazawa A., Ito S., Asano S., Tanaka I., Sato M., Kondo M., Hasegawa Y. Regulation of PD-L1 expression by matrix stiffness in lung cancer cells. Biochem. Biophys. Res. Commun. 2018;495:2344–2349. doi: 10.1016/j.bbrc.2017.12.115. [DOI] [PubMed] [Google Scholar]
- 10.Mittal P., Roberts C. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 2020;17:435–448. doi: 10.1038/s41571-020-0357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao D., Margolis C.A., Vokes N.I., Liu D., Taylor-Weiner A., Wankowicz S.M., Adeegbe D., Keliher D., Schilling B., Tracy A., Manos M., Chau N.G., Hanna G.J., Polak P., Rodig S.J., Signoretti S., Sholl L.M., Engelman J.A., Getz G., Jänne P.A., Haddad R.I., Choueiri T.K., Barbie D.A., Haq R., Awad M.M., Schadendorf D., Hodi F.S., Bellmunt J., Wong K.K., Hammerman P., Van Allen E.M. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 2018;50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J., Ju Z., Zhao W., Wang L., Peng Y., Ge Z., Nagel Z.D., Zou J., Wang C., Kapoor P., Ma X., Ma D., Liang J., Song S., Liu J., Samson L.D., Ajani J.A., Li G.M., Liang H., Shen X., Mills G.B., Peng G. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018;24:556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng J., Xu X., Fan X., Yi Q., Tang L. BAF57/SMARCE1 interacting with splicing factor srsf1 regulates mechanical stress-induced alternative splicing of cyclin D1. Genes (Basel) 2021;12:1–11. doi: 10.3390/genes12020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang L., Azzolin L., Di Biagio D., Zanconato F., Battilana G., Lucon X.R., Aragona M., Giulitti S., Panciera T., Gandin A., Sigismondo G., Krijgsveld J., Fassan M., Brusatin G., Cordenonsi M., Piccolo S. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature. 2018;563:265–269. doi: 10.1038/s41586-018-0658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barutcu A.R., Lajoie B.R., Fritz A.J., McCord R.P., Nickerson J.A., van Wijnen A.J., Lian J.B., Stein J.L., Dekker J., Stein G.S., Imbalzano A.N. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016;26:1188–1201. doi: 10.1101/gr.201624.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy A.T., Strother D.R., Judkins A.R., Burger P.C., Pollack I.F., Krailo M.D., Buxton A.B., Williams-Hughes C., Fouladi M., Mahajan A., Merchant T.E., Ho B., Mazewski C.M., Lewis V.A., Gajjar A., Vezina L.G., Booth T.N., Parsons K.W., Poss V.L., Zhou T., Biegel J.A., Huang A. Efficacy of high-dose chemotherapy and three-dimensional conformal radiation for atypical teratoid/rhabdoid tumor: a report from the children's oncology group trial ACNS0333. J. Clin. Oncol. 2020;38:1175–1185. doi: 10.1200/JCO.19.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi S.K., Kim M.J., You J.S. SMARCB1 acts as a quiescent gatekeeper for cell cycle and immune response in human cells. Int. J. Mol. Sci. 2020;21:1–14. doi: 10.3390/ijms21113969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmel-Gross I., Levy E., Armon L., Yaron O., Waldman B.H., Urbach A. Human pluripotent stem cell fate regulation by SMARCB1. Stem Cell Rep. 2020;15:1037–1046. doi: 10.1016/j.stemcr.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X., Ma L., Wu Y., Tang L. Micropillar-based culture platform induces epithelial-mesenchymal transition in the alveolar epithelial cell line. J. Biomed. Mater. Res. A. 2018;106:3165–3174. doi: 10.1002/jbm.a.36511. [DOI] [PubMed] [Google Scholar]
- 20.Park J., Kim D.H., Kim H.N., Wang C.J., Kwak M.K., Hur E., Suh K.Y., An S.S., Levchenko A. Directed migration of cancer cells guided by the graded texture of the underlying matrix. Nat. Mater. 2016;15:792–801. doi: 10.1038/nmat4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riegler J., Labyed Y., Rosenzweig S., Javinal V., Castiglioni A., Dominguez C.X., Long J.E., Li Q., Sandoval W., Junttila M.R., Turley S.J., Schartner J., Carano R. Tumor elastography and its association with collagen and the tumor microenvironment. Clin. Cancer Res. 2018;24:4455–4467. doi: 10.1158/1078-0432.CCR-17-3262. [DOI] [PubMed] [Google Scholar]
- 22.Liao Y., Feng J., Zhang Y., Tang L., Wu S. The mechanism of CIRP in inhibition of keratinocytes growth arrest and apoptosis following low dose UVB radiation. Mol. Carcinog. 2017;56:1554–1569. doi: 10.1002/mc.22597. [DOI] [PubMed] [Google Scholar]
- 23.Miskolczi Z., Smith M.P., Rowling E.J., Ferguson J., Barriuso J., Wellbrock C. Collagen abundance controls melanoma phenotypes through lineage-specific microenvironment sensing. Oncogene. 2018;37:3166–3182. doi: 10.1038/s41388-018-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang Z., Zhao G., Zhao Y., Lu H., Xiong W., Liang W., Sun J., Wang H., Zhu T., Rom O., Guo Y., Fan Y., Chang L., Yang B., Garcia-Barrio M.T., Lin J.D., Chen Y.E., Zhang J. BAF60a deficiency in vascular smooth muscle cells prevents abdominal aortic aneurysm by reducing inflammation and extracellular matrix degradation. Arterioscler. Thromb. Vasc. Biol. 2020;40:2494–2507. doi: 10.1161/ATVBAHA.120.314955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amelio I., Mancini M., Petrova V., Cairns R.A., Vikhreva P., Nicolai S., Marini A., Antonov A.A., Le Quesne J., Baena A.J., Dudek K., Sozzi G., Pastorino U., Knight R.A., Mak T.W., Melino G. p53 mutants cooperate with HIF-1 in transcriptional regulation of extracellular matrix components to promote tumor progression. Proc. Natl. Acad. Sci. USA. 2018;115:E10869–E10878. doi: 10.1073/pnas.1808314115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Lan J., Han C., Guo K., Wang G., Hu J., Gong J., Luo X., Cao Z. Brg1 promotes liver fibrosis via activation of hepatic stellate cells. Exp. Cell. Res. 2018;364:191–197. doi: 10.1016/j.yexcr.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Zhou B., Yan J., Guo L., Zhang B., Liu S., Yu M., Chen Z., Zhang K., Zhang W., Li X., Xu Y., Xiao Y., Zhou J., Fan J., Hung M.C., Li H., Ye Q. Hepatoma cell-intrinsic TLR9 activation induces immune escape through PD-L1 upregulation in hepatocellular carcinoma. Theranostics. 2020;10:6530–6543. doi: 10.7150/thno.44417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Zhang H., Wang Z., Wu P., Gong W. 5-Hydroxytryptamine1a receptors on tumour cells induce immune evasion in lung adenocarcinoma patients with depression via autophagy/pSTAT3. Eur. J. Cancer. 2019;114:8–24. doi: 10.1016/j.ejca.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Bu L.L., Yu G.T., Wu L., Mao L., Deng W.W., Liu J.F., Kulkarni A.B., Zhang W.F., Zhang L., Sun Z.J. STAT3 Induces Immunosuppression by Upregulating PD-1/PD-L1 in HNSCC. J. Dent. Res. 2017;96:1027–1034. doi: 10.1177/0022034517712435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Wang F., Du C; Guo H., Ma L., Liu X., Kornmann M., Tian X., Yang Y. BRM/SMARCA2 promotes the proliferation and chemoresistance of pancreatic cancer cells by targeting JAK2/STAT3 signaling. Cancer Lett. 2017;402:213–224. doi: 10.1016/j.canlet.2017.05.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this research are available from the corresponding authors upon reasonable request.