Abstract
This research deals with the development of asialoglycoprotein receptors (ASGPR) directed nanoliposomes incorporating a novel BRD4 (Bromodomain-containing protein 4) protein-targeted PROTAC (Proteolysis Targeting Chimera), ARV-825 (ARV) (GALARV), and to investigate the anticancer efficacy of GALARV for specific delivery in hepatocellular carcinoma. GALARV were prepared using the modified hydration method and characterized for their physicochemical properties as well as anticancer activity using 2D and 3D cell culture models. ARV and GALARV (93.83 ± 10.05 nm) showed significant in vitro cytotoxicity and apoptosis in hepatocellular carcinoma cells. GALARV also demonstrated a substantially higher intracellular concentration of ARV compared to non-targeted nanoliposomes (∼3 fold) and ARV alone (∼4.5 fold), showed good physical stability and negligible hemolysis. Immunoblotting results depicted substantial downregulation of target BRD4 protein, oncogenic c-Myc, apoptotic Bcl-2, and survivin proteins. Notably, GALARV treatment resulted in significant apoptosis and subsequent inhibition of the cell viability of 3D tumor spheroids of hepatocellular carcinoma. These results suggest that GALARV is a novel actively targeted PROTAC-based nanotherapeutic approach for hepatocellular carcinoma.
Keywords: ARV-825, BRD4, PROTAC, Galactosylated nanoliposomes, Active drug delivery, Hepatocellular carcinoma
Graphical abstract
Highlights
-
•
Hepatocellular carcinoma is a solid tumor with high BRD4 and c-Myc overexpression.
-
•
ARV is a novel PROTAC for target BRD4 degradation and c-Myc downregulation in HCC.
-
•
ASGPR-targeted GALARV demonstrated higher cellular uptake in hepatic cancer cells.
-
•
GALARV demonstrated potent anticancer activity in 2D and 3D hepatic cancer models.
-
•
GALARV is a unique PROTAC-based nanotherapy to target ‘undruggable’ c-Myc in HCC.
ARV-825; BRD4; PROTAC; Galactosylated nanoliposomes; Active drug delivery; Hepatocellular carcinoma.
1. Introduction
Hepatocellular carcinoma (HCC) is a common solid tumor and the fourth most common cause contributing to cancer-related deaths worldwide (Yang et al., 2019). In the past three decades, the incidence of liver cancer has increased by 75% globally and it has been estimated by the American Cancer Society that 42,810 new cases will be diagnosed and 30,160 deaths will occur due to liver cancer by 2020 in the United States (Singal et al., 2020). Earlier stages of liver cancer can be treated by local excision, surgery, or liver transplantation. However, HCC is usually detected at a very late stage in most cases. In that scenario, most patients require chemotherapy. Treatment strategy generally depends on various factors like tumor characteristics, liver dysfunction severity, and other underlying medical comorbidities (Yang et al., 2019). Sorafenib, Lenvatinib, Cabozantinib, and Regorafenib are United States Food and Drug Administration (USFDA) approved multiple kinase inhibitors used as the standard first- and second-line treatments for advanced HCC. Although these drugs improve the survival rate of HCC patients, sorafenib generally results in poor tumor response; while Lenvatinib and Regorafenib may potentially lead to drug resistance and intratumor hypoxia-related issues (Yin et al., 2019). Therefore, it is essential to device a novel targeted approach for HCC therapy.
It has been previously reported that c-Myc overexpression is commonly present in up to 70% of HCC, which is associated with liver carcinogenesis. Certain studies have shown that aberrantly high expression levels of oncogenic c-Myc play a key role in tumorigenesis and advancement of HCC. Researchers have indicated that c-Myc inactivation in cells with intact Rb, p16, p53, and Rb signaling leads to their senescence (Lin et al., 2010). Blocking c-Myc-mediated transactivation via small-molecule inhibitors has also developed as a c-Myc targeted therapeutic strategy. However, owing to their rapid metabolism, the application of these small molecule c-Myc inhibitors in vivo has been less promising (Guo et al., 2009). Bromodomain and extra-terminal (BET) proteins play an important role in the transcription of genes, such as c-Myc, BCL2, and BCL6, and their epigenetic deregulation has been found to be a critical factor for the development and metastasis of HCC. Bromodomain-containing protein 4 (BRD4), a BET family protein member, is found to be specifically involved in the transcription of these regulatory genes (Li et al., 2016). Accordingly, researchers have made efforts to target BRD4 for c-Myc inhibition via various small molecule inhibitors such as JQ1 and I-BET151, due to their encouraging anticancer activity in various preclinical cancer models. Nevertheless, they have limited efficacy in advanced solid tumors, and do not substantially inhibit cancer progression in most preclinical tumor models. Also, targeting BRD4 inhibition generally leads to weak anticancer activity in solid tumor cells due to feedback elevation of BRD4 protein (Fu et al., 2015).
One step ahead, we employed a BRD4 PROTAC (Proteolysis Targeting Chimera), ARV-825 (ARV), for selective degradation of BRD4 protein rather than its mere inhibition. PROTAC molecules comprise of two high-affinity binding ligands; one ligand binding to the target protein that is connected via a linker to another ligand binding to E3 ubiquitin ligase to eventually form a ternary complex for target protein degradation (Sun et al., 2019). ARV is a novel PROTAC composed of thienodiazepine moiety to target BRD4 protein and phthalimide moiety binding to E3 ubiquitin ligase cereblon (CRBN), linked via an ethoxy spacer. These two ligands upon binding to their respective receptors facilitate the recruitment of BRD4 protein to the E3 ubiquitin ligase cereblon, for its effective and prolonged degradation (Saraswat et al., 2020). Numerous researchers have demonstrated that ARV possesses significantly higher anticancer efficacy in in vitro and in vivo tumor models of pancreatic cancer, melanoma, Acute Myeloid Leukemia, prostate cancer, and Burkitt's Lymphoma as compared to small molecule BET inhibitors (Lu et al., 2015; Raina et al., 2016; Saenz et al., 2016; Saraswat et al., 2020). Therefore, instead of using a BRD4 inhibitor, we have identified the anticancer potential of a BRD4-targeted PROTAC, ARV, in HCC.
ARV is a weakly basic and a poorly water-soluble molecule that poses great difficulty for developing a clinically successful parenteral formulation (Rathod et al., 2019). Considering the potential of ARV in the treatment of HCC, there is a great need for its formulation development to facilitate its in vivo delivery. As it is widely known, nanoparticulate systems such as liposomes, lipid nanoparticles, and micelles tend to improve the bioavailability of therapeutic cargos and reduce their systemic toxicity. Among these, liposomes are spherical vesicles mainly composed of natural phospholipids. They serve as versatile drug delivery systems due to their biocompatibility, non-immunogenic nature and their biodegradability. Liposomes also have several advantages in terms of contributing to drug delivery; including enhancing drug solubility, enhancing circulation half-life of drugs, and serving as a sustained release system, while reducing their toxic effects of drugs and providing protection against drug degradation (Olusanya et al., 2018).
Various researchers have demonstrated that anticancer drug delivery in solid tumors is due to the enhanced permeability and retention (EPR) effect because of leaky tumor vasculature and poor lymphatic drainage system, resulting in passive targeting of chemotherapeutics. Furthermore, PEGylation of nanocarriers extends their systemic circulation by avoiding recognition and subsequent clearance by the reticuloendothelial system (RES) (Greish, 2010). To improve the selectivity towards liver cancer cells and reduce any off-target effects of the active moieties, some liver-targeted carriers have been established via their surface modification with galactose, cholic acid, glycyrrhetinic acid, etc. Among these, galactose can specifically recognize the asialoglycoprotein receptors (ASGPR) primarily expressed on the surfaces of hepatocytes for efficient liver-targeted delivery (Ding et al., 2019). In the present study, galactosyl ceramide would be utilized as an ASGPR targeting ligand to develop galactosylated nanoliposomes incorporating ARV (GALARV). There are no previous reports suggesting the potential use of ARV as a selective BRD4 protein degrader via an active targeting approach for the treatment of HCC. Furthermore, long circulation of nanoliposomes by PEGylation would facilitate their accumulation in the liver tumor matrix, and liver-specific delivery of ARV would minimize its off-target side effects. Therefore, specific objective of this research was the development and characterization of GALARV for its parenteral delivery. Targeted delivery of ARV as a novel PROTAC molecule has a great potential in exploiting the degradation of BRD4 protein to target the ‘undruggable’ c-Myc oncogene as an innovative approach towards HCC therapy.
2. Materials and methods
2.1. Materials
ARV-825 was purchased from MuseChem (NJ, USA). DMEM was purchased from Thermo Fisher Scientific Inc. (MA, USA), and fetal bovine serum (FBS) was obtained from Atlanta Biologicals (GA, USA). β-D-galactosylceramide (GC), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino (polyethylene glycol)-2000] (DSPE-PEG2000) and 1-oleoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3 phosphocholine (NBD-PC) were procured from Avanti (Alabaster, AL, USA). 1,2-Dioleoyl-sn-glycero-3 phosphocholine (DOPC) was procured from Cordenpharma (Liestal, Switzerland). Cholesterol, dimethyl sulfoxide (DMSO), sodium lauryl sulfate (SLS), and crystal violet were purchased from Sigma-Aldrich (MO, USA). Acetonitrile (ACN), MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide), citric acid, phosphate-buffered saline (PBS), and HPLC grade water were acquired from Thermo Fisher Scientific (NH, USA).
2.2. Cell culture
HepG2 and Hep3B human liver cancer cells were attained from American Type Culture Collection (VA, USA). The cell line was grown in high glucose DMEM media supplemented with 10% FBS, 1 mM sodium pyruvate, 2 mM L-glutamine, and antibiotics mixture at 37 °C in presence of 5% CO2.
2.3. HPLC analysis
The chromatographic detection of ARV was performed by using the Waters alliance system (Waters Corporation, MA, USA) (Rathod et al., 2019; Saraswat et al., 2020). The HPLC system consisted of a photodiode array (PDA) detector and InertSustain™ ODS C18 column with the dimensions of 150 mm × 4.6 mm and pore size of 5 μm (maintained at 25 °C). The mobile phase used was acetonitrile: potassium dihydrogen phosphate buffer (5 mM) pH 3.5 (70:30) with a flow rate of 0.7 mL/min and an injection volume of 10 μL. The retention time of ARV was found to be 5.6 ± 0.16 min, detected at 247 nm. Empower 3 software was used to detect the output signal.
2.4. Preparation and stability of galactose anchored ARV-loaded nanoliposomes (GALARV)
Modified hydration method was used to prepare galactose anchored nanoliposomes (GALARV). Briefly, ARV:DOPC:cholesterol:DSPE-PEG 2000:GC in a 1:51.1:16.2:1:0.4 M ratio were dissolved in chloroform. To prepare fluorescence-labeled liposomes, NBD-PC lipid (0.2 mol%) was added to the lipid mixture dissolved in chloroform. The lipid and drug solution was then added dropwise to parenteral-grade mannitol (200 μm) (maintained at 45 °C) with constant stirring followed by evaporation of chloroform. Resultant powder was dispersed in water with 0.05% w/v citric acid at 55 °C followed by probe sonication (30% amplitude) for 2 min. Similarly, non-targeted ARV-loaded liposomes (LARV) were prepared as per the aforementioned method without the incorporation of GC lipid.
Freshly prepared GALARV were then investigated for their physical stability in terms of particle size, zeta-potential as well as encapsulation efficiency (%) at different time points for a period of 6 months. During this period of 6 months, GALARV were stored at 4 °C.
2.5. Physicochemical characterization of ARV-loaded nanoliposomes
Average particle size, polydispersity index, and zeta potential of prepared liposomes were measured using a dynamic light scattering (DLS) particle size analyzer (Malvern Zetasizer Nano ZS, Royston, UK). Samples were diluted with deionized water and analyzed using folded capillary cells. HPLC analysis was performed to determine the encapsulation efficiency of ARV-loaded liposomes and was calculated using the following formula:
| Encapsulation efficiency (%) = (Amount of ARV in liposomes)/(Amount of ARV added) × 100% | (1) |
2.6. Microsomal enzyme assay
Human liver microsomal metabolism study of free ARV and GARLARV was performed as described previously (Patel et al., 2015; Rathod et al., 2019; Saraswat et al., 2020). ARV and GALARV stock solutions were prepared in Hank's balanced salt solution. Reaction samples were prepared by adding 2.5 μl of human liver microsomes (20 mg/ml) to 512.5 μl of prepared ARV and GALARV solution to achieve a final ARV concentration of 10 μM. Five microliter NADPH (50 mM) was then added to initiate the reaction which was carried out at 37 °C. Samples were withdrawn at specific time points and diluted with cold acetonitrile (ACN) to terminate the reaction. Following this, samples were centrifuged at 8400×g for 10 min after which HPLC analysis of the supernatant was performed to determine the ARV concentration. The percentage of ARV in the solution was plotted against time.
2.7. In vitro cytotoxicity assay
ARV, LARV, and GALARV were evaluated for their in vitro cytotoxicity in HepG2 and Hep3B human liver cancer cells. Briefly, 104 cells per well were seeded in 96-well plates and allowed to attach overnight. Cells were treated with various concentrations of ARV (in DMSO), LARV, and GALARV for a period of 72 h. Following this incubation, cell viability was analyzed by performing MTT colorimetric assay, and the drug concentration required to inhibit 50% growth (IC50) was calculated.
2.8. Cellular uptake and galactose competition assay
HepG2 cells were plated at a density of 1 × 104 cells/well in a 96-well plate and incubated overnight before treatment. For the galactose competition group, the cells were preincubated with 250 mM galactose as a competitive inhibitor of ASGPR for 30 min at 37 °C. Fluorescence-labeled non-targeted (LARV) and galactose anchored (GALARV) liposomes were incubated with cells for 2 h at 37 °C. Afterward, cells were washed with PBS and fixed with 4% v/v glutaraldehyde. The cellular uptake of LARV, GALARV in the absence and presence of galactose was observed for their fluorescence intensity using the EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific). Quantitative analysis of fluorescence intensity was performed by using ImageJ software.
For determining the cellular uptake of ARV, 4 × 105 HepG2 cells per well were seeded in a 6-well plate and then treated with ARV, LARV, and GALARV (40 μM) for 2 h at 37 °C. Afterward, the cells were washed twice with PBS and lysed by adding 0.5% sodium lauryl sulfate (SLS) to release the intracellular ARV. Samples were then diluted with acetonitrile, centrifuged at 13,300×g for 10 min and the supernatant was analyzed by HPLC to analyze the intracellular ARV concentration.
2.9. Flow cytometry for apoptosis analysis
Briefly, 2 × 105 HepG2 cells were seeded per well in a 12-well plate and were allowed to adhere overnight. Cells were treated with ARV, LARV, and GALARV (250 nM) for 48 h, and apoptosis was performed by using Muse Annexin V and Dead Cell Assay by following the manufacturer's protocol (MilliporeSigma, MA, USA). Following treatment, the cells were trypsinized and diluted to a concentration of 1 × 106 cells/ml with media containing 1% FBS and 1% bovine serum albumin (BSA). The cell suspension was diluted twice with MUSE Annexin V and dead cell reagent and incubated for 20 min, followed by its analysis using Muse® Cell Analyzer (MilliporeSigma).
2.10. In vitro hemolysis study
Mice red blood cells (RBCs) were used to carry out the in vitro hemolysis study of LARV and GALARV as described previously (Patel et al., 2016; Saraswat et al., 2020). C57BL/6 mice (5–6 weeks old) were received from Jackson laboratories (CT, USA). Briefly, mice were anesthetized by 2.5% isoflurane followed by a one-time blood collection using the cardiac puncture technique. Then, the animals were immediately euthanized by carbon dioxide. The experimental protocol was approved by the St. John's University Institutional Animal Care and Use Committee for the collection of blood from mice for laboratory use. Initially, centrifugation was performed at 450 x g for 10 min to separate the red blood cells from plasma and the cell pellet obtained was washed with and redispersed into a suitable volume of PBS to achieve the same hematocrit. LARV and GALARV were added to the RBC dispersion to achieve a 10 μg/ml ARV concentration. Following this, the samples were incubated for 30 min at 37 °C, after which they were centrifuged at 450×g for 10 min. The supernatant was diluted with PBS and analyzed for hemoglobin release using a UV spectrophotometer at 550 nm. Controls used for this experiment were PBS (negative control) and sodium lauryl sulfate (SLS) solution (positive control). Percentage hemolysis by the liposomal formulations was determined by the following formula:
| % Hemolysis = (absorbance of liposomal sample − absorbance of negative control)/(absorbance of positive control − absorbance of negative control) × 100% | (2) |
2.11. Formation and treatment of 3D multicellular liver tumor spheroids
For spheroid formation, HepG2 cells were seeded at a density of 1.5 × 103 cells/well in an ultra-low attachment 96-well plate (Corning Life Sciences, MA, USA), followed by centrifugation at 130×g for 10 min. Spheroid microplates were then cultured at 37 °C and 5% CO2 for a period of 72 h to form liver tumor spheroids of considerable integrity. Following this, they were treated with ARV, LARV, and GALARV (day 0) and were replaced with fresh treatment media every 48 h for up to 5 days. During the incubation with all the treatment groups, spheroids’ growth was observed in terms of their surface area and brightfield images were taken using an Evos imaging system (Thermo Fisher Scientific).
2.12. Cell viability within 3D multicellular liver tumor spheroids
Liver tumor spheroids were treated with ARV, LARV, and GALARV every alternate day for a period of 5 days, and on the 6th day of treatment, spheroids were stained with calcein-AM (μM) and Ethidium homodimer-1 (3 μM) using a Viability/Cytotoxicity Assay Kit (Biotium). Nuclei of cells were stained with DAPI (4 μM) (Thermo Fisher Scientific). All the three dyes were prepared in sterile PBS and added to the treated spheroids followed by a 3 h incubation at 37 °C for complete dye penetration before capturing fluorescent images using an Evos fluorescence microscope (Thermo Fisher Scientific).
2.13. Immunoblotting
HepG2 cells were treated with 0.5 μM ARV for 48 h and cells were lysed in modified RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% v/v NP40, 0.5% w/v deoxycholate, 0.1% w/v SDS, 10% v/v glycerol, 10 mM NaF, 0.4 mM EDTA) with protease inhibitors. Cell lysates were centrifuged at 4 °C for 10 min at 10,000 g. The supernatant was collected to which Laemmli sample buffer containing SDS and β-mercaptoethanol was added. Samples were denatured by heating at 95 °C for 10 min. Subsequently, samples were separated on polyacrylamide gels and transferred to PVDF membrane, and probed with primary antibodies BRD4 (13440), c-Myc (5605), Bcl-2 (3498), Survivin (2808) from Cell Signaling Technology, and β-actin antibody (66009) from Proteintech. HRP-conjugated secondary antibodies were used along with enhanced chemiluminescence substrate. Images were obtained with the Azure C500 imaging system and quantified using ImageJ 1.8.0 software.
2.14. Statistical analysis
Each experiment has been performed in triplicate and the data shown are reported as the mean ± standard deviation (SD). Student's t-test or one-way ANOVA followed by Bonferroni's or Tukey's multiple comparison test were used to perform all statistical analyses using GraphPad Prism7 (GraphPad Software, La Jolla, CA, USA) and Microsoft Excel. A p-value < 0.05 indicated a statistically significant difference between treatment groups.
3. Results
3.1. Physicochemical characterization and stability of ARV-loaded nanoliposomes
LARV and GALARV nanoliposomes were prepared by the modified hydration method. The physicochemical characterization of the non-targeted and targeted liposomes is presented in Figure 1. The particle size of both the nanoformulations was below 100 nm and they exhibited narrow particle size distributions, indicating the formation of uniform nano-sized particles. The non-targeted and targeted liposomal formulations were both negatively charged (−35.2 ± 3.41 mV, -27.30 ± 4.12 mV; respectively), suggesting that surface modification of liposomes with galactose did not alter their electrical potential. In addition, the encapsulation efficiency of these liposomes was similar and found to be >99%, indicating complete encapsulation of ARV within the lipid bilayers of the nanoliposomes formed.
Figure 1.
Physicochemical characterization ARV-loaded nanoliposomes. Dynamic light scattering graphs illustrating unimodal particle size distribution and zeta potential of (a) LARV and (b) GALARV.
GALARV were subjected to their stability study in liquid form for a period of 6 months. As depicted in Table 1, GALARV were found to be stable in terms of its physicochemical characterization and encapsulation efficiency. There was no statistically significant difference observed in the particle size, polydispersity index, zeta potential, and encapsulation efficiency of GALARV after 6 months in comparison to freshly prepared nanoliposomes. A narrow PDI and uniform particle size indicated that GALARV were stable, and that ARV was completely encapsulated within the liposomal lipid bilayer, preventing its precipitation or physicochemical degradation. The encapsulation efficiency was also found to be ∼100% in GALARV at the end of 6 months.
Table 1.
Physicochemical characterization of GALARV indicating their stability for 6 months.
| Time | Particle size (nm) | Polydispersity index | Zeta potential (mV) |
|---|---|---|---|
| 0 days | 93.83 ± 10.05 | 0.118 ± 0.054 | -27.30 ± 4.12 |
| 7 days | 90.07 ± 15.42 | 0.106 ± 0.027 | -28.54 ± 6.27 |
| 14 days | 92.05 ± 12.68 | 0.112 ± 0.012 | -26.40 ± 3.41 |
| 21 days | 89.86 ± 9.28 | 0.109 ± 0.025 | -25.85 ± 6.98 |
| 1 month | 90.54 ± 15.44 | 0.115 ± 0.026 | -27.69 ± 5.79 |
| 3 months | 94.78 ± 9.72 | 0.119 ± 0.047 | -28.42 ± 4.99 |
| 6 months | 92.66 ± 8.74 | 0.118 ± 0.034 | -26.86 ± 4.58 |
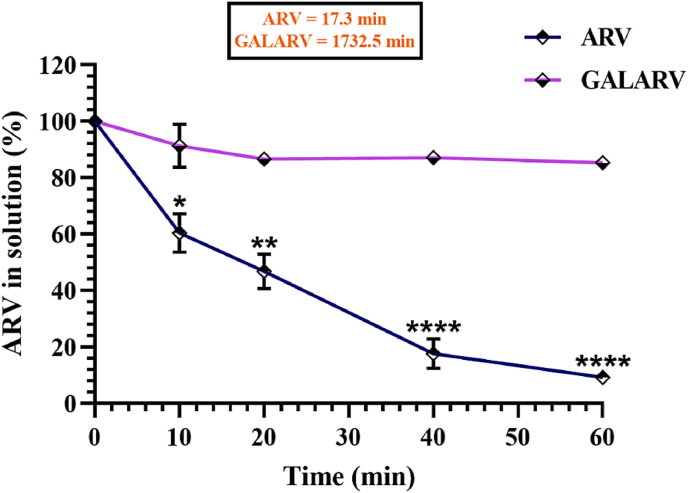
3.2. Microsomal enzyme assay
In Figure 2, a sharp decrease in ARV concentration with time was observed. Due to rapid metabolism by human liver microsomal enzyme, the half-life of ARV was around 17 min. While ARV-loaded nanoliposomes (GALARV) showed negligible ARV enzymatic metabolism compared with ARV alone at 60 min of the incubation period. This led to a ∼100-fold increase in the half-life of ARV encapsulated in the targeted nanoliposomes to about 1732 min, ascertaining the preclusion of its microsomal metabolism.
Figure 2.
Human liver microsomal assay results of ARV and GALARV. GALARV demonstrated a significant reduction in microsomal metabolism of ARV by nearly 80% in comparison to free ARV. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
3.3. In vitro cytotoxicity assay
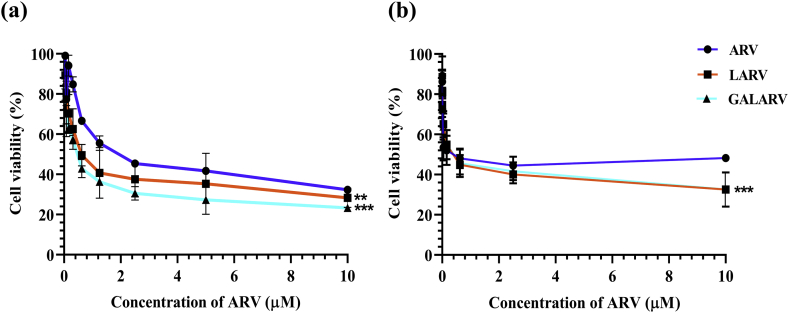
In vitro cytotoxicity of ARV, LARV and GALARV was evaluated in HepG2 and Hep3B cells. Table 2 summarizes the IC50 values of each treatment group. ARV and LARV showed comparable cytotoxicity with IC50 values of 0.82 μM and 0.76 μM in HepG2 cells while that of 0.65 μM and 0.40 μM in Hep3B cells, respectively. On the other hand, GALARV resulted in a reduced IC50 value of nearly 0.52 μM in HepG2 cells while that of 0.38 μM in Hep3B cells indicating higher cytotoxicity as compared to ARV alone. As shown in Figure 3, the order of cytotoxicity for the treatment groups in both the human hepatic cancer cell lines was as follows: ARV < LARV < GALARV.
Table 2.
IC50 (μM) of ARV, LARV, and GALARV in HepG2 and Hep3B liver cancer cells.
| Cell line | IC50 (μM) of various treatment groups |
||
|---|---|---|---|
| ARV | LARV | GALARV | |
| HepG2 | 0.82 ± 0.11 | 0.76 ± 0.08 | 0.52 ± 0.05 |
| Hep3B | 0.65 ± 0.24 | 0.40 ± 0.15 | 0.38 ± 0.18 |
Figure 3.
In vitro cytotoxicity assay of ARV, LARV and GALARV treatment in (a) HepG2 and (b) Hep3B cells (∗∗p < 0.01, ∗∗∗p < 0.001).
3.4. Cellular uptake and galactose competition assay
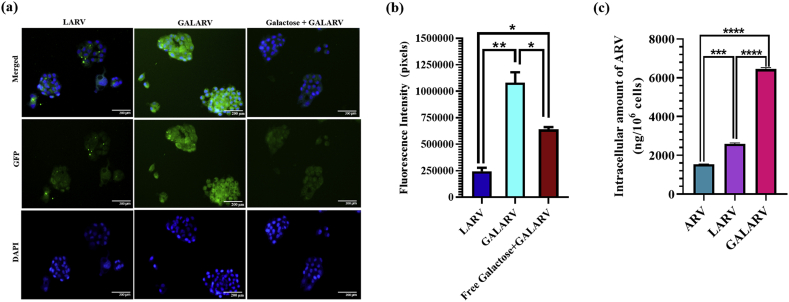
The ligand-conjugated drug delivery system is one of the most explored approaches for achieving liver-specific drug delivery. To achieve a highly effective liver-targeted delivery system, GALARV was developed as mentioned before, and evaluated for their targeted delivery efficiency in HepG2 human liver cancer cells. As depicted by the fluorescence images and quantitative analysis of fluorescence intensity (Figure 4 a and b), cellular uptake of GALARV was found to be significantly higher in HepG2 cells than that of LARV (fluorescence intensity ratio of 4.5:1), indicating that surface modification of liposomes with galactose could promote their recognition by hepatic cancer cells via ligand-receptor based interaction. The fluorescence intensity of GALARV in HepG2 was about 4.5 times higher than that of LARV, indicating the improvement in hepatoma cell-specific targeting ability of galactosyl-modified liposomes. This is due to the reason that the galactosyl group of GC present in GALARV can specifically recognize ASGPR overexpressed on the surface of liver cancer cells (Yousef et al., 2018). Moreover, the fluorescence intensity of cells treated with GALARV significantly reduced (∼2 times) when preincubated with galactose, further justifying their liver-targeted cellular uptake via ASGPR overexpressed on the surface of HepG2 cells. These results revealed the ASGPR-selective uptake of GALARV in liver cancer cells.
Figure 4.
Qualitive cellular uptake and galactose competition assay results. (a) Fluorescence microscopy images of HepG2 cells incubated with LARV and GALARV in presence and absence of galactose for 2 h at 37 °C. (b) Fluorescence intensity quantification of each GFP image as calculated by Image J software. Scale bar: 200 μm (∗p < 0.05, ∗∗p < 0.01). (c) Quantitative analysis of intracellular amount of ARV in HepG2cells incubated with ARV, LARV and GALARV in for 2 h at 37 °C. ((∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
To confirm the targeting efficiency of GALARV, intracellular drug uptake was also analyzed in HepG2 cells. As seen in Figure 4c, the intracellular amount of ARV was significantly higher in cells treated with both the nanoliposomes, LARV (∼1.6 fold) and GALARV (∼4.3 fold), in comparison to ARV alone (<1500 ng/106 cells). Moreover, the cellular uptake of ARV was much higher in GALARV (∼2.6 fold) as compared to LARV. This could be due to the high targeting efficiency of GALARV via modification with galactose, which acts as a targeting ligand for the overexpressed ASGPR on the surface of liver cancer cells. Hence, our results obtained from the cellular uptake study and galactose competition assay corroborated that developed GALARV were ASGPR-selective, which accounted for their enhanced uptake in liver cancer cells.
3.5. In vitro apoptosis assay
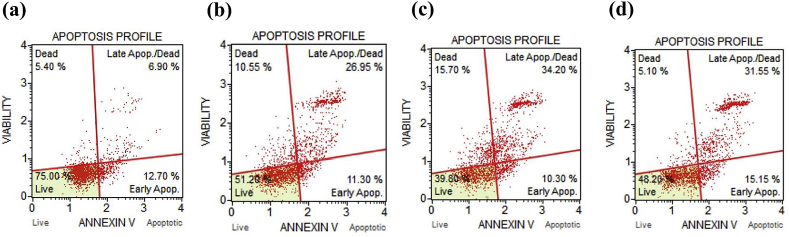
Annexin V apoptosis assay is a well-established technique for the identification and quantitative analysis of apoptotic population (Rathod et al., 2019; Saraswat et al., 2020; Wlodkowic et al., 2009). Hence, it was used to calculate the percentage of the apoptotic and dead population in HepG2 cells on exposure to ARV, LARV, and GALARV. As shown in Figure 5, ARV resulted in 38.25%, 44.50%, and 46.70% cell apoptosis for ARV, LARV, and GALARV, respectively. However, there was no significant difference observed between the LARV and GALARV induced apoptotic population. Overall, the in vitro apoptosis assay results signified that the anti-proliferative activity of ARV in liver cancer cells was exerted through induction of apoptosis.
Figure 5.
Quantification of apoptosis in HepG2 cells by flow cytometry for various treatment groups: (a) Control, (b) ARV, (c) LARV, and (d) GALARV. Significant apoptotic effect of ARV and ARV-loaded liposomes resulting in >35% and >40% apoptotic cell population, respectively following their treatment.
3.6. In vitro hemolysis study
Numerous factors are responsible for the hemocompatibility of nanoformulations including their size, shape, surface modification as well as the charge on the surface of the nanocarrier (Mukhopadhyay et al., 2019; Sharma et al., 2012). It is therefore very imperative to analyze that the surface of developed nanoliposomes is not toxic to the membrane of RBCs on systemic administration. For this purpose, the effect of LARV and GALARV on mice red blood cells was evaluated by performing the in vitro hemolysis study. As shown in Figure 6, LARV and GALARV, both exhibited negligible hemolysis (<5%) even at a 10 μg/ml concentration of ARV, in comparison to the positive control (sodium lauryl sulfate) which showed complete hemolysis. Notably, rapid and complete redispersion of RBCs following centrifugation implied that the nanoliposomes did not change the surface characteristics of RBCs.
Figure 6.
In vitro hemolysis study results of LARV and GALARV. Both the nanoformulations demonstrated negligible hemolysis even at 10 μg/mL ARV concentration.
3.7. Cell viability within 3D multicellular liver tumor spheroids
Three-dimensional cell culture models are widely used in investigations of cancer cells, intracellular interactions and for evaluation of toxicity and efficacy of potential chemotherapeutic and tumor microenvironment modulating drugs, and therefore show promise in filling the gap between 2D culturing and experiments with animals. To our comprehension, this is the very first report demonstrating the anticancer efficacy of a novel BRD4 PROTAC molecule – ARV, in 3D multicellular liver tumor spheroids. As indicated in Figure 7a, spheroids treated with ARV, LARV, and GALARV showed a gradual reduction in the diameter and area of the tumor spheroids, while a continued growth was observed in the control spheroids for up to 5 days. Moreover, the morphology of ARV and liposomal formulations treated spheroids was also different from the control group. The control spheroids showed a dark and dense core with a small number of apoptotic cells on the periphery. The treatment groups, on the other hand, showed only the dense core and irregular surface of the periphery due to the presence of a higher population of apoptotic cells as a result of significant cell killing. Amongst the treatment groups, liposomes treated groups showed a smaller and more dense spheroids when compared to ARV treated spheroids. This could be due to the significantly higher killing of cancer cells on the periphery while inhibiting the growth of spheroids during the treatment.
Figure 7.
Results for cell viability within 3D multicellular liver tumor spheroids. (a) Representative images of spheroids treated with control, ARV, LARV, and GALARV on days 0–5 of treatment. (b) Evaluation of the area of spheroids treated with various treatment groups in comparison to control during the 5 days of treatment. Significant difference depicted in the graph is in comparison to control group (c) Area of spheroids treated with all treatment groups as compared to control on the 5th day of treatment. (d) Fluorescence images depicting apoptosis of spheroids treated with various treatment groups. Composite images of DAPI (blue), calcein-AM (green) and the-1 (red) (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Scale bars, 400 μm.
Figure 7b represents the area of spheroids treated with ARV, LARV, and GALARV as a function of time. It can be observed that all treatment groups inhibited the growth of spheroids during the 5 days of treatment, resulting in a significant reduction in the area of spheroids treated with ARV, LARV, and GALARV when compared to control on the 5th day of treatment. Moreover, treatment with liposomes led to significantly higher inhibition in the growth of spheroids compared to ARV alone. As depicted in Figure 7c, treatment with liposomal formulations revealed even smaller spheroids in terms of their area as compared to ARV treatment alone. This could be due to the higher penetration of liposomes within the spheroids because of their high lipophilicity. Moreover, the galactose modification in GALARV would cause a higher penetration across the spheroids by recognizing the overexpressed ASGPR on the surface of HepG2 cells. Therefore, we believe that similar hepatoma-targeted cytotoxicity and efficacy would be achieved for GALARV in vivo.
The Live/Dead Cell Assay Kit was used to determine cell viability within the liver spheroids, which was further analyzed by fluorescence microscopy. Calcein AM stain emitted green fluorescence indicative of metabolically viable cells, while red fluorescence of ethidium homodimer-1 stain indicated dead cells with a compromised cell membrane that binds to intracellular nucleic acids. Blue fluorescence observed in the fluorescence images was due to DAPI staining of the cell nuclei. As seen in Figure 7d, spheroids treated with ARV as well as LARV and GALARV comprised of a higher number of dead cells (high red fluorescence) as compared to the control (high green fluorescence). The liposomal formulations, especially GALARV treated spheroids exhibited even a higher red fluorescence indicative of a higher cytotoxic effect by apoptosis as compared with ARV alone.
3.8. Western blot analysis
Expression of ARV target protein BRD4 and its regulated protein c-Myc was significantly reduced in ARV-treated cells by 2-fold and 3-fold, respectively (Figure 8, Figure S3). In addition, levels of an apoptosis inhibitor protein, survivin, and Bcl-2, an antiapoptotic protein, were significantly decreased by 4-fold and 5-fold, respectively, in the ARV-treatment group compared with control. The results show that ARV treatment induces apoptosis in human liver cancer HepG2 cells.
Figure 8.
ARV treatment induces apoptosis in HepG2 cells. (a) ARV was treated at 0.5 μM concentration in HepG2 cells for 48 h. Representative Western blots for protein expression of BRD-4, c-Myc, Bcl-2, and Survivin are shown (Supplementary material Figure S3). β-actin was used as an internal control. (b) Quantification of the Western blot images.
4. Discussion
c-Myc oncogene overexpression is often responsible for cancer cell proliferation, differentiation, and apoptosis in various types of cancers (Dang et al., 2006). HCC frequently overexpresses c-Myc, moreover, its amplification heralds a more advanced and aggressive form of HCC (C.-P. Lin et al., 2010). Given the significance of c-Myc in HCC tumorigenesis, it is a well-known target for developing novel therapeutic approaches for HCC. Shachaf et al. (2004), provided the very first evidence that down-regulation of c-Myc inhibited the growth of liver tumors and caused rapid loss of expression of the tumor markers; yet could not eradicate the tumor completely. Moreover, c-Myc reactivation immediately restored the neoplastic features of HCC. These results show how oncogenic c-Myc inactivation may reverse tumorigenesis in HCC (Shachaf et al., 2004). Simile et al. (2004), also showed that downregulation of c-Myc via antisense oligonucleotide inhibits HCC growth in vitro (Simile et al., 2004). Hence, the c-Myc oncogene is a well-validated but currently ‘undruggable’ driver in HCC. Therefore, there is a need to recognize novel molecules that target the ‘undruggable’ c-Myc for its effective inhibition and successive treatment of HCC.
BRD4, a BET family protein, is a key driver of oncogenesis that activates c-Myc transcription. It has previously been investigated as a therapeutic target in Myc-driven cancers (Devaiah et al., 2020). As suggested by Lin et al. (2007), a small molecule c-Myc inhibitor, 10058-F4, inhibited the proliferation of HCC cells in vitro to further sensitize the chemotherapeutic agents against HCC (Lin et al., 2007). Various researchers have also targeted the transcription and expression of c-Myc via inhibition of BET proteins. For instance, Li et al. (2016), demonstrated that siRNA or JQ1 mediated suppression of BRD4 protein could reduce tumor cell proliferation and induce apoptosis in hepatic cancer cell lines in vitro and also retarded the growth of HCC tumor xenograft in vivo (G.-Q. Li et al., 2016). Yin et al. (2019), proved that c-Myc is upregulated in liver cancer cells and JQ1 treatment resulted in a more potent anticancer activity as compared to sorafenib in c-Myc-positive HCC cells (Yin et al., 2019). Zhang et al. (2018), demonstrated that a cyclin-dependent kinase inhibitor, flavopiridol, when combined with JQ-1, downregulated Mcl-1 to induce apoptosis in multiple HCC cell lines (Zhang et al., 2018). However, BET inhibitors lack the selectivity for individual BET proteins, thereby limiting their scope in selectively targeting the physiologically relevant BRD4 protein to efficiently inhibit c-Myc expression (Zengerle et al., 2015). In this research work, we propose the use of a novel class of anticancer molecule, ARV, which selectively degrades the target BRD4 protein for sustained inhibition of c-Myc expression in HCC. Further, we have developed and characterized a galactosylated nanoformulation (GALARV) for targeted delivery of ARV and explored its anticancer activity in 2D and 3D in vitro cell culture models of human liver cancer. To our knowledge, this is the first study signifying the application of targeted nanoformulation of a novel class of BRD4 – PROTAC molecule as a treatment strategy for HCC.
As depicted by the preformulation studies of ARV performed by Rathod et al. (2019), it is a highly lipophilic and a high molecular weight compound with very poor aqueous solubility (Rathod et al., 2019). Therefore, there is a need to develop a stable formulation for its parenteral delivery. For this purpose, we formulated galactosyl anchored ARV-loaded nanoliposomes (GALARV) by modified hydration method, which is previously reported to produce liposomes with better physical stability and higher entrapment efficiency in comparison to conventional methods (Fu et al., 2019; Patel et al., 2016). GALARV was developed as a liver tumor-specific nanocarrier to improve the therapeutic potential of ARV in HCC by targeting ASGPR overexpressed on human hepatocellular carcinoma cells. To evaluate the efficacy of targeted nanocarriers in vitro, we developed galactose anchored nanoliposomes (GALARV) as well as non-targeted nanoliposomes (LARV) to encapsulate ARV. Both the formulations resulted in average particle size of about 100 nm, a negative surface charge, and uniform particle size distribution. Generally, the particle size of <200 nm is suitable for exploiting the passive targeting approach for nanocarriers in solid tumors via the EPR effect (Blanco et al., 2015). Also, the addition of PEG chains to the surface of our nanoliposomes will help bypass the reticuloendothelial system (RES), thereby reducing their clearance on systemic administration (Saraswat and Maher, 2020). This leads to the establishment of a steric barrier surrounding the liposomes to improve the efficacy of encapsulated ARV by harnessing its in vivo opsonization; prolonging blood circulation and providing accumulation at the target site while also attenuating possible side effects (Sercombe et al., 2015). We also found that GALARV were physically and chemically stable in liquid form for a period of 6 months at 4 °C. Our human liver microsomal enzyme study indicated that the half-life of ARV was prolonged by ∼100-fold after its encapsulation in nanoliposomal carrier GALARV, which reduced its enzymatic metabolism by approximately 80%. This suggests that the protection of ARV via its incorporation in the lipophilic phospholipid bilayers of liposomes would prevent its degradation in vivo. In vitro, drug release studies were also performed to predict the in vivo behavior of GALARV. ARV release from GALARV followed zero-order kinetics and exhibited a sustained release behavior without showing any burst release of ARV from the liposomes for up to 24 h (Figure S1). Therefore, we predict that ARV will be restricted within the liposomal bilayers of GALARV on systemic administration and will subsequently release the drug upon internalization by the hepatic cancer cells further contributing to its in vivo cytotoxicity. Also, our in vitro hemolysis study results suggested negligible hemolysis by GALARV even at a higher concentration of ARV, indicating its blood compatibility for intravenous administration. Similar results were also obtained for LARV, confirming the systemic safety of our developed nanoformulation. Furthermore, our in vitro biosafety results depicted that both GALARV and ARV were not toxic to Human embryonic kidney 293 (HEK 293) cells at any tested concentrations, implying its safety and specificity towards hepatic cancer cells (Figure S2).
The fluorescence images obtained from the cellular uptake study indicated significantly higher fluorescence intensity of GALARV (∼4.5 fold) as compared to LARV; obviously indicating their higher uptake in HepG2 cells. Moreover, the fluorescence intensity produced by GALARV significantly reduced (∼2 fold) when preincubated with galactose. On preincubation, galactose is identified and taken up by the ASGPR present on the surface of HepG2 cells to further inhibit the ASGPR-selective cellular uptake of GALARV. This confirms the competitive and liver-targeted cellular uptake of GALARV via ASGPR-mediated endocytosis. Consecutively, the drug uptake of GALARV was also found to be significantly higher than LARV (∼2.6 fold) and ARV (∼4.3 fold) alone, advocating the higher cellular uptake of actively targeted nanoformulation. Asialoglycoprotein is a receptor present on the surface of hepatocytes capable of specifically recognizing the terminals of β-d-galactose (Wu et al., 2002). One of the human hepatocellular carcinoma cell lines, HepG2, displays approximately 225,000 ASGPR per cell, leading to a high binding affinity for the galactosyl-modified nanocarriers (Singh and Ariatti, 2003). Therefore, ASGPR-targeted delivery is being actively explored for drug and gene targeting into liver cancer cells. This leads to higher cellular uptake of galactose conjugated nanocarriers and subsequent drug delivery of encapsulated cargos in hepatocellular carcinoma. Zhang et al. (2021), demonstrated the successful uptake of lupeol-loaded galactosylated liposomes by HepG2 cells, followed by efficacious in vitro and in vivo antitumor effects in the HCC xenograft model (Zhang et al., 2021). As previously shown by Nair et al. (2019), gemcitabine-loaded galactosylated chitosan nanoparticles resulted in liver tumor-specific delivery and enhanced the anti-HCC efficacy of gemcitabine in vivo (Nair et al., 2019). Similarly, Abd-Rabou et al., 2020, showed that viramidine-encapsulated galactosyl-terminating solid lipid nanoparticles exhibited higher cytotoxicity, apoptotic effect, and anti-angiogenic activity compared to the free drug; with confirmed specificity against liver cancer HepG2 cells (Abd-Rabou et al., 2020). Wei et al. (2015), also demonstrated that lactoferrin-modified doxorubicin nanoparticles exhibited higher anti-HCC efficacy in tumor xenograft model as compared to non-targeted nanoparticles and free doxorubicin (Wei et al., 2015). Our results are in accordance with previous studies where researchers demonstrated that higher fluorescence intensity obtained with targeted formulations could be an advantageous feature in attaining tumor cell-specific cytotoxic effect. Hence, our qualitative and quantitative cellular uptake study results showed that the targeted nanoliposomes (GALARV) were ideal for successful targeting of ARV specifically to the liver. Additionally, we have added DSPE-PEG 2000 (1 mol%) for steric stabilization of GALARV to prevent their macrophage-mediated uptake and RES clearance from the systemic circulation. Some researchers have indicated reduced uptake of galactosylated sterically stabilized liposomes by Kupffer cells, thus supporting our hypothesis that GALARV would achieve hepatocyte-specific delivery of ARV to exert its therapeutic effect in vivo (Nag and Ghosh, 1999; Samuelsson et al., 2017; Zhang et al., 2016). The enhanced permeability and retention (EPR) effect is a renowned passive targeting approach for tumor accumulation of nanoparticles with an average size beyond the renal clearance threshold, to extravasate from leaky tumor vessels (Shi et al., 2020). Therefore, we used a liver tumor-specific active targeting strategy to complement EPR based passive targeting of ARV-loaded nanoliposomes for their enhanced tumor accumulation and retention.
The results of in vitro cytotoxic activity of ARV in human liver cancer cells were very encouraging, indicating that ARV could be a potential candidate for HCC therapy. The IC50 values of GALARV were found to be significantly lower than ARV and LARV in both HepG2 and Hep3B cells. We expect that the in vivo behavior of GALARV might be more prominent considering the preferential distribution of actively targeted nanoliposomes in liver tumors and their extended systemic circulation due to PEGylation. Apoptosis assay was also performed to further evaluate the anticancer activity of ARV and developed nanoformulations in liver cancer. On exposure to ARV, LARV, and GALARV, a large population of cells exhibited early/late apoptosis in HepG2 cells. Also, the liposomal nanoformulations showed a higher percentage of apoptotic cells compared with free ARV; while the total apoptotic population resulted by GALARV was still higher than LARV. This could be justified by our Western blot results illustrating that ARV treatment significantly reduces the levels of anti-apoptotic proteins including Bcl-2 and survivin, signifying its anti-proliferative activity in HepG2 cells via apoptosis. Hence, the results obtained from in vitro cytotoxicity and apoptosis studies suggest that ARV is efficient in inhibiting the proliferation of liver cancer cells and its encapsulation in GALARV further enhanced its anti-hepatic cancer potential.
Promising anticancer activity of ARV in liver cancer is attributed to the target BRD4 protein degradation. Previously, investigators have reported that BRD4 is overexpressed in most HCC tumor tissues and induces epithelial-mesenchymal transition (EMT) phenotypes to cause proliferation and progression of HCC (Li et al., 2016; Zhang et al., 2015). This suggests that BRD4 protein could be a promising target-directed towards the treatment and management of HCC. Accordingly, BRD4 protein has been frequently targeted using small molecule BET inhibitors like JQ1 and I-BET151; which demonstrated potential anti-cancer activity in solid tumors, largely through the suppression of c-Myc oncogene (Filippakopoulos et al., 2010; Klingbeil et al., 2016). However, a short half-life and higher drug concentrations required to ensure sufficient inhibitory activity of such small molecule inhibitors can lead to toxicity and development of resistance (French, 2016). Hence, we used a direct protein degradation strategy by using a novel BRD4-PROTAC molecule, ARV, to recruit targeted BRD4 proteins to the E3 ubiquitin ligase for its complete degradation rather than its mere inhibition. From our Western blot results, we confirmed that the anticancer activity of ARV in HCC cells was via downregulation of target BRD4 protein and c-Myc oncogene. It has previously been demonstrated that c-Myc plays a critical role in the transcriptional regulation of survivin in different types of cancers including breast cancer, liver cancer, and leukemia (Fang et al., 2009; Galuppo et al., 2014; Papanikolaou et al., 2011; Warrier et al., 2020). Hence, c-Myc is a crucial transcription factor that activates survivin, a downstream target oncogene, to prevent apoptosis. This is in accordance with our Western blot results where treatment of human liver cancer cells with ARV significantly reduced the levels of oncogenic c-Myc via target BRD4 protein degradation to further downregulate the expression of anti-apoptotic protein survivin.
Three-dimensional (3D) growth of immortalized established cell lines is considered as a more illustrative model for screening anticancer drugs in vitro (Thoma et al., 2014). Kimlin et al. (2013), suggested that 3D cell culture models possess several in vivo tumor features like intracellular interaction, penetration, and resistance of therapeutic cargos, as well as tumor extracellular matrix production (Kimlin et al., 2013). Therefore, 3D in vitro models could fill the gap between conventional 2D in vitro studies and tumor xenograft models to study cancer cell dynamics and better investigate the efficacy of anticancer therapeutics (Zanoni et al., 2016). We developed 3D multicellular liver tumor spheroids and analyzed the cell viability within the spheroids using Viability/Cytotoxicity Assay Kit by fluorescent microscopy. Our results suggested that ARV as well as its liposomal formulations, LARV and GALARV, induced significant apoptosis in the HepG2 spheroids based on the high red fluorescence produced by apoptotic/dead cells in comparison to the green fluorescence of live cells produced by the control spheroids. Also, reduction in the spheroid size after treatment with ARV and both liposomal formulations suggested that permeation of lipophilic ARV in spheroids led to the substantial killing of the cells. While ARV and its liposomal formulations, both kill liver cancer cells in 2D cell culture models, their cytotoxic effect will highly depend on their permeability through the developed 3D liver tumor spheroids. On the 5th day of treatment, we observed that GALARV led to the highest reduction in spheroid size compared to other treatment groups while the control showed continued growth. It could be postulated that the galactose groups present on the surface of GALARV allowed their higher uptake via overexpressed ASGPR in HCC cells in addition to the highly lipophilic phospholipid bilayers of nanoliposomes which could cross the cell membrane to cause release and diffusion of encapsulated ARV through the tightly bound cells in 3D spheroids for effective cell killing. Thus, we think that the targeted nanoliposomes of ARV (GALARV) would be a promising anticancer therapy for the treatment of HCC, with exceptional apoptotic activity in the in the vivo tumor microenvironment.
5. Conclusion
Our study confirms the anticancer activity of galactosylated nanoliposomes incorporating BRD4-targeted PROTAC, ARV, against liver cancer. GALARV were developed as a combination of active and passive targeting approach for parenteral delivery of lipophilic ARV. It demonstrated higher hepatic uptake via ASGPR and showed encouraging apoptosis and significant cytotoxicity in 2D cell culture models as well as in the 3D multicellular tumor spheroids model of liver cancer cells. Target protein BRD4 degradation and suppression of oncogenic c-Myc expression by ARV was also confirmed by Western blot analysis. The very first liver-specific delivery of ARV via its incorporation in a stable galactose anchored nanoliposomal system makes it a strong anticancer candidate for HCC therapy. Hence, our research work elicits robust anti-HCC activity of GALARV and strongly suggests that BRD4 degradation using PROTAC technology and its delivery via targeted nanoformulation could be a unique therapeutic approach for treatment and management of HCC.
Declarations
Author contribution statement
Aishwarya Saraswat: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Hari Priya Vemana: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Vikas V Dukhande: Conceived and designed the experiments; Analyzed and interpreted the data.
Ketan Patel: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the College of Pharmacy and Health Sciences, St. John’s University, Queens, NY.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Abd-Rabou A.A., Bharali D.J., Mousa S.A. Viramidine-loaded galactosylated nanoparticles induce hepatic cancer cell apoptosis and inhibit angiogenesis. Appl. Biochem. Biotechnol. 2020;190:305–324. doi: 10.1007/s12010-019-03090-2. [DOI] [PubMed] [Google Scholar]
- Blanco E., Shen H., Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V., O’Donnell K.A., Zeller K.I., Nguyen T., Osthus R.C., Li F. The c-Myc target gene network. Semin. Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Devaiah B.N., Mu J., Akman B., Uppal S., Weissman J.D., Cheng D., Baranello L., Nie Z., Levens D., Singer D.S. MYC protein stability is negatively regulated by BRD4. Proc. Natl. Acad. Sci. U.S.A. 2020;117:13457–13467. doi: 10.1073/pnas.1919507117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., Li Z., Wang J., Zhu X., Zhao Z., Wang M. Design and synthesis of galactose-biotin lipid materials for liposomes to promote the hepatoma cell–targeting effect. J. Pharm. Sci. 2019;108:3074–3081. doi: 10.1016/j.xphs.2019.04.007. [DOI] [PubMed] [Google Scholar]
- Fang Z.H., Dong C.L., Chen Z., Zhou B., Liu N., Lan H.F., Liang L., Liao W. Bin, Zhang L., Han Z.C. Transcriptional regulation of survivin by c-Myc in BCR/ABL-transformed cells: implications in anti-leukaemic strategy. J. Cell Mol. Med. 2009;13:2039–2052. doi: 10.1111/j.1582-4934.2008.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., Philpott M., Munro S., McKeown M.R., Wang Y., Christie A.L., West N., Cameron M.J., Schwartz B., Heightman T.D., La Thangue N., French C.A., Wiest O., Kung A.L., Knapp S., Bradner J.E. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C.A. Small-molecule targeting of BET proteins in cancer. Adv. Cancer Res. 2016;131:21–58. doi: 10.1016/bs.acr.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Fu L., Tian M., Li X., Li J., Huang J., Ouyang L., Zhang Y., Liu B. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget. 2015;6:5501–5516. doi: 10.18632/oncotarget.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Rathod D., Abo-Ali E.M., Dukhande V.V., Patel K. EphA2-Receptor targeted PEGylated nanoliposomes for the treatment of BRAF(V600E) mutated parent- and vemurafenib-resistant melanoma. Pharmaceutics. 2019;11 doi: 10.3390/pharmaceutics11100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galuppo R., Maynard E., Shah M., Daily M.F., Chen C., Spear B.T., Gedaly R. Synergistic inhibition of HCC and liver cancer stem cell proliferation by targeting RAS/RAF/MAPK and WNT/β-catenin pathways. Anticancer Res. 2014;34:1709–1713. [PMC free article] [PubMed] [Google Scholar]
- Greish K. In: Enhanced Permeability and Retention (EPR) Effect for Anticancer Nanomedicine Drug Targeting BT - Cancer Nanotechnology: Methods and Protocols. Grobmyer S.R., Moudgil B.M., editors. Humana Press; Totowa, NJ: 2010. pp. 25–37. [DOI] [PubMed] [Google Scholar]
- Guo J., Parise R.A., Joseph E., Egorin M.J., Lazo J.S., Prochownik E.V., Eiseman J.L. Efficacy, pharmacokinetics, tisssue distribution, and metabolism of the Myc-Max disruptor, 10058-F4 [Z,E]-5-[4-ethylbenzylidine]-2-thioxothiazolidin-4-one, in mice. Cancer Chemother. Pharmacol. 2009;63:615–625. doi: 10.1007/s00280-008-0774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimlin L.C., Casagrande G., Virador V.M. In vitro three-dimensional (3D) models in cancer research: an update. Mol. Carcinog. 2013;52:167–182. doi: 10.1002/mc.21844. [DOI] [PubMed] [Google Scholar]
- Klingbeil O., Lesche R., Gelato K.A., Haendler B., Lejeune P. Inhibition of BET bromodomain-dependent XIAP and FLIP expression sensitizes KRAS-mutated NSCLC to pro-apoptotic agents. Cell Death Dis. 2016;7:e2365. doi: 10.1038/cddis.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.-Q., Guo W.-Z., Zhang Y., Seng J.-J., Zhang H.-P., Ma X.-X., Zhang G., Li J., Yan B., Tang H.-W., Li S.-S., Wang L.-D., Zhang S.-J. Suppression of BRD4 inhibits human hepatocellular carcinoma by repressing MYC and enhancing BIM expression. Oncotarget. 2016;7:2462–2474. doi: 10.18632/oncotarget.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-P., Liu C.-R., Lee C.-N., Chan T.-S., Liu H.E. Targeting c-Myc as a novel approach for hepatocellular carcinoma. World J. Hepatol. 2010;2:16–20. doi: 10.4254/wjh.v2.i1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-P., Liu J.-D., Chow J.-M., Liu C.-R., Liu H.E. Small-molecule c-Myc inhibitor, 10058-F4, inhibits proliferation, downregulates human telomerase reverse transcriptase and enhances chemosensitivity in human hepatocellular carcinoma cells. Anti Cancer Drugs. 2007;18:161–170. doi: 10.1097/CAD.0b013e3280109424. [DOI] [PubMed] [Google Scholar]
- Lu J., Qian Y., Altieri M., Dong H., Wang J., Raina K., Hines J., Winkler J.D., Crew A.P., Coleman K., Crews C.M. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Veroniaina H., Chimombe T., Han L., Zhenghong W., Xiaole Q. Synthesis and compatibility evaluation of versatile mesoporous silica nanoparticles with red blood cells: an overview. RSC Adv. 2019;9:35566–35578. doi: 10.1039/c9ra06127d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Ghosh P.C. Assessment of targeting potential of galactosylated and mannosylated sterically stabilized liposomes to different cell types of mouse liver. J. Drug Target. 1999;6:427–438. doi: 10.3109/10611869908996849. [DOI] [PubMed] [Google Scholar]
- Nair A.B., Shah J., Al-Dhubiab B.E., Patel S.S., Morsy M.A., Patel V., Chavda V., Jacob S., Sreeharsha N., Shinu P., Attimarad M., Venugopala K.N. Development of asialoglycoprotein receptor-targeted nanoparticles for selective delivery of gemcitabine to hepatocellular carcinoma. Molecules. 2019;24:4566. doi: 10.3390/molecules24244566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olusanya T.O.B., Haj Ahmad R.R., Ibegbu D.M., Smith J.R., Elkordy A.A. Liposomal drug delivery systems and anticancer drugs. Molecules. 2018;23:907. doi: 10.3390/molecules23040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolaou V., Iliopoulos D., Dimou I., Dubos S., Kappas C., Kitsiou-Tzeli S., Tsezou A. Survivin regulation by HER2 through NF-κB and c-myc in irradiated breast cancer cells. J. Cell Mol. Med. 2011;15:1542–1550. doi: 10.1111/j.1582-4934.2010.01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K., Chowdhury N., Doddapaneni R., Boakye C.H.A., Godugu C., Singh M. Piperlongumine for enhancing oral bioavailability and cytotoxicity of docetaxel in triple-negative breast cancer. J. Pharm. Sci. 2015;104:4417–4426. doi: 10.1002/jps.24637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K., Doddapaneni R., Chowdhury N., Boakye C.H., Behl G., Singh M. Tumor stromal disrupting agent enhances the anticancer efficacy of docetaxel loaded PEGylated liposomes in lung cancer. Nanomedicine. 2016;11:1377–1392. doi: 10.2217/nnm.16.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina K., Lu J., Qian Y., Altieri M., Gordon D., Rossi A.M.K., Wang J., Chen X., Dong H., Siu K., Winkler J.D., Crew A.P., Crews C.M., Coleman K.G. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. 2016;113:7124. doi: 10.1073/pnas.1521738113. LP – 7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod D., Fu Y., Patel K. BRD4 PROTAC as a novel therapeutic approach for the treatment of vemurafenib resistant melanoma: preformulation studies, formulation development and in vitro evaluation. Eur. J. Pharmaceut. Sci. 2019;138:105039. doi: 10.1016/j.ejps.2019.105039. [DOI] [PubMed] [Google Scholar]
- Saenz D.T., Fiskus W., Raina K., Manshouri T., Coleman K., Winkler J., Qian Y., Crew A., Shen A., Mill C.P., Sun B., Verstovsek S., Crews C., Bhalla K.N. Superior lethal activity of novel BET protein proteolysis targeting Chimera (BETP-PROTACs) versus betp bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm (MPN) secondary (s) AML cells. Blood. 2016;128:747. doi: 10.1038/leu.2016.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson E., Shen H., Blanco E., Ferrari M., Wolfram J. Contribution of Kupffer cells to liposome accumulation in the liver. Colloids Surf. B Biointerfaces. 2017;158:356–362. doi: 10.1016/j.colsurfb.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraswat A., Patki M., Fu Y., Barot S., Dukhande V.V., Patel K. Nanoformulation of PROteolysis TArgeting Chimera targeting ‘undruggable’ c-Myc for the treatment of pancreatic cancer. Nanomedicine. 2020;15:1761–1777. doi: 10.2217/nnm-2020-0156. [DOI] [PubMed] [Google Scholar]
- Saraswat A.L., Maher T.J. Development and optimization of stealth liposomal system for enhanced in vitro cytotoxic effect of quercetin. J. Drug Deliv. Sci. Technol. 2020;55 [Google Scholar]
- Sercombe L., Veerati T., Moheimani F., Wu S.Y., Sood A.K., Hua S. Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 2015;6:286. doi: 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachaf C.M., Kopelman A.M., Arvanitis C., Karlsson A., Beer S., Mandl S., Bachmann M.H., Borowsky A.D., Ruebner B., Cardiff R.D., Yang Q., Bishop J.M., Contag C.H., Felsher D.W. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Sharma A., Madhunapantula S.V., Robertson G.P. Toxicological considerations when creating nanoparticle-based drugs and drug delivery systems. Expet Opin. Drug Metabol. Toxicol. 2012;8:47–69. doi: 10.1517/17425255.2012.637916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., van der Meel R., Chen X., Lammers T. The EPR effect and beyond: strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics. 2020 doi: 10.7150/thno.49577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simile M.M., De Miglio M.R., Muroni M.R., Frau M., Asara G., Serra S., Muntoni M.D., Seddaiu M.A., Daino L., Feo F., Pascale R.M. Down-regulation of c-myc and Cyclin D1 genes by antisense oligodeoxy nucleotides inhibits the expression of E2F1 and in vitro growth of HepG2 and Morris 5123 liver cancer cells. Carcinogenesis. 2004;25:333–341. doi: 10.1093/carcin/bgh014. [DOI] [PubMed] [Google Scholar]
- Singal A.G., Lampertico P., Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J. Hepatol. 2020;72:250–261. doi: 10.1016/j.jhep.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Ariatti M. Targeted gene delivery into HepG2 cells using complexes containing DNA, cationized asialoorosomucoid and activated cationic liposomes. J. Contr. Release. 2003;92:383–394. doi: 10.1016/s0168-3659(03)00360-2. [DOI] [PubMed] [Google Scholar]
- Sun X., Gao H., Yang Y., He M., Wu Y., Song Y., Tong Y., Rao Y. PROTACs: great opportunities for academia and industry. Signal Transduct. Target. Ther. 2019;4:64. doi: 10.1038/s41392-019-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma C.R., Zimmermann M., Agarkova I., Kelm J.M., Krek W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv. Drug Deliv. Rev. 2014;69–70:29–41. doi: 10.1016/j.addr.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Warrier N.M., Agarwal P., Kumar P. Emerging importance of survivin in stem cells and cancer: the development of new cancer therapeutics. Stem Cell Rev. Rep. 2020;16:828–852. doi: 10.1007/s12015-020-09995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Guo X., Tu L., Zou Q., Li Q., Tang C., Chen B., Xu Y., Wu C. Lactoferrin-modified PEGylated liposomes loaded with doxorubicin for targeting delivery to hepatocellular carcinoma. Int. J. Nanomed. 2015;10:5123–5137. doi: 10.2147/IJN.S87011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodkowic D., Skommer J., Darzynkiewicz Z. Flow cytometry-based apoptosis detection. Methods Mol. Biol. 2009;559:19–32. doi: 10.1007/978-1-60327-017-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Nantz M.H., Zern M.A. Targeting hepatocytes for drug and gene delivery: emerging novel approaches and applications. Front. Biosci. 2002;7:d717–d725. doi: 10.2741/A806. [DOI] [PubMed] [Google Scholar]
- Yang J.D., Hainaut P., Gores G.J., Amadou A., Plymoth A., Roberts L.R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Sun M., Zhan X., Wu C., Geng P., Sun X., Wu Y., Zhang S., Qin J., Zhuang Z., Liu Y. EGFR signaling confers resistance to BET inhibition in hepatocellular carcinoma through stabilizing oncogenic MYC. J. Exp. Clin. Cancer Res. 2019;38:1–15. doi: 10.1186/s13046-019-1082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef S., Alsaab H.O., Sau S., Iyer A.K. Development of asialoglycoprotein receptor directed nanoparticles for selective delivery of curcumin derivative to hepatocellular carcinoma. Heliyon. 2018;4 doi: 10.1016/j.heliyon.2018.e01071. e01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni M., Piccinini F., Arienti C., Zamagni A., Santi S., Polico R., Bevilacqua A., Tesei A. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016;6:19103. doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengerle M., Chan K.-H., Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.-P., Li G.-Q., Zhang Y., Guo W.-Z., Zhang J.-K., Li J., Lv J.-F., Zhang S.-J. Upregulation of Mcl-1 inhibits JQ1-triggered anticancer activity in hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2018;495:2456–2461. doi: 10.1016/j.bbrc.2017.12.153. [DOI] [PubMed] [Google Scholar]
- Zhang J., Hu X., Zheng G., Yao H., Liang H. In vitro and in vivo antitumor effects of lupeol-loaded galactosylated liposomes. Drug Deliv. 2021;28:709–718. doi: 10.1080/10717544.2021.1905749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Dong Z., Cai J., Zhang C., Shen Z., Ke A., Gao D., Fan J., Shi G. BRD4 promotes tumor growth and epithelial-mesenchymal transition in hepatocellular carcinoma. Int. J. Immunopathol. Pharmacol. 2015;28:36–44. doi: 10.1177/0394632015572070. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-N., Poon W., Tavares A.J., McGilvray I.D., Chan W.C.W. Nanoparticle–liver interactions: cellular uptake and hepatobiliary elimination. J. Contr. Release. 2016;240:332–348. doi: 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.