On 24 November, a new detected variant B.1.1.529 of SARS-CoV-2 by South Africa was reported to WHO. After only 2 days, this variant was designated as “variant of concern” (VOC) and named as Omicron. In the past few weeks, Omicron had reported from more than 80 countries. It has been reported as the dominant SARS-CoV-2 in U.S. due to the rapid spread of Omicron. A new wave of infection driven by Omicron is in progress.
The major reason that Omicron raise a great concern is its accumulated mutations, including more than 30 of those in the spike (S) protein. More importantly, 15 of those mutations occurs on receptor-binding domain (RBD) (Fig. 1a), which is not only the vital binding site to the host receptor angiotensin-converting enzyme 2 (ACE2) for the entry of SARS-CoV-2, but also the key target of neutralizing antibodies produced by immune response and therapeutics antibodies. By contrast, other VOCs including Alpha, Beta, Gamma, and Delta possess 9–12 mutations on their S protein regions. Even so, some crucial mutations like D614G, N501Y, K417N, and E484A in these known VOCs have been reported about the effect on viral infectivity and transmission [1–4]. Except for these shared mutations mentioned above, with additional mutations on Omicron, there is a pressing need for more evidence directed at the evaluation of the synergistic effect.
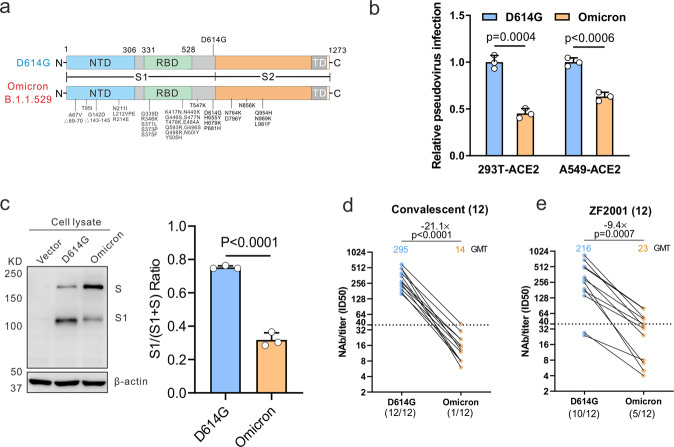
Fig. 1.
The effect of the Omicron variant on viral infectivity and immune escape. a Mutational landscapes in variants Omicron and D614G. b HEK293T-hACE2 cells or A549-hACE2 cells were infected with pseudotyped Omicron and D614G particles bearing all S mutations to determine viral infectivity using a luciferase assay. c HEK293T cells were transfected with the spike-expressing plasmids of Omicron and D614G variants and the immunoblots were probed with the indicated antibodies; the cleavage quantified by densitometry using ImageJ. d, e Pseudovirus-based neutralizing assays were performed to detect neutralizing activity of the sera from the convalescents (n = 12, sampled at around 30 days after symptom onset) (d) and vaccinated individuals with RBD subunit vaccine ZF2001 (n = 12, sampled at 15-60 days post the 3rd dose) (e) against SARS-CoV-2 Omicron and D614G variants. The half-maximal inhibitory dose (ID50) was indicated by geometric mean titers (GMT). The threshold of ID50 detection was 1:40. Statistical data analysis was performed using GraphPad Prism version 8.0 software. Student’s t-tests were used to compare between Omicron and D614G variants. Statistical significance was determined using ANOVA for multiple comparisons. When P values < 0.05, differences were deemed as the statistically significant
To determine the effect of Omicron S protein mutations on its capacity of infectivity and immune evasion, a previously described method was used to construct Omicron pseudotyped virus [5]. After codon optimization according to the sequence from GISAID (EPI_ISL_6640917), the synthesized S gene of Omicron was constructed into a luciferase-expressing pseudotyping lentiviral system based on HIV-1 backbone. Pseudotyped Omicron particles bearing all S mutations were used to infect the HEK293T cells or A549 cells expressing ACE2 receptor for the measurement of viral infectivity in a luciferase assay. A previously constructed SARS-CoV-2 pseudovirus with the ancestral D614G mutation in S protein was also applied as the reference, since the D614G mutation has been reported among almost all SARS-CoV-2 variants. As shown in Fig. 1b, the lower entry efficiency of Omicron than D614G was observed with 49% and 37% decreased luciferase activity in HEK293T-hACE2 and A549-hACE2 cells at 72 h post infection, suggesting that Omicron S protein mutations lead to its reduced infectivity. Furthermore, we transfected HEK293T cells with the S-expressing plasmids of Omicron and D614G variants to examine protein expression and cleavage. The immunoblot analysis displayed S proteins cleaved into two major bands denoting full-length S and S1 subunit by the cellular proteases. Compared with the expression level of the two proteins in the D614G variant, Omicron showed a reduced level of S1 subunit (Fig. 1c). It indicates less proteases cleavage of the Omicron variant, consistent with another report [6].
On the other hand, pseudotype-based neutralizing assay was performed as previously described to analyze neutralizing antibodies (NAbs) elicited by previously infection or the RBD-based protein subunit vaccine ZF2001 against the Omicron variant [7]. Sera sampled at around one month after symptom onset from 12 convalescents who were previously infected by SARS-CoV-2 original strain shows a more than 20-fold decrease of neutralizing activity against Omicron variant, when compared to D614G variant (Fig. 1d). Only one of these 12 individuals remains slight neutralizing effect on the Omicron variant. Among 12 individuals vaccinated by RBD-based protein subunit vaccine ZF2001, 58.3% (7/12) sera sampled at 15-60 days after 3rd-dose vaccination did not neutralize Omicron. Geometric mean titers (GMTs, 50% inhibitory dose [ID50]) of these sera against Omicron were 9.4-fold lower than against D614G (Fig. 1e).
Here we have shown that the new SARS-CoV-2 variant Omicron S protein with a large number of mutations has an outstanding effect on the viral infectivity and immune escape ability. Unexpectedly, different with several previous reports, compared to other VOCs, reduced entry efficiency and less cleavage ability were observed in our study. It makes sense that a reduced cleavage efficiency leads to the decrease of viral entry. The reduced infection was also observed in human lung epithelia-derived CaLu-3 cells [6] and in an ex vivo model of human lung tissue with authentic virus [8]. Seventy-fold faster infection in human bronchus with the low lung infection has been thought as an indicator of the fast spread of Omicron with lower disease severity [8]. However, using SARS-CoV-2 virus-like particles, omicron with mutations of all four structural proteins displayed increased infectivity [9]. The contrary results may be caused by the diverse entry efficiency of the Omicron variant generated by the pseudovirus or the virus-like particles system in distinct cell lines. Due to the limitation of the pseudoviruses with S protein, further investigation using authentic virus should be addressed to validate and explain the mechanisms about Omicron’s infectivity and fast transmission.
Consistent with other studies, we have observed that protective immunity after previous infection could barely neutralize Omicron. Even worse, almost all vaccines that have extensively used exhibit the remarkable reduced neutralization against Omicron [6, 10, 11]. However, it is worth noting that administration of a booster dose and vaccination of individuals with previous infection present a better neutralizing response [10]. In our study, the better neutralization of third-dose RBD subunit vaccine sera against Omicron displays a lower fold-change (9.4 folds) than convalescent sera (21.1 folds). We hypothesize that antibody affinity maturation induced by vaccines with multiple doses will be benefit for increased neutralization against future variants like Omicron [12].
Taken together the results suggested a higher risk of Omicron breakthrough infections and reduced efficiency of the protective immunity elicited by existing vaccines. There are important implications about the modification and optimization of the current epidemic prevention and control including vaccine strategies and therapeutic antibodies against the new SARS-CoV-2 variant Omicron.
Supplementary information
Increased immune escape of the new SARS-CoV-2 variant of concern Omicron
Acknowledgements
We acknowledge funding support from the China National Natural Science Foundation (grant no. U20A20392), the 111 Project (No. D20028), the Research Fund Program of the Key Laboratory of Molecular Biology for Infectious Diseases, CQMU (No. 202105, 202102), the Emergency Project from the Science & Technology Commission of Chongqing (cstc2020jscx-fyzx0053), the Emergency Project for Novel Coronavirus Pneumonia from the Chongqing Medical University (CQMUNCP0302), the Leading Talent Program of CQ CSTC (CSTCCXLJRC201719), and a Major National Science & Technology Program grant (2017ZX10202203) from the Science & Technology Commission of China, National Natural Science Foundation of China (Grant No.82102361), China Postdoctoral Science Foundation (2021M693924), Natural Science Foundation of Chongqing, China (cstc2021jcyj-bshX0115) and Chongqing Postdoctoral Science Special Foundation (2010010005216630).
Author contributions
AH, NT, K Wang, PP, and JH developed the conceptual ideas and designed the study. JH, PP, and K Wu performed the experiments and statistical analysis. QL and JC provided the samples. All authors provided scientific expertise and the interpretation of data for the work. PP drafted the manuscript. All authors contributed to critical revision of the manuscript for important intellectual content. All authors reviewed and approved the final version of the report.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jie Hu, Pai Peng, Xiaoxia Cao.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00836-z.
References
- 1.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–21. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Liu J, Plante KS, Plante JA, Xie X, Zhang X, et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2021; 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed]
- 3.Jhun H, Park H-Y, Hisham Y, Song C-S, Kim S. SARS-CoV-2 Delta (B.1.617.2) variant: a unique T478K mutation in receptor binding motif (RBM) of spike gene. Immune Netw. 2021;21:e32. doi: 10.4110/in.2021.21.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell host microbe. 2021;29:477–88. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J, Gao Q, He C, Huang A, Tang N, Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020;7:551–7. doi: 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong Z, Evans JP, Qu P, Faraone J, Zheng YM, Carlin C, et al. Neutralization and Stability of SARS-CoV-2 Omicron Variant. bioRxiv, 2021; Preprint at 10.1101/2021.2012.2016.472934.
- 7.Peng P, Hu J, Deng HJ, Liu BZ, Fang L, Wang K, et al. Changes in the humoral immunity response in SARS-CoV-2 convalescent patients over 8 months. Cell Mol Immunol. 2021;18:490–1. doi: 10.1038/s41423-020-00605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan CW, Hui PY, Tam WC, Poon LLM, Nicholls J. Omicron SARS-CoV-2 can infect faster and better than Delta in human bronchus but with less severe infection in lung. In press (December 15th, 2021). https://www.med.hku.hk/en/news/press/20211215-omicron-sars-cov-2-infection.
- 9.Syed F, Li W, Relich RF, Russell PM, Zhang S, Zimmerman MK, et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. medRxiv, 2021; Preprint at 10.1101/2021.12.20.21268048.
- 10.Liu L, Iketani S, Guo Y, Chan JFW, Wang M, Liu L, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021; 10.1038/d41586-021-03826-3. [DOI] [PubMed]
- 11.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, et al. Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. Nature. 2021;596:276–80. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 12.Tang J, Grubbs G, Lee Y, Huang C, Ravichandran S, Forgacs D, et al. Antibody affinity maturation and cross-variant activity following SARS-CoV-2 mRNA vaccination: Impact of prior exposure and sex. EBioMedicine. 2021;74:103748. doi: 10.1016/j.ebiom.2021.103748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Increased immune escape of the new SARS-CoV-2 variant of concern Omicron