ABSTRACT
The ongoing pandemic of coronavirus (CoV) disease 2019 (COVID-19) continues to exert a significant burden on health care systems worldwide. With limited treatments available, vaccination remains an effective strategy to counter transmission of severe acute respiratory syndrome CoV 2 (SARS-CoV-2). Recent discussions concerning vaccination strategies have focused on identifying vaccine platforms, number of doses, route of administration, and time to reach peak immunity against SARS-CoV-2. Here, we generated a single-dose, fast-acting vesicular stomatitis virus (VSV)-based vaccine derived from the licensed Ebola virus (EBOV) vaccine rVSV-ZEBOV, expressing the SARS-CoV-2 spike protein and the EBOV glycoprotein (VSV-SARS2-EBOV). Rhesus macaques vaccinated intramuscularly (i.m.) with a single dose of VSV-SARS2-EBOV were protected within 10 days and did not show signs of COVID-19 pneumonia. In contrast, intranasal (i.n.) vaccination resulted in limited immunogenicity and enhanced COVID-19 pneumonia compared to results for control animals. While both i.m. and i.n. vaccination induced neutralizing antibody titers, only i.m. vaccination resulted in a significant cellular immune response. RNA sequencing data bolstered these results by revealing robust activation of the innate and adaptive immune transcriptional signatures in the lungs of i.m. vaccinated animals only. Overall, the data demonstrate that VSV-SARS2-EBOV is a potent single-dose COVID-19 vaccine candidate that offers rapid protection based on the protective efficacy observed in our study.
KEYWORDS: VSV, vesicular stomatitis virus, SARS-CoV-2, i.m., i.n., rhesus macaques
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense, single-stranded RNA virus first isolated from a patient with severe respiratory illness in Wuhan, China (1). SARS-CoV-2 infection manifests as a clinical syndrome termed coronavirus disease 2019 (COVID-19), which can lead to respiratory failure (2). In addition to respiratory distress, other clinical manifestations associated with SARS-CoV-2 infection include cardiac pathology, gastrointestinal disease, coagulopathy, and hyperinflammatory syndrome (3–5). Patients with an increased risk of severe clinical manifestation include the elderly, the immunocompromised, and individuals with comorbidities (obesity, diabetes, hypertension, etc.) (6). Virtually every country has been affected, with over 250 million infections to date and an estimated case fatality rate of ∼2% (https://coronavirus.jhu.edu/map.html). The widespread morbidity, mortality, and socioeconomic impact of COVID-19 emphasize the urgent need for the development and deployment of countermeasures, including vaccines.
The COVID-19 pandemic has made the development of a vaccine a global priority (7–9). An ideal vaccine candidate is safe, effective, fast acting, and rapidly deployable and requires only a single immunization. Most of the current vaccine candidates encode the trimeric SARS-CoV-2 spike (S) protein as the primary antigen. S is essential for SARS-CoV-2 infectivity since it binds the angiotensin-converting enzyme 2 (ACE2) receptor and promotes virus-cell membrane fusion (10). It is also the main target for virus neutralization (11). The route of vaccination can greatly influence the local immune environment at the vaccination and infection sites. Recently, the comparison of intramuscular (i.m.) and intranasal (i.n.) vaccinations of mice with a chimpanzee adenoviral-vector-based vaccine revealed an increase in stimulation of local mucosal immunity and generation of antigen-specific IgA and lung-resident T cells after i.n. vaccination. The local mucosal immunity was improved by the generation of antigen-specific IgA and lung-resident T cell generation after i.n. vaccination (12). Prior to progression to human clinical trials, several COVID-19 vaccine candidates were i.m. administered to nonhuman primates (NHPs) to evaluate their efficacy (13–16).
The recombinant vesicular stomatitis virus (rVSV) vaccine platform has previously been utilized in vaccines against multiple viral pathogens, such as Ebola virus (EBOV) and the Marburg, Nipah, and Lassa viruses (17–19). VSV-based vaccines elicit a robust and rapid immune response to the encoded antigen(s) after a single immunization (20). The time to immunity has been demonstrated to be within 7 to 10 days for a number of pathogens in preclinical and clinical studies, greatly reducing the time needed between vaccination and protection (21–24). Multiple routes of VSV-based vaccination, such as the i.m. and i.n. routes, have been shown to be efficacious (21, 22, 25). Furthermore, the general population is predominantly seronegative for VSV, circumventing preexisting immunity neutralizing the vaccine virus (20). These unique attributes—robust immune stimulation and time to immunity—make this an attractive vaccine platform for SARS-CoV-2. However, the immunogenicity and efficacy of an i.m. or i.n. administered COVID-19 VSV-based vaccine have not been tested in the NHP model.
In the present study, we developed VSV-based vectors expressing SARS-CoV-2 S alone (VSV-SARS2) or in combination with the EBOV glycoprotein (GP) (VSV-SARS2-EBOV). Only the VSV-SARS2-EBOV vaccine protected hamsters from pneumonia within 10 days after i.m. administration (26). Therefore, we utilized the NHP challenge model and examined the VSV-SARS2-EBOV vaccine’s efficacy with this short time to challenge in tandem with the optimal route of immunization. We demonstrate that i.m. vaccinated NHPs developed no or mild lesions of COVID-19 with varied immunopathologies, whereas i.n. vaccination resulted in immune-enhanced disease with interstitial pneumonia in NHPs. i.m. vaccination resulted in robust and rapid humoral and cellular immune responses, while i.n. vaccination did not. Transcriptional analysis of the lungs supports our immunological findings by revealing greater expression of innate and adaptive immune genes in the i.m. vaccination group.
RESULTS
Vaccine construction.
The VSV backbone encoding the EBOV Kikwit GP, VSV-EBOV (also known as rVSV-ZEBOV), was used as a parental vector to construct this COVID-19 vaccine. First, we generated a vector replacing the EBOV GP with SARS-CoV-2 S lacking the 21 C-terminal amino acids (VSV-SARS2), as described by others (27). We also generated a VSV construct coexpressing the EBOV GP and the full-length SARS-CoV-2 S protein (VSV-SARS2-EBOV) by adding full-length codon-optimized SARS-CoV-2 S upstream of the EBOV GP into the existing VSV vector (see Fig. S1A in the supplemental material) (26). The constructs were recovered from plasmid by following previously established protocols (28), and expression of antigens (Fig. S1B) as well as viral growth kinetics were assessed (Fig. S1C). We performed efficacy studies with hamsters, demonstrating that vaccination with VSV-SARS2-EBOV prevented pneumonia in hamsters with higher efficacy than did vaccination with VSV-SARS2 (26). Therefore, we moved forward with the VSV-SARS2-EBOV vaccine.
Schematic and characterization of VSV-based vaccines. (A) Schematic illustrating vaccine vector design. T7 promotor; N, nucleoprotein; P, phosphoprotein; M, matrix protein; EBOV GP, Ebola virus glycoprotein; L, RNA-dependent RNA polymerase; SARS2-S, full-length SARS-CoV-2 S. (B) Western blot analysis of cell supernatant samples containing VSV vaccines probed for SARS-CoV-2 S (left), VSV M (right), or EBOV GP (right). Lane 1, wild-type VSV; lane 2, VSV-EBOV; lane 3, VSV-SARS2-EBOV. (C) Viral growth kinetics performed in triplicates on Vero E6 cells. Geometric means and SDs are depicted. Download FIG S1, PDF file, 0.06 MB (71.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Efficacy in NHPs.
A previously published study demonstrated that mucosal immunization with the parental vector VSV-EBOV protected NHPs from EBOV disease (25). In order to determine the efficacy of mucosal vaccination against COVID-19 in NHPs, we compared the efficacies of i.m. and i.n. vaccinations in the rhesus macaque model (29). Groups of 6 NHPs were either i.n. or i.m. vaccinated with VSV-SARS2-EBOV, while control animals received a single dose of VSV-EBOV i.n. (n = 2) or i.m. (n = 2) (Fig. S2A). All NHPs were observed for potential adverse effects, particularly after i.n. vaccination, as this is not the standard route of administration for this vaccine platform, but no clinical changes were noted. After 10 days, all NHPs were challenged with SARS-CoV-2 as previously described (29). On days postchallenge (dpc) 0, 1, 3, 5, and 7, a clinical exam, including blood draw, thoracic radiographs, and nasal swab collection, was performed; in addition, the dpc 3 exam included a bronchoalveolar lavage (BAL). On dpc 7, all NHPs were euthanized and samples were collected for analysis. None of the vaccinated animals displayed clinical signs of disease after challenge, and blood cell levels remained mostly unchanged over the course of the study (Fig. S2B).
NHP study outline and hematology. (A) Schematic outline of the rhesus macaque study. (B) Levels of white blood cell (WBCs), neutrophils, lymphocytes, red blood cells (RBCs), platelets, reticulocytes, hemoglobin, and hematocrit in the blood of vaccinated and challenged NHPs. Download FIG S2, PDF file, 0.07 MB (94.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
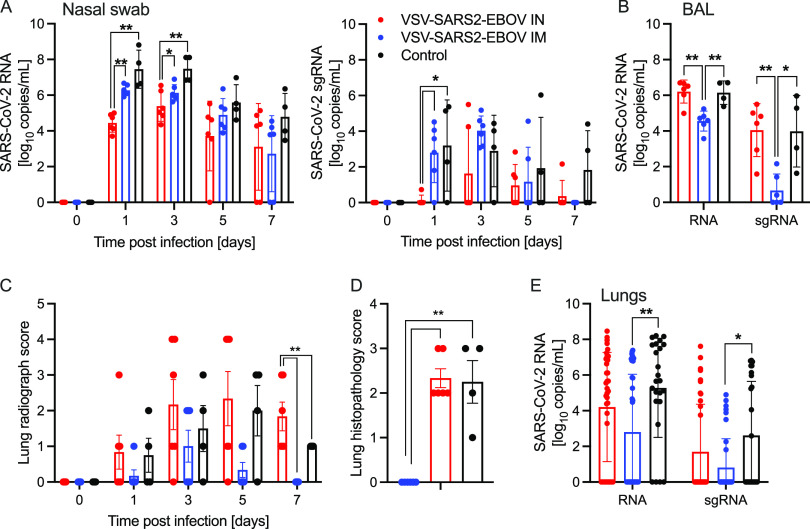
We determined differences in total SARS-CoV-2 RNA and subgenomic RNA (sgRNA) in the nasal swabs of the animals throughout the study. Interestingly, i.n. vaccination resulted in significantly lower levels of nasal viral RNA on dpc 1 than did i.m. challenge, indicating better local control of virus replication (Fig. 1A). However, on dpc 3, only the total SARS-CoV-2-specific RNA levels were significantly different (Fig. 1A). In contrast, both total and sgRNA levels in the BAL fluid were significantly lower for the i.m. vaccination group than for the i.n. vaccination and control groups (Fig. 1B). This finding is supported by our observation that the i.n. vaccinated NHPs had more lung infiltrates than the i.m. vaccination and control groups at the time of euthanasia (Fig. 1C; Fig. S3). Additionally, only i.n. vaccinated NHPs exhibited lung lesions consistent with coronavirus-induced broncho-interstitial pneumonia (Fig. S4). Of note, gross lung lesions are inconsistently present in this model system (29). Interestingly, i.m. vaccinated NHPs had profound interstitial infiltration of lymphocytes that frequently formed moderately large cuffs around blood vessels, leading to increased interstitial lung pathology scores relative to those of the i.n. vaccinated NHPs. However, epithelialized cellular debris, the presence of degenerate leukocytes, and expansion of septa with edema fluid was almost exclusively observed in i.n. vaccinated and control NHPs (Fig. 1D). This was accompanied by a significant reduction of total RNA and sgRNA in i.m. vaccinated, but not i.n. vaccinated, NHPs compared to levels in controls (Fig. 1E). The comparison of RNA levels in individual lung lobes and other examined tissue samples did not reveal any significant differences (Fig. S5). Virus isolation and titration from tissue samples yielded inconsistent results (data not shown).
FIG 1.
SARS-CoV-2 loads in vaccinated NHPs. Groups of 6 NHPs were intranasally (IN) or intramuscularly (IM) vaccinated with a single dose of VSV-SARS2-EBOV; 4 control animals received VSV-EBOV. (A) Total SARS-CoV-2-specific RNA (left) and subgenomic RNA (sgRNA) (right) in nasal swabs collected from NHPs. (B) Total SARS-CoV-2-specific RNA and sgRNA in bronchoalveolar lavage (BAL) samples collected on day 3. (C) Lung radiograph scores after challenge. (D) Lung histopathology score (presence of septal edema/infiltrates) at the time of euthanasia. (C, D) Means and standard deviations (SDs) are shown. (E) Total SARS-CoV-2-specific RNA and sgRNA in lung samples collected on day 7. (A, B, E) Geometric means and geometric SDs are depicted. Statistical significance is indicated.
Radiographs of NHPs. Thoracic radiographs taken before SARS-CoV-2 infection (day 0) and at the time of euthanasia (day 7) are shown for each NHP. Download FIG S3, PDF file, 0.9 MB (13.1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gross lung pathology of NHPs. NHPs were vaccinated 10 days before challenge with SARS-CoV-2. Representative pictures of NHP lungs with lesions on day 7 after challenge are shown. IM, intramuscular vaccination; IN, intranasal vaccination. Lesions are circled. Download FIG S4, PDF file, 0.3 MB (317.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Virus load in NHP tissue samples on day 7. (A) Total SARS-CoV-2-specific RNA (left) and subgenomic RNA (sgRNA) (right) in lung samples collected from NHPs. RLU, right lobe upper; RLM, right lobe middle; RLL, right lobe lower; LLU, left lobe upper; LLM, left lobe middle; LLL, left lobe lower. (B) Total SARS-CoV-2-specific RNA in tissue samples (right) from NHPs. LN, lymph node; R, right; L, left. Download FIG S5, PDF file, 0.06 MB (69.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Histopathology in NHPs.
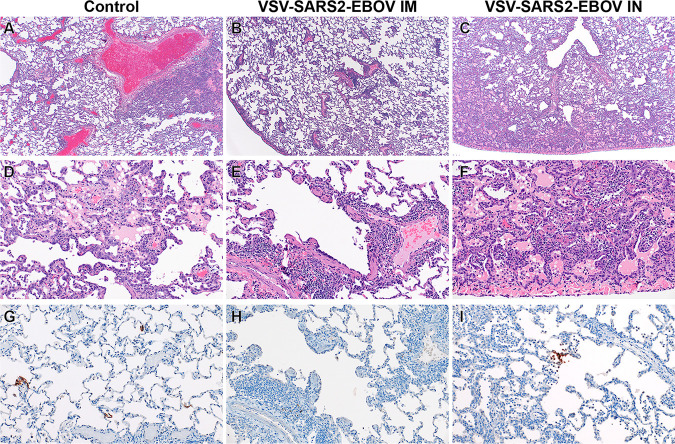
Histopathologic analysis of the collected lung samples revealed pulmonary pathology consistent with that of the previously described rhesus macaque model of SARS-CoV-2 infection in the control group regardless of i.m. or i.n. administration of the control vaccine (Fig. 2A and D) (29). In the i.m. vaccination group, pulmonary lesions consisted of low-to-moderate numbers of eosinophils multifocally infiltrating bronchiolar mucosa, excess mucus accumulation in the lumens of bronchi and bronchioles, and profound perivascular lymphocytic cuffing (interpreted as immune pathology) disseminated throughout all lung lobes (Fig. 2B and E). In combination, these lesions are suggestive of a localized hypersensitivity response. Hypersensitivity lesion location mirrored that of what has previously been described for this NHP model of COVID-19 (29), with a high proportion of lesions located at the periphery of the lung and increased lesion severity in lower lung lobes. Limited evidence of type I pneumocyte damage was present in rare foci and was characterized by the lining of alveoli by type II pneumocytes and a scant amount of proteinaceous fluid within alveolar spaces (Fig. 2B and E). Histopathologic lesions in the i.n. vaccination group mirrored the enhanced gross lesion severity and histologically consisted of an immune-enhanced disease with evidence of classic moderate-to-severe SARS-CoV-2 pulmonary pathology and a moderate hypersensitivity response (Fig. 2C and F). The hypersensitivity response was similar to that observed in the i.m. vaccinated group but was more severe with the addition of eosinophil spillover into bronchiolar lumens and moderate numbers of alveolar spaces. SARS-CoV-2 nucleoprotein immunoreactivity was observed in type I pneumocytes and macrophages of both the control and the i.n. vaccination group but not in the i.m. vaccination group (Fig. 2G to I).
FIG 2.
Histopathology and immunohistochemistry of NHP lungs. (A) Pulmonary lesions depicting typical coronavirus respiratory pathologies, including locally extensive regions of bronchointerstitial pneumonia and proteinaceous fluid accumulation in adjacent alveoli (magnification, ×40; H&E). (B) Disseminated immunopathology with prominent perivascular lymphocytic cuffing and multifocal involvement at terminal airways (magnification, ×40; H&E). (C) i.n. vaccination shows pulmonary pathology characterized by a combination of interstitial pneumonia and immunopathology (magnification, ×40; H&E). (D) Foci of interstitial pneumonia are characterized by prominent type II pneumocyte hyperplasia, leukocyte infiltration, expansion of alveolar septa, and accumulation of low numbers of macrophages, neutrophils, and proteinaceous fluid in alveolar spaces (magnification, ×200; H&E). (E) Terminal airways and medium- to small-caliber blood vessels are cuffed by moderate numbers of lymphocytes with scattered eosinophils (magnification, ×200; H&E). (F) Foci of interstitial pneumonia show pronounced type II pneumocyte hyperplasia, thickening of alveolar septa by an infiltration of leukocytes, and leukocyte spillover into adjacent alveolar spaces, with moderate numbers of alveolar eosinophils noted and multifocal fibrin mats filling alveolar spaces (magnification, ×200; H&E). (G) Low numbers of type I pneumocytes in regions lacking pathology are immunoreactive for SARS-CoV-2 antibody (magnification, ×200; immunohistochemistry [IHC]). (H) SARS-CoV-2-specific immunoreactivity was not observed in evaluated sections of the i.m. vaccinated group (magnification, ×200; IHC). (I) Low numbers of type I pneumocytes and alveolar macrophages are immunoreactive for SARS-CoV-2 in select foci of interstitial pneumonia (magnification, ×200; IHC).
Immune responses in NHPs.
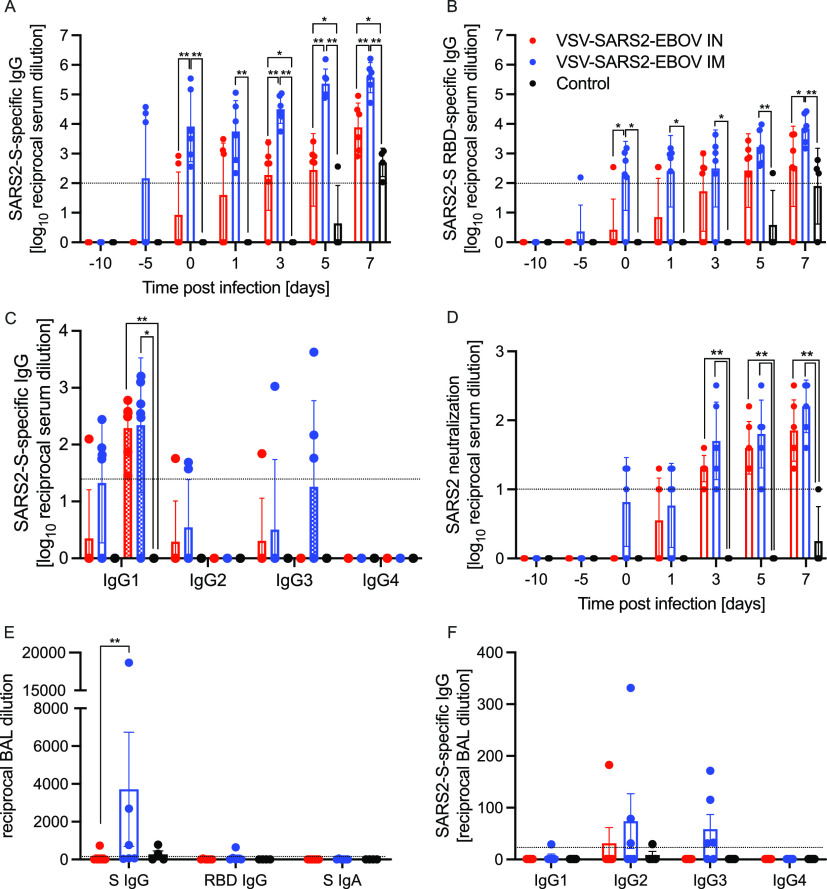
We next analyzed the peripheral humoral response. IgG responses to full-length SARS-CoV-2 S, the S receptor binding domain (RBD), and the EBOV GP were determined following vaccination and challenge (Fig. 3 and Fig. S6). We demonstrated that the i.m. vaccinated NHPs attained significantly higher SARS-CoV-2 S-specific IgG levels starting 10 days after vaccination and following challenge than levels in i.n. vaccinated NHPs and controls (Fig. 3A). Similarly, the IgG response to the SARS-CoV-2 S RBD was higher at day 10 postvaccination (0 dpc) in the i.m. vaccination group than in the i.n. vaccination and control groups (Fig. 3B). Analysis of the IgG subclasses in serum on dpc 0 and 7 showed that both vaccination routes resulted in predominantly IgG1, IgG2, and IgG3 antibodies, with no significant difference between the groups (Fig. 3C). Only IgG1 significantly increased after challenge in both vaccine groups compared to levels in controls (Fig. 3C). Interestingly, on dpc 7, both IgG2 and IgG3 antibodies were largely absent in sera from i.n. vaccinated NHPs (Fig. 3C). IgG responses specific to EBOV GP support the finding that i.m. vaccination appears more immunogenic than i.n. vaccination even though the data are significantly different only on dpc 0 and 3 (Fig. S6A). Measurable SARS-CoV-2-neutralizing titers were detected as early as 10 days following vaccination for the i.m. vaccination group and 11 days for the i.n. vaccination group (Fig. 3D). On dpc 3, significantly higher titers were observed in i.m. vaccinated NHPs than in controls only (Fig. 3D). At the time of euthanasia, the neutralizing titers in i.m. and i.n. vaccinated NHPs were comparable but significantly higher than those observed in control animals (Fig. 3D).
FIG 3.
Humoral immune responses in NHPs. (A to C) Serum samples collected throughout the study from all NHPs were examined for SARS-CoV-2 S-specific IgG (A), SARS-CoV-2 S receptor binding domain (RBD)-specific IgG (B), or IgG subclasses specific to SARS-CoV-2 S by ELISA (C). (D) Titers neutralizing SARS-CoV-2 were determined. (E) Bronchoalveolar lavage (BAL) samples were analyzed for SARS-CoV-2 S-specific IgG (S IgG) or IgA (S IgA) and SARS CoV-2 S RBD-specific IgG (RBD IgG) by ELISA. (A to D) Geometric means and geometric SDs are depicted. (F) IgG subclasses specific to SARS-CoV-2 S in BAL samples were analyzed by ELISA. (E, F) Means and SDs are depicted. Statistical significance is indicated.
EBOV GP-specific antibodies in NHPs after vaccination and challenge. (A) Serum samples collected throughout the study and (B) bronchoaveolar lavage (BAL) samples collected on day 3 were analyzed by ELISA for EBOV GP-specific IgG. Statistical significance is indicated. Download FIG S6, PDF file, 0.04 MB (46.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Next, we investigated the humoral responses in the BAL fluid obtained on dpc 3. We detected SARS-CoV-2 S-specific IgG in 3 of the 6 NHPs in the i.m. vaccinated group but in only 1 of 6 NHPs in the i.n. vaccinated group (Fig. 3E). Only the i.m. vaccinated NHP with the highest titer of SARS-CoV-2 S-specific IgG had anti-SARS-CoV-2 S RBD IgG (Fig. 3E). SARS-CoV-2 S-specific IgA was not detected in any of the BAL samples (Fig. 3E). Unlike in serum, no IgG1 was detected; however, IgG2 and IgG3 were readily detected in several i.m. vaccinated NHPs (Fig. 3F). Unexpectedly, i.m. vaccination resulted in higher humoral responses in the lung to all antigens, including EBOV GP (Fig. S6B).
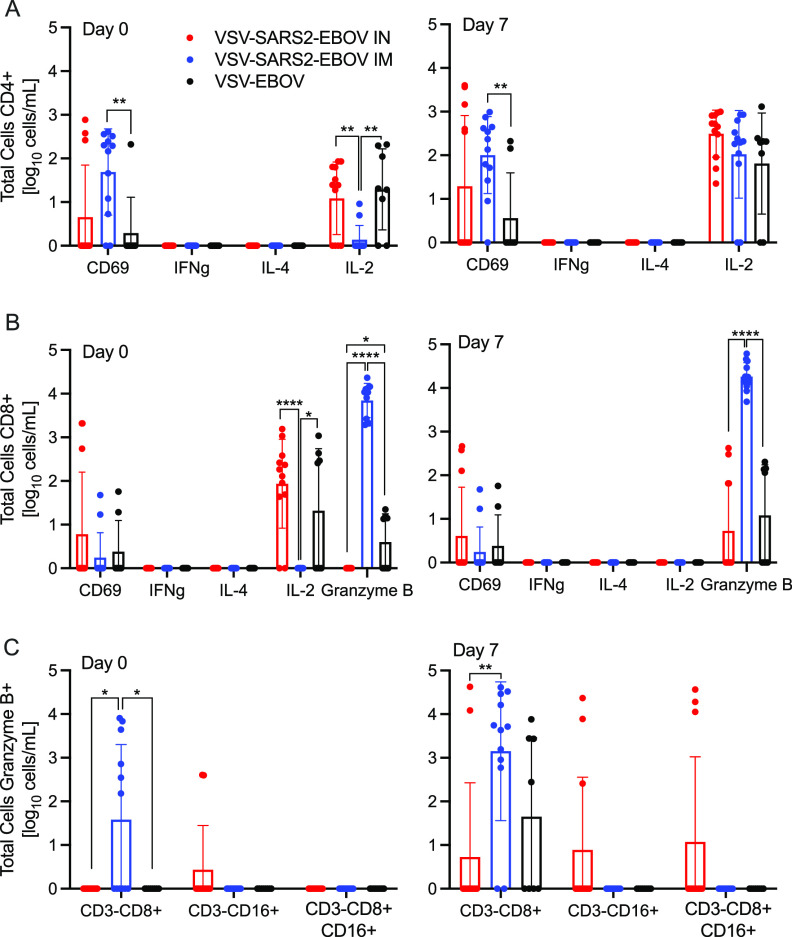
We analyzed the peripheral cellular response even though T cell responses have been shown to play only a limited role in mediating protection using the VSV-EBOV vaccine (30, 31). Peripheral blood mononuclear cells (PBMCs) were stimulated with a peptide pool spanning the entire length of the SARS-CoV-2 S protein, and the antigen-specific T cells were identified using intracellular cytokine staining (Fig. 4A and B). While there was minimal cytokine production by CD4+ T cells, a significant increase in the CD69 activation marker was seen in i.m. vaccinated NHPs on dpc 0 and 7 relative to levels in controls (Fig. 4A). Similarly, a significantly higher portion of the CD8+ T cells from the i.m. vaccinated group produced granzyme B on dpc 0 and 7 than the portions that did so in the control and i.n. vaccinated groups (Fig. 4B). Interestingly, numbers of IL-2+ CD4+ and CD8+ T cells were significantly lower in the i.m. vaccinated group on dpc 0 than in the i.n. vaccinated and control groups and comparable to numbers in the i.n. vaccinated and control groups by dpc 7 (Fig. 4A and B). Additionally, a greater number of granzyme B+ NK cells was measured on dpc 0 and 7 in i.m. vaccinated animals than in i.n. vaccinated and control animals (Fig. 4C).
FIG 4.
Peripheral cellular immune responses postchallenge. (A) CD4+ T cells and PBMCs were stained for expression of the early activation marker CD69, and intracellular cytokine staining (ICS) was performed for IFN-γ, IL-4, and IL-2 on dpc 0 and 7. (B) CD8+ T cells from PBMCs were phenotyped for the expression of the early activation marker CD69, and ICS was performed for IFN-γ, IL-4, IL-2, and granzyme B on dpc 0 and 7. (C) NK cell subpopulations were stained for the expression of granzyme B on dpc 0 and 7. Data were measured in duplicate for all animals. Geometric means and SDs are depicted. Statistical significance is indicated.
Finally, we monitored levels of systemic and BAL fluid cytokines and chemokines. We found a significant increase in MCP-1 on dpc 1 and 3 in i.n. vaccinated animals compared to levels in i.m. vaccinated animals. A similar trend was observed for IL-18 on dpc 3 and 5 (Fig. S7A). Levels of MIP-1β were significantly lower at all dpc in i.m. vaccinated animals than in the control and i.n. vaccinated animals (Fig. S7A). Analysis of cytokine and chemokine levels in BAL fluid revealed that MCP-1 levels were significantly decreased in i.n. vaccinated NHPs (Fig. S7B). All other investigated cytokines did not show significant differences.
Serum cytokine levels in NHPs after vaccination and challenge. (A and B) Serum samples collected throughout the study (A) and BAL samples collected on day 3 (B) were analyzed. Statistical significance is indicated. Download FIG S7, PDF file, 0.1 MB (113.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transcriptional analysis of BAL samples.
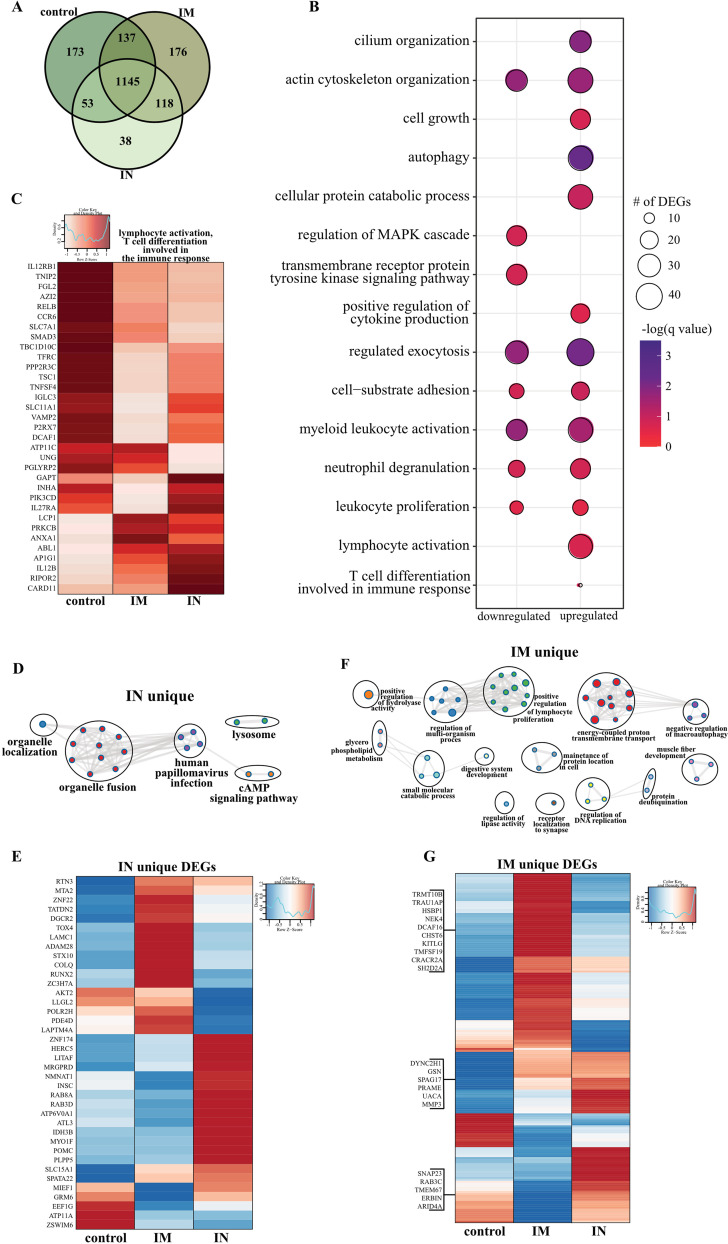
To better understand the molecular underpinnings of differential vaccine responses, we profiled the host transcriptional response in BAL fluid (dpc 3) samples. Principal-component analysis (PCA) of BAL fluid samples indicated a distinct separation of uninfected/naive (historical data) and the three challenged groups, with no clear distinction between the challenged groups (Fig. S8A). Therefore, the samples from control, i.m. vaccinated, or i.n. vaccinated animals were compared to the uninfected samples. Over 1,000 differentially expressed genes (DEGs) were detected in the challenged groups compared to genes in the uninfected animals, with most DEGs being upregulated (Fig. S8B). The majority of DEGs were shared among the three challenged groups (Fig. 5A). Functional enrichment showed that upregulated and downregulated DEGs shared by all three challenged groups play a role in regulating cell structure (e.g., “actin cytoskeleton organization”) and innate immunity (e.g., “positive regulation of cytokine production,” “myeloid leukocyte activation”) (Fig. 5B). However, only upregulated DEGs associated with gene ontology (GO) terms having to do with adaptive immunity (e.g., “lymphocyte activation,” “T cell differentiation”) (Fig. 5B).
FIG 5.
BAL fluid RNA sequencing. (A) Venn diagram of DEGs expressed dpc 3 with SARS-CoV-2. Animals received either a control, intramuscular (IM), or intranasal (IN) vaccination. (B) Bubble plot representing functional enrichmentof DEGs shared by all infected groups at dpc 3. The color intensity of each bubble represents the negative logarithm of the P value, and the relative size of each bubble represents the number of DEGs belonging to the specified gene ontology (GO) term. MAPK, mitogen-activated protein kinase. (C) Heatmap representing shared upregulated DEGs associated with the GO terms “lymphocyte activation” and “T cell differentiation involved in the immune response.” Expression is represented as the normalized number of reads per kilobase of transcript per million mapped reads [rpkm], where each column represents the median rpkm of the given group. The range of colors is based on a scale and centered rpkm values of the represented DEGs. (D and F) GO term network depicting functional enrichment of DEGs unique to i.n. (D) and i.m. (F) vaccinated groups using Mediascape. Color-coded clustered nodes correspond to one GO term or KEGG pathway. Node size represents the number of DEGs associated with the indicated term or pathway. Gray lines represent shared interactions between terms/pathways, with density and number indicating the strength of connections between closely related terms/pathways. cAMP, cyclic AMP. (E and G) Heatmaps representing DEGs unique to i.n. (E) and i.m. (G) vaccinated animals. Exemplar DEGs are annotated. Red represents upregulation, and blue presents downregulation. Each column represents the median number of rpkm of the given group. For all heatmaps, the range of colors is based on a scale and centered rpkm values of the represented DEGs.
BAL fluid RNA sequencing. (A) Principal-component analysis of BAL samples from uninfected animals and vaccinated animals dpc 3 (control, intramuscular vaccination, or intranasal vaccination). (B) Down- and up-regulated differentially expressed genes (DEGs). (C to E) Heatmaps representing DEGs shared by all infected groups and associated with the gene ontology (GO) terms “myeloid cell activation” and “neutrophil degranulation” for upregulated DEGs (C) and downregulated DEGs (D) and those associated with “autophagy” and “positive regulation of cytokine production” for upregulated DEGs (E). Each column represents the median number of rpkm of the given group. The range of colors is based on a scale and centered rpkm values of the represented DEGs. Red represents upregulated DEGs; blue represents downregulated DEGs. (F) Digital cell quantitation using the ImmQuant IRIS database comparing challenged groups to uninfected controls. Red represents upregulation; blue represents downregulation. Each column represents the average relative predicted frequency of the given cell type. P values are calculated relative to values for the uninfected animals. Statistical significance is indicated. For all heatmaps, the range of colors is based on a scale and centered rpkm values of the represented DEGs. Download FIG S8, PDF file, 0.3 MB (310.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Further analysis of shared DEGs showed that genes involved in proinflammatory pathways (e.g., RELB, MFHAS1, IL12RB1, TNFSF4, TRAF2, C5AR1, and TOLLIP) were more highly expressed in the control group than in the i.m. and i.n. vaccinated groups (Fig. S8C and S8D). A second cluster of inflammation-related genes was induced to a greater extent in the i.n. vaccinated group (IFT88, IL12B, CLEC9, and IL27RA), which is in line with the greater inflammatory response observed in these animals. On the other hand, DEGs implicated in T and B cell-mediated immunity (e.g., LCP1, PRKCB) were induced to a greater extent in i.m. vaccinated NHPs (Fig. 5C). Genes associated with the GO term “myeloid cell activation/neutrophil downregulation” were suppressed to a greater extent in the i.m. vaccinated cohort, consistent with the lower inflammation profile observed in this group (Fig. S8E).
We next analyzed vaccine-specific transcriptional responses in the BAL samples of the i.n. and i.m. vaccinated groups to elucidate unique molecular responses (Fig. 5D to G). DEGs unique to the i.n. vaccinated group associated with GO terms related to organelle dynamics, such as “organelle localization” as well as antiviral immunity (“human papillomavirus infection”) (Fig. 5D). Genes important for vesicular mobilization (RAB8A and RABD3D) as well as antiviral HERC5 were highly expressed in i.n. vaccinated NHPs, whereas those associated with signaling (e.g., AKT2, PDE4D) were more downregulated (Fig. 5E) than in the i.m. vaccinated NHPs. DEGs uniquely upregulated in the i.m. group have roles in protein synthesis and folding (e.g., TRMT10B, TRNAU1AP, HSPB1), cell proliferation (e.g., KITLF, TM4SF19), and T cell activation (e.g., CRACR2A, SH2D2A) (Fig. 5G).
Due to limited sample availability, we were unable to perform phenotyping of immune cells in the BAL fluid. Therefore, we performed digital cell quantitation to infer changes in cell frequencies based on the transcriptional landscape (Fig. S8F). This analysis predicted significant increases in the levels of monocytes, NK cells, and stimulated CD4 Th2 cells, while frequencies of naive and plasma B cells, CD4+ and CD8+ T cells, and stimulated dendritic cells and neutrophils were predicted to decline for all three challenged groups. On the other hand, plasma cells and monocytes were predicted to be induced to a lower magnitude in controls than in the i.m. and i.n. vaccinated groups (Fig. S8F).
Transcriptional analysis of lung samples.
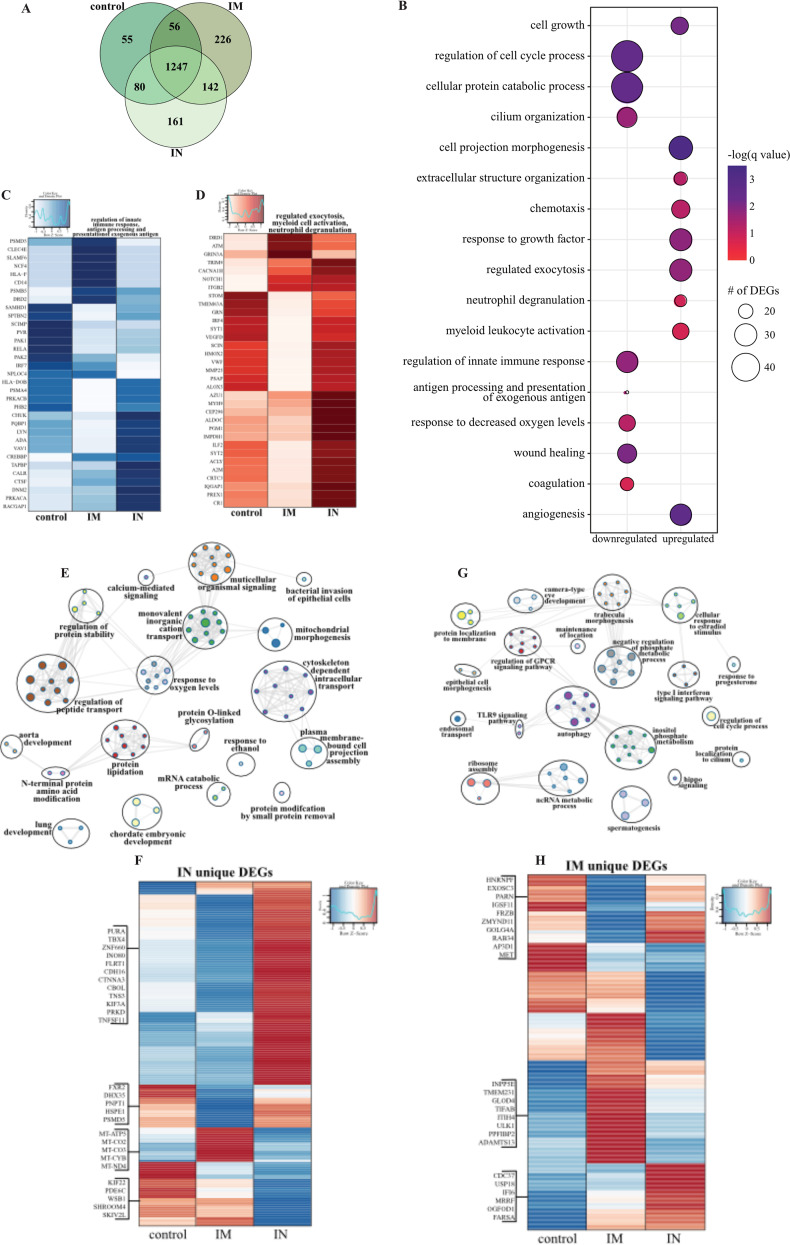
As described for BAL samples, we observed a clear distinction between lung samples of uninfected or naive and vaccinated and challenged NHPs, with no clear separation between the three challenged groups; therefore, we employed the same strategy as described above for BAL fluid (Fig. S9A). A robust transcriptional response to SARS-CoV-2 infection was evident in all three vaccinated groups compared to that of naive animals (Fig. S9B), with the majority of DEGs shared among the three groups (Fig. 6A). Downregulated DEGs shared by all three vaccinated groups associated with GO terms involved primarily in innate immunity (e.g., “regulation of innate immune response,” “antigen processing and presentation”), cellular stress (e.g., “coagulation,” “response to decreased oxygen levels,” “wound healing”), and the cell cycle (e.g., “regulation of cell cycle process”) (Fig. 6B). Genes playing roles in protein folding and turnover (CALR and CTSF), immune activation (e.g., LYN and ADA), coagulation (e.g., PLAT, SIRT2, FERMT3), fluid homeostasis (e.g., ADM, SERPINA5), and cell morphogenesis (e.g., NOTCH4, TEK) were more suppressed in the i.n. vaccinated group (Fig. 6C, S9C). Shared upregulated DEGs associated with GO terms reflecting innate immune processes (e.g., “regulated exocytosis,” “myeloid leukocyte activation,” “neutrophil degranulation”), as well as cell migration (e.g., “chemotaxis”) and extracellular structural dynamics (e.g., “extracellular structural organization,” “cell projection morphogenesis”) (Fig. 6B). Overall, a large portion of these genes were induced to a greater extent in the i.n. vaccinated group, notably those that play a role in neutrophil activation (e.g., GRN, AZU1, CR1), cell metabolism (e.g., ALDOC, PGM1, IMPDH1), chemotaxis (e.g., CCL20, CCL8, CCL13, ICAM3), and extracellular matrix remodeling (e.g., MMP25, MMP16) (Fig. 6D; Fig. S9D). Additionally, genes important for angiogenesis and apoptosis were more upregulated in the i.n. vaccinated group (e.g., VEGFD, PRKD1, SHC1, DAB2IP) (Fig. S9E).
FIG 6.
Lung RNA sequencing. (A) Venn diagram of DEGs expressed dpc 7 with SARS-CoV-2. Animals received either a control, intramuscular (IM), or intranasal (IN) vaccination. (B) Bubble plot representing functional enrichment of DEGs shared by all infected groups at dpc 7. The color intensity of each bubble represents the negative logarithm of a P value, and the relative size of each bubble represents the number of DEGs belonging to the specified GO term. (C and D) Heatmaps representing the shared GO terms “regulation of innate immune response” and “antigen processing and presentation of exogenous antigen” for downregulated DEGs (C) and “regulated exocytosis,” “myeloid leukocyte activation,” and “neutrophil degranulation” for upregulated DEGs (D). Expression is represented as the normalized number of rpkm, where each column represents the median rpkm of the given group. The range of colors is based on a scale and centered rpkm values of the represented DEGs. (E and G) GO term network depicting functional enrichment of DEGs unique to i.n. (E) and i.m. (G) vaccinated groups using Mediascape. Color-coded clustered nodes correspond to one GO term or KEGG pathway. Node size represents the number of DEGs associated with the indicated term or pathway. Gray lines represent shared interactions between terms/pathways, with density and number indicating the strength of connections between closely related terms/pathways. GPCR, G protein-coupled receptor; ncRNA, noncoding RNA. (F and H) Heatmaps representing DEGs unique to i.n. (F) and i.m. (H) vaccinated groups. Exemplar DEGs are annotated. Red represents upregulation, and blue presents downregulation. Each column represents the median number of rpkm of the given group. For all heatmaps, the range of colors is based on a scale and centered rpkm values of the represented DEGs.
Lung RNA sequencing. (A) Principal-component analysis of lower left lung (LLL) samples from uninfected animals and vaccinated animals dpc 7 (control, intramuscular vaccination, or intranasal vaccination). (B) Down- and up-regulated DEGs. (C to E) Heatmaps representing DEGs shared by all infected groups and associated with the GO terms “coagulation,” “response not decreased oxygen levels,” and “wound healing” for downregulated DEGs (C), “chemotaxis”, “cell projection morphogenesis,” and “extracellular structure organization” for upregulated DEGs (D), and “angiogenesis” for upregulated DEGs (E). Each column represents the median number of rpkm of the given group. The range of colors is based on a scale and centered rpkm values of the represented DEGs. Red represents upregulated DEGs; blue represents downregulated DEGs. (F) Digital cell quantitation using the ImmQuant IRIS database comparing challenged groups to uninfected controls. Red represents upregulation; blue represents downregulation. Each column represents the average relative predicted frequency of the given cell type. P values are calculated relative to values for the uninfected animals. Statistical significance is indicated. For all heatmaps, the range of colors is based on a scale and on centered rpkm values of the represented DEGs. Download FIG S9, PDF file, 0.3 MB (296.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We next analyzed vaccine route-specific DEGs to identify differences in the molecular responses to vaccination and challenge (Fig. 6E to H). Enrichment of the 161 DEGs unique to the i.n. vaccinated group revealed enrichment in GO terms suggesting tissue injury (e.g., “lung development”), metabolism (e.g., “protein modification by small protein removal”), and signaling (e.g., “calcium-mediated signaling”) (Fig. 6F). Notably, downregulated DEGs in this group include components of the mitochondrial cellular respiration complex (e.g., MT-CYB, MT-CO2) and cellular homeostasis (e.g., SKIV2L, PDE6C, SKIV2L, SHROOM4) (Fig. 6F).
In contrast, the 226 DEGs unique to the i.m. vaccinated group associated with GO terms related to cellular defense (e.g., “type I interferon signaling pathway,” “TLR9 signaling,” “autophagy”), morphogenesis (e.g., “epithelial cell morphogenesis”), and membrane dynamics (e.g., “protein localization to membrane”) (Fig. 6G). The higher expression of DEGs related to the type I interferon response (e.g., CDC37, USP18, IFI6) and ribosome assembly (e.g., MRRF, OGFOD1, FARSA) in the i.m. vaccinated and control groups suggests greater mobilization of host defense processes (Fig. 6H). Additionally, genes associated with cilium formation (e.g., INPP5E, TMEM231, PPFIBP2) and regulation of inflammation (e.g., GLOD5, ITIH5, TIFAB) were highly expressed in the i.m. vaccinated group, in line with the reduced damage in this group (Fig. 6H). Digital cell quantitation analysis indicated that these transcriptional changes were consistent with increases in the levels of neutrophils, monocytes, NK cells, dendritic cells, and naive lymphocytes in all the infected animals, while frequencies of activated monocytes were predicted to decrease relative to levels in tissue from naive animals (Fig. S9F).
DISCUSSION
Many vaccine platforms have been utilized to develop a COVID-19 vaccine quickly (13–16), with several approved for human use within a year of SARS-CoV-2 emergence. However, many of these vaccines require 2 doses to elicit protection and are delivered i.m. rather than at the site of infection. Therefore, we developed a single-dose, fast-acting, VSV-based vaccine against COVID-19 which is based on the VSV-EBOV (also known as rVSV-ZEBOV) vaccine approved by the U.S. Food and Drug administration (FDA) and the European Medicines Agency (EMA) for human use.
We generated two vaccine candidates, one expressing SARS-CoV-2 S alone (VSV-SARS2) and the second coexpressing SARS-CoV-2 S and EBOV GP (VSV-SARS2-EBOV). Growth kinetics on Vero E6 cells revealed slower replication kinetics of VSV-SARS2 than of VSV-EBOV and VSV-SARS2-EBOV, potentially negatively impacting future vaccine production. In contrast, VSV-SARS2-EBOV grew similarly to the parental vaccine, VSV-EBOV. A hamster experiment revealed the superior protective efficacy of a single i.m. dose of VSV-SARS2-EBOV compared to that of VSV-SARS2 after 10 days. Therefore, we dropped the VSV-SARS2 vaccine and focused on comparing the protective efficacies of i.m. and i.n. delivery of only the VSV-SARS2-EBOV vaccine in the rhesus macaque model (29). Recently, Merck, a recipient of funding for a VSV-based COVID-19 vaccine through Operation Warp Speed (32), announced the discontinuation of the development of their version of a VSV-SARS2 vaccine due to limited immunogenicity (33). This turn of events highlights the problems associated with the VSV-SARS2 vaccine but also emphasizes the potential of the VSV-SARS2-EBOV vaccine, as it demonstrated superior efficacy in our hamster studies (26).
NHPs are valuable animal models for infectious diseases, as they often faithfully recapitulate clinical signs and pathogenesis observed in humans (34). The rhesus macaque model for COVID-19 was first described to have a relatively consistent disease phenotype early after infection, with rapid disease recovery among the small study cohort (29); however, subsequent studies highlighted its limitations, as clinical disease was variable and rather mild, with limited lung pathology (13, 35–37). Many factors, like the ages and origins of the NHPs, SARS-CoV-2 isolate used, and routes of exposure may have contributed to the heterogeneity in disease presentation, as has been observed with other macaque species (38, 39). Here, we faced some of these limitations. We followed the schedule of previously published SARS-CoV-2 vaccine efficacy studies in rhesus macaques (13, 37) and terminated the experiment 7 dpc for determination of virus loads and changes in lung pathology. While our data show a significant reduction of disease burden in the i.m. vaccinated NHPs, disease severity in all groups was variable and rather mild.
A single-dose, i.m., but not i.n., delivered vaccine protected NHPs from COVID-19 pneumonia within 10 days postvaccination. This short time to immunity is a tremendous advantage and highlights its potential to be used rapidly during a public health crisis, particularly in emergency situations when many people are exposed at once. Interestingly, i.m. vaccination resulted in a superior immune response compared to that of i.n. vaccinated animals, as evidenced by the significantly higher SARS-CoV-2 S-specific antibody titers and lower viral loads in this group. None of the animals in this study showed overt clinical signs of disease, including changes in blood cell counts, regardless of their vaccination status. However, histological examination of the lung tissue revealed immunopathology that was most significant in i.n. vaccinated animals. The observed immunopathology was not consistent with a classic hypersensitivity response or immune-enhanced disease, as the lesions were limited to the periphery of lung lobes and almost exclusively observed in lower lung lobes, as previously reported for SARS-CoV-2 infection (29). Importantly, i.m. vaccinated animals did not develop signs of interstitial pneumonia, nor could we detect SARS-CoV-2 antigen in the lungs. Indeed, at the time of euthanasia, lung lesions were apparent in animals from the i.n. vaccinated and controls groups but not from the i.m. vaccinated group. This is surprising given that mucosal vaccination for other respiratory pathogens has been demonstrated to be superior or comparable to i.m. vaccination (40, 41). It possible that the immunopathology observed in the vaccinated animals is due to the short duration between vaccination and challenge (10 days) and that it might also occur with other SARS-CoV-2 vaccine platforms, as this change was clinically silent in our model.
In line with the observations above, the transcriptional analysis of lung samples showed a divergence of antiviral states between the i.n. vaccinated, i.m. vaccinated, and control groups. i.n. vaccination induced transcriptional changes associated with cell metabolism, apoptosis, angiogenesis, and neutrophil activation processes. Some notable DEGs include genes associated with neutrophil activation and the formation of azurophil granules (e.g., GRN, CR1, and AZU1). While neutrophils play a role in protecting the host, sustained neutrophil activation has been shown to directly correlate with more severe COVID-19 cases (42). In contrast, i.m. vaccination induced transcriptional changes playing a role in cilium formation, inflammation regulation, and type I interferon (IFN). A majority of these genes are responsible for the regulation and control of early innate inflammation, such as USP18, which disrupts the JAK-STAT pathway downstream of the IFN receptor (43), and TIFAB, which inhibits the activation of the NF-κB pathway (44). Collectively, these findings support the significant decrease in virus replication between the i.n. and i.m. vaccination groups. Transcriptional analysis of acute BAL samples also demonstrated a divergence of antiviral states evidenced by T cell differentiation genes upregulated after i.m. vaccination, compared to innate antiviral posttranslational modifications after i.n. vaccination. While variable immunopathology was observed in all VSV-SARS2-EBOV-vaccinated NHPs regardless of the route, transcriptional analysis of the BAL fluid demonstrated an upregulation of antiviral genes, such as HERC5, in the i.n. vaccinated group only, in line with enhanced viral loads in this group. HERC5 is responsible for the production of HECT-type E3 protein ligase, a facilitator of the IFN-stimulated gene (ISG) conjugation system of interferon-stimulated gene 15 (45), possibly contributing to the enhanced immunopathology observed in the i.n. vaccinated group.
Since the importance of a cellular immune response has recently been highlighted in COVID-19 patients (46), we assessed the development of both innate and adaptive cellular responses following each vaccination strategy. Our analysis showed a higher frequency of granzyme B+ NK cells after i.m. vaccination. While there was minimal antigen-specific cytokine production from the CD4+ T cells, a significant increase in the early activation marker CD69 was observed on 0 and 7 dpc. In addition, an increase in interleukin 2 (IL-2) production was observed only 7 dpc, indicative of the priming of cytotoxic CD8+ T cells (47). Transcriptional analysis revealed that i.m. vaccination induced upregulation of CRACR2A and SH2D2a. SH2D2a encodes a T cell-specific adaptor protein which facilitates the formation and maintenance of the immunological synapse between antigen-presenting cells and the T cell receptors, allowing for a more robust antigen-specific stimulation (48). CRACR2A also has a role in the maintenance of the immunological synapse and promotes downstream signaling, which results in an increased Th1 response and Th17 effector functions, which is supported by the significant decrease of IL-2 on 0 dpc (49).
Serum cytokine analyses demonstrated a significant increase in IL-18 and the innate chemokines MCP-1 and MIP-1β circulating in animals that presented with severe pathology. The increased expression of IL-18 may indicate a priming of the infiltrating immune cells in the lungs to a proinflammatory state that results in the observed tissue destruction. However, we did not observe an increase of IL-18 in BAL samples from 3 dpc. The most striking observation was the significant downregulation of MIP-1β on all dpc measured in i.m. vaccinated animals. MIP-1β and MCP-1 have previously been demonstrated to be indicators of severe COVID-19 pathogenesis by transcriptomic profiling of human patients (50). A decrease in MIP-1β and MCP-1 may contribute to the lack of immune cell infiltration in the lungs of the i.m. vaccinated group. Predicted digital cell quantitation data from BAL fluid on 3 dpc showed a decrease in naive and plasma B cells, CD4+ and CD8+ T cells, and stimulated dendritic cells and neutrophils, supporting this hypothesis. Furthermore, digital cell quantitation data from the lungs on 7 dpc were indicative of a decrease in activated monocytes for all SARS-CoV-2-infected NHPs. Further immune cell characterization from BAL fluid and within the lungs is needed to expand upon our results and confirm this hypothesis.
In summary, in this study, we generated a potent single-dose, fast-acting vaccine for COVID-19. This vaccine grows to high titers, like the parent VSV-EBOV vector, and to higher titers than those of a VSV vaccine expressing SARS-CoV-2 S alone (51). Several important questions remain to be addressed in future studies. An extension of the time between vaccination and challenge might overcome the observed difference in protection between the vaccination routes and might eliminate the signs of immunopathology. This aspect will be investigated in future studies in conjunction with assessing the durability of SARS-CoV-2-specific immunity and a possible dose reduction of the vaccine, as has been described for VSV-EBOV, the parental vaccine (52). VSV-EBOV has been shown to elicit a durable humoral response, which lasts for at least 2 years in humans (53). We will also investigate the addition of another SARS-CoV-2 antigen into the vaccine to promote a stronger T cell response, as these responses are typically longer lasting. Furthermore, we will analyze if preexisting immunity to EBOV impacts the immunogenicity of this bivalent vaccine. For now, the VSV-SARS2-EBOV vaccine presents a vaccine with high potential as a boosting option after the already-approved mRNA-based vaccine, because VSV-SARS2-EBOV elicits primarily a humoral response, in contrast to the predominantly T cell-driven immune response after mRNA vaccination (16).
MATERIALS AND METHODS
Ethics statement.
All infectious work with SARS-CoV-2 was performed in the containment laboratories at the Rocky Mountain Laboratories (RML), Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The RML are an institution accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). All procedures followed standard operating procedures (SOPs) approved by the RML Institutional Biosafety Committee (IBC). Animal work was performed in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, the Office of Animal Welfare (54), and the Animal Welfare Act, U.S. Department of Agriculture (55). The studies were approved by the RML Animal Care and Use Committee (ACUC). Procedures were conducted with animals anesthetized by trained personnel under the supervision of veterinary staff. All efforts were made to maintain animal welfare and minimize animal suffering in accordance with the Weatherall report on the use of nonhuman primates in research (56). Animals were housed in adjoining individual primate cages that enabled social interactions, under controlled conditions of humidity, temperature, and light (12-h light/dark cycles). Food and water were available ad libitum. Animals were monitored and fed commercial monkey chow, treats, and fruit at least twice a day by trained personnel. Environmental enrichment consisted of commercial toys, music, videos, and social interaction.
Animal studies.
Sixteen female rhesus macaques (3.5 to 10 years of age; 4.5 to 10 kg, Indian origin) were used in this study. The NHPs were randomly selected for two vaccine groups (n = 6) and one control group (n = 4). On day 10, NHPs received a single vaccine dose of 1 × 107 PFU of VSV-SARS2-EBOV by the i.m. (injection in the caudal thigh) or i.n. (dropping vaccine into each nostril) route. Control animals received the same dose of a control vaccine (VSV-EBOV) by the i.m. (n = 2) or i.n. (n = 2) route (see Fig. S3A in the supplemental material). On dpc 0, animals were challenged with SARS-CoV-2 as previously described (29). On dpc 0, 1, 3, 5, and 7, a clinical exam including thoracic radiograph and nasal swab collection was performed. The dpc 3 exam included bronchoalveolar lavage (BAL), which was performed by insertion of an endotracheal tube and bronchoscope into the trachea past the 3rd bifurcation and subsequent installation of 10 ml of sterile saline. Manual suction was applied to retrieve the BAL sample. On day 7, all animals were euthanized for sample collection.
Cells and viruses.
Huh7 and Vero E6 cells were grown at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum (FBS) (Wisent Inc., St. Bruno, Canada), 2 mM l-glutamine (Thermo Fisher Scientific, Waltham, MA), 50 U/mL penicillin (Thermo Fisher Scientific), and 50 μg/mL streptomycin (Thermo Fisher Scientific). BHK-T7 (baby hamster kidney) cells expressing T7 polymerase were grown at 37°C and 5% CO2 in minimum essential medium (Thermo Fisher Scientific) containing 10% tryptose phosphate broth (Thermo Fisher Scientific), 5% FBS, 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/mL streptomycin. SARS-CoV-2 isolate nCoV-WA1-2020 (GenBank accession number MN985325.1) (57) was used for the animal challenge studies and neutralization test.
Generation of VSV-based vaccine candidates.
The SARS-CoV-2 S open reading frame (ORF) was PCR amplified from an expression plasmid encoding the codon-optimized (human) gene based on GenBank accession number MN908947, which was kindly provided by Vincent Munster (NIAID). Full-length SARS-CoV-2 S was cloned into the pATX-VSV-EBOV plasmid upstream of the EBOV Kikwit GP, resulting in VSV-SARS2-EBOV (26) according to a previously successful strategy (58). The 21-amino-acid deletion in the cytoplasmic tail was introduced by PCR and was cloned into the pATX-VSV plasmid, resulting in VSV-SARS2. The replication-competent recombinant VSVs were recovered in BHK-T7 cells as described previously (28). VSV-SARS2 was propagated on Vero E6 cells; VSV-SARS2-EBOV was propagated on Huh7 cells. The complete sequence of the viruses was confirmed by Sanger sequencing. The titer of the virus stocks was quantified using a standard plaque assay of Vero E6 cells.
Western blot analysis.
Supernatant samples containing VSV were mixed 1:1 with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer containing 20% β-mercaptoethanol and heated to 99°C for 10 min. SDS-PAGE and transfer to Trans-Blot polyvinylidene difluoride membranes (Bio-Rad Laboratories) of all samples was performed as described elsewhere (22). Protein detection was performed using anti-SARS-CoV-2 S RBD (1:1,000; Sino Biological), anti-EBOV GP (ZGP 12/1.1, 1 μg/ml; kindly provided by Ayato Takada, Hokkaido University, Japan), or anti-VSV M (23H12, 1:1,000; Kerafast Inc.). After horseradish peroxidase (HRP)-labeled secondary antibody staining using either anti-mouse IgG (1:10,000) or anti-rabbit IgG (1:5,000) (Jackson ImmunoResearch), the blots were imaged using the SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific) and an iBright CL1500 imaging system (Thermo Fisher Scientific).
Hematology.
The total white blood cell, neutrophil, lymphocyte, platelet, reticulocyte, and red blood cell counts, as well as hemoglobin and hematocrit values, were determined from EDTA blood with the IDEXX ProCyte DX analyzer (IDEXX Laboratories, Westbrook, ME).
RNA extraction and RT-qPCR.
RNA from blood, BAL fluid, and nasal swabs was extracted using the QIAamp viral RNA minikit (Qiagen) according to manufacturer specifications. RNA from tissues, a maximum of 30 mg each, was processed and extracted using the RNeasy minikit (Qiagen) according to manufacturer specifications. One-step reverse transcription-quantitative PCR (RT-qPCR) for both genomic and subgenomic viral RNAs was performed using specific primer-probe sets and the QuantiFast probe RT-PCR plus ROX vial kit (Qiagen) in Rotor-Gene Q (Qiagen) as described previously (13). Five microliters of each RNA extraction was run alongside dilutions of SARS-CoV-2 standards with a known concentration of RNA copies.
Virus loads.
Virus loads were determined from lung and BAL samples on Vero E6 cells (mycoplasma negative). To this end, cells were seeded in 96-well plates the day before titration. At the day of titration, BAL samples and lung tissue samples were thawed. Tissues were homogenized in 1 ml plain DMEM, and the supernatant was cleared by centrifugation. For all samples, 10-fold serial dilutions were prepared in DMEM supplemented with 2% FBS. Medium was removed from cells, and triplicate wells were inoculated with each dilution. Cells were monitored for cytopathic effect (CPE), and the 50% tissue culture infectious dose (TCID50) was calculated for each sample.
ELISAs.
Serum and BAL samples from SARS-CoV-2-infected animals were inactivated by gamma irradiation and used under biosafety level 2 (BSL2) conditions according to IBC-approved SOPs. Nunc MaxiSorp Immuno plates were coated with 50 μL of 1-μg/mL recombinant SARS-CoV-2 spike (S1 plus S2), the SARS-CoV-2 RBD (Sino Biological), or EBOV GP antigen at 4°C overnight and then washed three times with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST). The plates were blocked with 3% skim milk in PBS for 3 h at room temperature, followed by three additional washes with PBST. The plates were incubated with 50 μl of serial dilutions of the samples in PBS containing 1% skim milk for 1 h at room temperature. After 3 washes with PBST, the bound antibodies were labeled using 50 μL of 1:2,500 HRP-labeled anti-monkey IgG (H+L) (SeraCare Life Sciences) diluted in 1% skim milk in PBST. For the IgG subclass enzyme-linked immunosorbent assays (ELISAs), the plates were incubated with samples at 4°C overnight. After three washes with PBST, 50 μL of 1-μg/mL anti-rhesus IgG1 (ena), IgG2 (dio), IgG3 (tria), or IgG4 (tessera) (NHPRR) diluted in 1% skim milk in PBST was added, and the mixture was incubated for 1 h at room temperature. After 3 washes with PBST, the bound antibodies were labeled using 50 μL of 1:10,000 HRP-labeled anti-mouse IgG (H+L) (SeraCare Life Sciences) diluted in 1% skim milk in PBST. For all ELISAs, after incubation for 1 h at room temperature and 3 washes with PBST, 50 μL of the KPL ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] peroxidase substrate solution mix (SeraCare Life Sciences) was added to each well, and the mixture was incubated for 30 min at room temperature. The optical density (OD) at 405 nm was measured using a GloMax explorer (Promega). The OD values were normalized to the baseline samples obtained on day −10, and the cutoff value was set as the mean OD plus the standard deviation (SD) of the blank.
Virus neutralization assay.
The day before the virus neutralization assay, Vero E6 cells were seeded in 96-well plates. Serum samples were heat inactivated for 30 min at 56°C, and 2-fold serial dilutions were prepared in DMEM with 2% FBS. Next, 100 TCID50 of SARS-CoV-2 were added, and the mixture was incubated for 1 h at 37°C and 5% CO2. Finally, medium was removed from cells, and the mixture was added to Vero E6 cells and incubated at 37°C and 5% CO2 for 6 days. Then the CPE was documented, and the virus neutralization titer was expressed as the reciprocal value of the highest dilution of the serum which inhibited virus replication (no CPE).
Flow cytometry.
Rhesus macaque PBMCs were isolated from EDTA whole blood by overlay on a Histopaque-1077 density cushion and separated according to the manufacturers’ instructions. Isolated PBMCs were resuspended in FBS with 10% dimethyl sulfoxide (DMSO) and frozen at −80°C until analysis. For analysis of T cell intracellular cytokine production, cells were stimulated for 6 h with 1-μg/mL the SARS-CoV-S peptide pool, media, and cell stimulation cocktail (containing phorbol myristate acetate [PMA]-ionomycin; BioLegend) or a Lassa virus (LASV) GP peptide pool with 5 μg/mL brefeldin A (BioLegend). Following surface staining with LIVE/DEAD-allophycocyanin (APC)-Cy7, CD3-fluorescein isothiocyanate (FITC), CD4-Alexa700, CD8-phycoerythrin (PE)-Texas Red, CD56-BV421, and CD69-PE-Cy7, cells were fixed with 4% paraformaldehyde (PFA) and stained intracellularly with IFN-γ–BV605, IL-4–APC, and IL-2–PerCPCy5.5 diluted in perm-wash buffer (BioLegend). For analysis of NK cell intracellular cytokine production, cells were stimulated as described above. Following surface staining with LIVE/DEAD-APC-Cy7, CD3-FITC, CD4-PerCPCy5.5, CD8-PE-Texas Red, CD16-Alexa700, and CD56-BV421, cells were fixed with 4% PFA and stained intracellularly with granzyme B-APC.
Sample acquisition was performed on a CytoFLEX S flow cytometer (Beckman Coulter), and data were analyzed in FlowJo v10 (TreeStar). Antigen-specific T cells were identified by gating on LIVE/DEAD-negative, doublet-negative (side scatter height [SSC-H] versus side scatter area [SSC-A]), CD3+, CD56−, CD4+, or CD8+ cells and determination of cytokine positivity. Three NK cell subpopulations were identified by gating on LIVE/DEAD-negative, doublet-negative (SSC-H versus SSC-A), CD3−, CD56−, and CD8+ or CD16+ cells or on CD8+ CD16+ doubly positive cells. Cytokine responses for each subpopulation were identified by gating on the population and then granzyme B+ cells. Cytokine-positive responses are presented after subtraction of the background responses detected in the LASV GP peptide-stimulated samples.
Cytokine analysis.
Macaque serum and BAL samples were inactivated by gamma irradiation and used under BSL2 conditions according to IBC-approved SOPs. Samples were then diluted 1:2 in serum matrix for analysis with the Milliplex nonhuman primate magnetic bead panel as per the manufacturer’s instructions (Millipore Corporation). Concentrations for each cytokine were determined for all samples using the Bio-Plex 200 system (Bio-Rad Laboratories Inc.).
Thoracic radiographs.
Ventrodorsal and right/left lateral radiographs were taken on clinical exam days prior to any other procedures (e.g., bronchoalveolar lavage) and were evaluated and scored for the presence of pulmonary infiltrates by two board-certified clinical veterinarians according to a standard scoring system (59). Radiographs taken prior to SARS-CoV-2 inoculation at 0 dpc served as a baseline for each animal, with scores for all lung lobes set to 0, i.e., normal examination. All subsequent radiographs were compared to these radiographs and evaluated for changes from baseline. Each lung lobe (upper left, middle left, lower left, upper right, middle right, lower right) was scored individually based on the following scores: 0, normal examination; 1, mild interstitial pulmonary infiltrates; 2, moderate interstitial pulmonary infiltrates, perhaps with partial cardiac border effacement and small areas of pulmonary consolidation (alveolar patterns and air bronchograms); and 3, pulmonary consolidation as the primary lung pathology, seen as a progression from grade 2 lung pathology. At study completion, thoracic radiograph findings were reported as a single radiograph score for each animal on each exam day. To obtain this score, the scores assigned to each of the six lung lobes were added together and recorded as the radiograph score for each animal on each exam day. Scores may range from 0 to 18 for each animal on each exam day.
Histology and immunohistochemistry.
Necropsies and tissue sampling were performed according to IBC-approved SOPs. Lungs were perfused with 10% formalin and processed for histologic review. Harvested tissues were fixed for 8 days in 10% neutral buffered formalin, embedded in paraffin, processed using a VIP-6 Tissue Tek (Sakura Finetek, USA) tissue processor, and embedded in Ultraffin paraffin polymer (Cancer Diagnostics, Durham, NC). Samples were sectioned at 5 μm and dried overnight at 42°C, and the resulting slides were stained with hematoxylin and eosin (H&E). Specific anti-CoV immunoreactivity was detected using an in-house SARS-CoV-2 nucleocapsid protein (U864YFA140-4/CB2093) rabbit antibody (GenScript) at a 1:1,000 dilution. The immunohistochemistry (IHC) assay was carried out on a Discovery Ultra automated staining instrument (Roche Tissue Diagnostics) with a Discovery ChromoMap DAB (Ventana Medical Systems) kit. All tissue slides were evaluated by a board-certified veterinary pathologist. Sections taken at 3 levels from each lung lobe, in total, 18 sections, were evaluated for each animal; a representative lesion from each group was selected for Fig. 2. Lung sections were analyzed for evidence of septal edema and infiltrates representing interstitial pneumonia and were assigned the following scores: 0, normal; 1, minimal; 2, mild; 3, moderate; and 4, severe.
Library construction and sequencing.
The quality and quantity of RNA from BAL fluid and the lower left lung (LLL) were determined using an Agilent 2100 bioanalyzer. cDNA libraries were constructed using the NEB Next Ultra II Direction RNA library prep kit (Thermo Fischer). RNA was treated with RNase H and DNase I following depletion of rRNA. Adapters were ligated to cDNA products, and the subsequent ∼300-bp amplicons were PCR amplified and selected by size exclusion. cDNA libraries were assessed for quality and quantity prior to 150-bp single-end sequencing using the Illumina NovaSeq platform.
Bioinformatic analysis.
Preliminary data analysis was performed with the transcriptome sequencing (RNA-Seq) workflow module of systemPipeR, developed by Backman and Girke (60). RNA-Seq reads were demultiplexed, quality filtered, and trimmed using Trim Galore (average Phred score cutoff, 30; minimum length of 50 bp). FastQC was used to generate quality reports. Hisat2 was used to align reads to the reference genome of Macaca mulatta (Macaca_mulatta.Mmul_8.0.1.dna.toplevel.fa), and Macaca_mulatta.Mmul_8.0.1.97.gtf was used for annotation. For viral read quantification, RNA-Seq reads were separately aligned to the severe acute respiratory syndrome coronavirus 2 Wuhan isolate genome (NC_045512.2), and the GCF_009858895.2_ASM985889v3_genomic.gff annotation file was used. Raw expression values (gene-level read counts) were generated using the summarizeOverlaps function and normalized (reads per kilobase of transcript per million mapped reads [rpkm]) using the edgeR package. Statistical analysis with edgeR was used to determine DEGs meeting the following criteria: genes with a median rpkm of ≥5, a false-discovery rate (FDR)-corrected P value of ≤0.05, and a log2 fold change of ≥1 compared to values for uninfected tissues. The number of total viral reads was determined as the total number of normalized read counts mapping to all viral genes.
Functional enrichment of DEGs was performed using Metascape to identify relevant gene ontology (GO) biological process terms and KEGG pathways. Digital cell quantitation was performed using ImmQuant with the IRIS database. Heatmaps, bubble plots, Venn diagrams, and violin plots were generated using the R packages ggplot2 and VennDiagram. GO network plots were generated in Cytoscape (version 3.5.1). Graphs were generated using GraphPad Prism software (version 8).
Statistical analyses.
All statistical analysis was performed in Prism 8 (GraphPad). The in vitro growth kinetics of recombinant VSVs (Fig. S1C) and hematology data (Fig. S2B) were examined using two-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test to evaluate statistical significance at all time points. Bioinformatics data were analyzed using one-way ANOVA with multiple comparisons, and comparisons were made to either uninfected animals or control vaccinated animals. The two-tailed Mann-Whitney rank or Wilcoxon test was conducted to compare differences between groups for all other data. A Bonferroni correction was used to control for type I error rate, where required. Statistically significant differences are indicated as follows: a P of <0.0001 (****), a P of <0.001 (***), a P of <0.01 (**), and a P of <0.05 (*).
Data availability.
RNA sequencing data are accessible at BioProject accession number PRJNA750507.
ACKNOWLEDGMENTS
We thank the Rocky Mountain Veterinary Branch, NIAID, for supporting the animal studies and Anita Mora (NIAID) for assistance generating the pathology figures. We also thank Jonathan Schulz (NIAID) for assistance with molecular assays.
The study was funded by the Intramural Research Program, NIAID, NIH. RNA sequencing was funded by the National Center for Research Resources and the National Center for Advancing Translational Sciences, NIH, through grant UL1 TR001414 awarded to I.M.
A.M. conceived the idea. A.M. and I.M. secured funding. W.F. and A.M. designed the studies. W.F., K.S., A.J.G., F.F., A.O., T.G., J.L., P.W.H., T.T., C.S.C., K.L.O., and A.M. conducted the studies, processed the samples, and acquired the data. A.N.P. and A.J. performed the transcriptomics work. W.F., K.S., A.N.P., C.S.C., I.M., K.L.O., and A.M. analyzed and interpreted the data. W.F., I.M., and A.M. prepared the manuscript. All authors approved the manuscript.
We declare no conflicts of interest.
Contributor Information
Andrea Marzi, Email: marzia@niaid.nih.gov.
Anne Moscona, Columbia University Medical College.
REFERENCES
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for Covid-19. 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung EW, Zachariah P, Gorelik M, Boneparth A, Kernie SG, Orange JS, Milner JD. 2020. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in New York City. JAMA 324:294–296. doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao R, Qiu Y, He JS, Tan JY, Li XH, Liang J, Shen J, Zhu LR, Chen Y, Iacucci M, Ng SC, Ghosh S, Chen MH. 2020. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wichmann D, Sperhake JP, Lutgehetmann M, Steurer S, Edler C, Heinemann A, Heinrich F, Mushumba H, Kniep I, Schroder AS, Burdelski C, de Heer G, Nierhaus A, Frings D, Pfefferle S, Becker H, Bredereke-Wiedling H, de Weerth A, Paschen HR, Sheikhzadeh-Eggers S, Stang A, Schmiedel S, Bokemeyer C, Addo MM, Aepfelbacher M, Puschel K, Kluge S. 2020. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med 173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. 2020. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK, Washington State 2019-nCoV Case Investigation Team. 2020. First case of 2019 novel coronavirus in the United States. N Engl J Med 382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L. 2020. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 12.Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, Chen RE, Winkler ES, Wessel AW, Case JB, Kashentseva E, McCune BT, Bailey AL, Zhao H, VanBlargan LA, Dai YN, Ma M, Adams LJ, Shrihari S, Danis JE, Gralinski LE, Hou YJ, Schafer A, Kim AS, Keeler SP, Weiskopf D, Baric RS, Holtzman MJ, Fremont DH, Curiel DT, Diamond MS. 2020. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell 183:169–184.e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, Avanzato VA, Bushmaker T, Flaxman A, Ulaszewska M, Feldmann F, Allen ER, Sharpe H, Schulz J, Holbrook M, Okumura A, Meade-White K, Perez-Perez L, Edwards NJ, Wright D, Bissett C, Gilbride C, Williamson BN, Rosenke R, Long D, Ishwarbhai A, Kailath R, Rose L, Morris S, Powers C, Lovaglio J, Hanley PW, Scott D, Saturday G, de Wit E, Gilbert SC, Munster VJ. 2020. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Van Ry A, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin Z, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan H, Cai Y, Chen B, Schmidt AG, Wesemann DR, Baric RS, et al. 2020. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, He X, Martinez DR, Rutten L, Bos R, van Manen D, Vellinga J, Custers J, Langedijk JP, Kwaks T, Bakkers MJG, Zuijdgeest D, Rosendahl Huber SK, Atyeo C, Fischinger S, Burke JS, Feldman J, Hauser BM, Caradonna TM, Bondzie EA, Dagotto G, Gebre MS, Hoffman E, Jacob-Dolan C, Kirilova M, Li Z, Lin Z, Mahrokhian SH, Maxfield LF, Nampanya F, Nityanandam R, Nkolola JP, Patel S, Ventura JD, Verrington K, Wan H, Pessaint L, Van Ry A, Blade K, Strasbaugh A, Cabus M, et al. 2020. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O'Connell S, Bock KW, Minai M, Nagata BM, Andersen H, Martinez DR, Noe AT, Douek N, Donaldson MM, Nji NN, Alvarado GS, Edwards DK, Flebbe DR, Lamb E, Doria-Rose NA, Lin BC, Louder MK, O'Dell S, Schmidt SD, Phung E, Chang LA, Yap C, Todd JM, Pessaint L, Van Ry A, Browne S, Greenhouse J, Putman-Taylor T, Strasbaugh A, Campbell TA, Cook A, Dodson A, Steingrebe K, Shi W, Zhang Y, Abiona OM, Wang L, Pegu A, Yang ES, Leung K, Zhou T, Teng IT, Widge A, et al. 2020. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med 383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzi A, Feldmann H, Geisbert TW, Falzarano D. 2011. Vesicular stomatitis virus-based vaccines for prophylaxis and treatment of filovirus infections. J Bioterror Biodef S1:2157-2526-S1-004. doi: 10.4172/2157-2526.S1-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mire CE, Geisbert JB, Agans KN, Versteeg KM, Deer DJ, Satterfield BA, Fenton KA, Geisbert TW. 2019. Use of single-injection recombinant vesicular stomatitis virus vaccine to protect nonhuman primates against lethal Nipah virus disease. Emerg Infect Dis 25:1144–1152. doi: 10.3201/eid2506.181620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Safronetz D, Mire C, Rosenke K, Feldmann F, Haddock E, Geisbert T, Feldmann H. 2015. A recombinant vesicular stomatitis virus-based Lassa fever vaccine protects guinea pigs and macaques against challenge with geographically and genetically distinct Lassa viruses. PLoS Negl Trop Dis 9:e0003736. doi: 10.1371/journal.pntd.0003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fathi A, Dahlke C, Addo MM. 2019. Recombinant vesicular stomatitis virus vector vaccines for WHO blueprint priority pathogens. Hum Vaccin Immunother 15:2269–2285. doi: 10.1080/21645515.2019.1649532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marzi A, Robertson SJ, Haddock E, Feldmann F, Hanley PW, Scott DP, Strong JE, Kobinger G, Best SM, Feldmann H. 2015. Ebola vaccine. VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 349:739–742. doi: 10.1126/science.aab3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuyama W, Reynolds P, Haddock E, Meade-White K, Quynh Le M, Kawaoka Y, Feldmann H, Marzi A. 2020. A single dose of a vesicular stomatitis virus-based influenza vaccine confers rapid protection against H5 viruses from different clades. NPJ Vaccines 5:4. doi: 10.1038/s41541-019-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown KS, Safronetz D, Marzi A, Ebihara H, Feldmann H. 2011. Vesicular stomatitis virus-based vaccine protects hamsters against lethal challenge with Andes virus. J Virol 85:12781–12791. doi: 10.1128/JVI.00794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, Carroll MW, Dean NE, Diatta I, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Gsell PS, Hossmann S, Watle SV, Konde MK, Keita S, Kone S, Kuisma E, Levine MM, Mandal S, Mauget T, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Rottingen JA, Kieny MP. 2017. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial. Lancet 389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu X, Fernando L, Alimonti JB, Melito PL, Feldmann F, Dick D, Stroher U, Feldmann H, Jones SM. 2009. Mucosal immunization of cynomolgus macaques with the VSVDeltaG/ZEBOVGP vaccine stimulates strong Ebola GP-specific immune responses. PLoS One 4:e5547. doi: 10.1371/journal.pone.0005547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Donnell KL, Griffin AJ, Shifflett K, Gourdine T, Thomas T, Long CM, Furuyama W, Marzi A. 2021. Optimization of single dose VSV-based COVID-19 vaccination in hamsters. BioRxiv doi: 10.1101/2021.09.03.458735. [DOI] [PMC free article] [PubMed]
- 27.Case JB, Rothlauf PW, Chen RE, Liu Z, Zhao H, Kim AS, Bloyet LM, Zeng Q, Tahan S, Droit L, Ilagan MXG, Tartell MA, Amarasinghe G, Henderson JP, Miersch S, Ustav M, Sidhu S, Virgin HW, Wang D, Ding S, Corti D, Theel ES, Fremont DH, Diamond MS, Whelan SPJ. 2020. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emanuel J, Callison J, Dowd KA, Pierson TC, Feldmann H, Marzi A. 2018. A VSV-based Zika virus vaccine protects mice from lethal challenge. Sci Rep 8:11043. doi: 10.1038/s41598-018-29401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Perez-Perez L, Schulz J, Meade-White K, Okumura A, Callison J, Brumbaugh B, Avanzato VA, Rosenke R, Hanley PW, Saturday G, Scott D, Fischer ER, de Wit E. 2020. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzi A, Engelmann F, Feldmann F, Haberthur K, Shupert WL, Brining D, Scott DP, Geisbert TW, Kawaoka Y, Katze MG, Feldmann H, Messaoudi I. 2013. Antibodies are necessary for rVSV/ZEBOV-GP-mediated protection against lethal Ebola virus challenge in nonhuman primates. Proc Natl Acad Sci USA 110:1893–1898. doi: 10.1073/pnas.1209591110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menicucci AR, Versteeg K, Woolsey C, Mire CE, Geisbert JB, Cross RW, Agans KN, Jankeel A, Geisbert TW, Messaoudi I. 2017. Transcriptome analysis of circulating immune cell subsets highlight the role of monocytes in Zaire Ebola virus Makona pathogenesis. Front Immunol 8:1372. doi: 10.3389/fimmu.2017.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiland N, Sanger DE. 27 July 2020. Trump administration selects five coronavirus vaccine candidates as finalists. New York Times; https://www.nytimes.com/2020/06/03/us/politics/coronavirus-vaccine-trump-moderna.html. [Google Scholar]
- 33.Merck. 25 January 2021. Merck discontinues development of SARS-CoV-2/COVID-19 vaccine candidates; continues development of two investigational therapeutic candidates. Merck, Kenilworth, NJ. https://www.merck.com/news/merck-discontinues-development-of-sars-cov-2-covid-19-vaccine-candidates-continues-development-of-two-investigational-therapeutic-candidates/. [Google Scholar]
- 34.Estes JD, Wong SW, Brenchley JM. 2018. Nonhuman primate models of human viral infections. Nat Rev Immunol 18:390–404. doi: 10.1038/s41577-018-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munster VJ, Flagg M, Singh M, Yinda CK, Williamson BN, Feldmann F, Perez-Perez L, Schulz J, Brumbaugh B, Holbrook MG, Adney DR, Okumura A, Hanley PW, Smith BJ, Lovaglio J, Anzick SL, Martens C, van Doremalen N, Saturday G, de Wit E. 2021. Subtle differences in the pathogenicity of SARS-CoV-2 variants of concern B.1.1.7 and B.1.351 in rhesus macaques. Sci Adv 7:eabj3627. doi: 10.1126/sciadv.abj3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasenkrug KJ, Feldmann F, Myers L, Santiago ML, Guo K, Barrett BS, Mickens KL, Carmody A, Okumura A, Rao D, Collins MM, Messer RJ, Lovaglio J, Shaia C, Rosenke R, van Doremalen N, Clancy C, Saturday G, Hanley P, Smith BJ, Meade-White K, Shupert WL, Hawman DW, Feldmann H. 2021. Recovery from acute SARS-CoV-2 infection and development of anamnestic immune responses in T cell-depleted rhesus macaques. mBio 12:e01503-21. doi: 10.1128/mBio.01503-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassan AO, Feldmann F, Zhao H, Curiel DT, Okumura A, Tang-Huau TL, Case JB, Meade-White K, Callison J, Chen RE, Lovaglio J, Hanley PW, Scott DP, Fremont DH, Feldmann H, Diamond MS. 2021. A single intranasal dose of chimpanzee adenovirus-vectored vaccine protects against SARS-CoV-2 infection in rhesus macaques. Cell Rep Med 2:100230. doi: 10.1016/j.xcrm.2021.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, Andersen H, Baric RS, Carroll MW, Cavaleri M, Qin C, Crozier I, Dallmeier K, de Waal L, de Wit E, Delang L, Dohm E, Duprex WP, Falzarano D, Finch CL, Frieman MB, Graham BS, Gralinski LE, Guilfoyle K, Haagmans BL, Hamilton GA, Hartman AL, Herfst S, Kaptein SJF, Klimstra WB, Knezevic I, Krause PR, Kuhn JH, Le Grand R, Lewis MG, Liu WC, Maisonnasse P, McElroy AK, Munster V, Oreshkova N, Rasmussen AL, Rocha-Pereira J, Rockx B, Rodriguez E, Rogers TF, Salguero FJ, Schotsaert M, Stittelaar KJ, Thibaut HJ, Tseng CT, et al. 2020. Animal models for COVID-19. Nature 586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cleary SJ, Pitchford SC, Amison RT, Carrington R, Robaina Cabrera CL, Magnen M, Looney MR, Gray E, Page CP. 2020. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br J Pharmacol 177:4851–4865. doi: 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Thorson L, Stokes RW, Santosuosso M, Huygen K, Zganiacz A, Hitt M, Xing Z. 2004. Single mucosal, but not parenteral, immunization with recombinant adenoviral-based vaccine provides potent protection from pulmonary tuberculosis. J Immunol 173:6357–6365. doi: 10.4049/jimmunol.173.10.6357. [DOI] [PubMed] [Google Scholar]
- 41.Neutra MR, Kozlowski PA. 2006. Mucosal vaccines: the promise and the challenge. Nat Rev Immunol 6:148–158. doi: 10.1038/nri1777. [DOI] [PubMed] [Google Scholar]
- 42.Cavalcante-Silva LHA, Carvalho DCM, Lima EA, Galvao J, da Silva JSF, Sales-Neto JM, Rodrigues-Mascarenhas S. 2021. Neutrophils and COVID-19: the road so far. Int Immunopharmacol 90:107233. doi: 10.1016/j.intimp.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Francois-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, Piehler J, Pellegrini S, Uze G. 2011. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One 6:e22200. doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumura T, Semba K, Azuma S, Ikawa S, Gohda J, Akiyama T, Inoue J. 2004. TIFAB inhibits TIFA, TRAF-interacting protein with a forkhead-associated domain. Biochem Biophys Res Commun 317:230–234. doi: 10.1016/j.bbrc.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 45.Wong JJ, Pung YF, Sze NS, Chin KC. 2006. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci USA 103:10735–10740. doi: 10.1073/pnas.0600397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, Lopez-Camacho C, Slon-Campos J, Zhao Y, Stuart DI, Paesen GC, Grimes JM, Antson AA, Bayfield OW, Hawkins D, Ker DS, Wang B, Turtle L, Subramaniam K, Thomson P, Zhang P, Dold C, Ratcliff J, Simmonds P, de Silva T, Sopp P, Wellington D, Rajapaksa U, Chen YL, Salio M, Napolitani G, Paes W, Borrow P, Kessler BM, Fry JW, Schwabe NF, Semple MG, Baillie JK, Moore SC, Openshaw PJM, Ansari MA, Dunachie S, Barnes E, Frater J, Kerr G, Goulder P, ISARIC4C Investigators, et al. 2020. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol 21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyman O, Sprent J. 2012. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol 12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 48.Abrahamsen G, Sundvold-Gjerstad V, Habtamu M, Bogen B, Spurkland A. 2018. Polarity of CD4+ T cells towards the antigen presenting cell is regulated by the Lck adapter TSAd. Sci Rep 8:13319. doi: 10.1038/s41598-018-31510-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo JS, Srikanth S, Kim KD, Elsaesser H, Lu J, Pellegrini M, Brooks DG, Sun Z, Gwack Y. 2018. CRACR2A-mediated TCR signaling promotes local effector Th1 and Th17 responses. J Immunol 201:1174–1185. doi: 10.4049/jimmunol.1800659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, Guo D, Hu W, Yang J, Tang Z, Wu H, Lin Y, Zhang M, Zhang Q, Shi M, Liu Y, Zhou Y, Lan K, Chen Y. 2020. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Case JB, Rothlauf PW, Chen RE, Kafai NM, Fox JM, Smith BK, Shrihari S, McCune BT, Harvey IB, Keeler SP, Bloyet LM, Zhao H, Ma M, Adams LJ, Winkler ES, Holtzman MJ, Fremont DH, Whelan SPJ, Diamond MS. 2020. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe 28:465–474.e4. doi: 10.1016/j.chom.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marzi A, Reynolds P, Mercado-Hernandez R, Callison J, Feldmann F, Rosenke R, Thomas T, Scott DP, Hanley PW, Haddock E, Feldmann H. 2019. Single low-dose VSV-EBOV vaccination protects cynomolgus macaques from lethal Ebola challenge. EBioMedicine 49:223–231. doi: 10.1016/j.ebiom.2019.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huttner A, Agnandji ST, Combescure C, Fernandes JF, Bache EB, Kabwende L, Ndungu FM, Brosnahan J, Monath TP, Lemaître B, Grillet S, Botto M, Engler O, Portmann J, Siegrist D, Bejon P, Silvera P, Kremsner P, Siegrist C-A, Krishna S, Addo MM, Becker S, Krähling V, Njuguna P, Kieny M-P, Ahmed R, Anderson J, Auderset F, Borgianni L, Ciabattini A, Haks MC, Harandi A, Heppner DG, Gerlini A, Medaglini D, Ottenhoff THM, Pejoski D, Page M, Pozzi G, Santoro F, Dubey S, Fernandes JF, Nakaya H, Orourke F, Pozzi G, Rothenberger S. 2018. Determinants of antibody persistence across doses and continents after single-dose rVSV-ZEBOV vaccination for Ebola virus disease: an observational cohort study. Lancet Infect Dis 18:738–748. doi: 10.1016/S1473-3099(18)30165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. The National Academies Press, Washington, DC. https://grants.nih.gov/grants/olaw/guide-for-the-care-and-use-of-laboratory-animals.pdf. [Google Scholar]
- 55.National Agricultural Library, USDA. 2015. Chapter 54—Transportation, sale, and handling of certain animals. 7 US Code. https://www.govinfo.gov/content/pkg/USCODE-2015-title7/html/USCODE-2015-title7-chap54.htm.
- 56.The Royal Society. 2006. The Weatherall report on the use of non-human primates in research. The Royal Society, London, United Kingdom. https://royalsociety.org/policy/publications/2006/weatherall-report/. [Google Scholar]
- 57.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD, Thornburg NJ. 2020. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis 26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsuda Y, Safronetz D, Brown K, LaCasse R, Marzi A, Ebihara H, Feldmann H. 2011. Protective efficacy of a bivalent recombinant vesicular stomatitis virus vaccine in the Syrian hamster model of lethal Ebola virus infection. J Infect Dis 204(Suppl 3):S1090–S1097. doi: 10.1093/infdis/jir379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brining DL, Mattoon JS, Kercher L, LaCasse RA, Safronetz D, Feldmann H, Parnell MJ. 2010. Thoracic radiography as a refinement methodology for the study of H1N1 influenza in cynomolgus macaques (Macaca fascicularis). Comp Med 60:389–395. [PMC free article] [PubMed] [Google Scholar]
- 60.Backman TWH, Girke T. 2016. systemPipeR: NGS workflow and report generation environment. BMC Bioinformatics 17:388. doi: 10.1186/s12859-016-1241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic and characterization of VSV-based vaccines. (A) Schematic illustrating vaccine vector design. T7 promotor; N, nucleoprotein; P, phosphoprotein; M, matrix protein; EBOV GP, Ebola virus glycoprotein; L, RNA-dependent RNA polymerase; SARS2-S, full-length SARS-CoV-2 S. (B) Western blot analysis of cell supernatant samples containing VSV vaccines probed for SARS-CoV-2 S (left), VSV M (right), or EBOV GP (right). Lane 1, wild-type VSV; lane 2, VSV-EBOV; lane 3, VSV-SARS2-EBOV. (C) Viral growth kinetics performed in triplicates on Vero E6 cells. Geometric means and SDs are depicted. Download FIG S1, PDF file, 0.06 MB (71.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
NHP study outline and hematology. (A) Schematic outline of the rhesus macaque study. (B) Levels of white blood cell (WBCs), neutrophils, lymphocytes, red blood cells (RBCs), platelets, reticulocytes, hemoglobin, and hematocrit in the blood of vaccinated and challenged NHPs. Download FIG S2, PDF file, 0.07 MB (94.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Radiographs of NHPs. Thoracic radiographs taken before SARS-CoV-2 infection (day 0) and at the time of euthanasia (day 7) are shown for each NHP. Download FIG S3, PDF file, 0.9 MB (13.1MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gross lung pathology of NHPs. NHPs were vaccinated 10 days before challenge with SARS-CoV-2. Representative pictures of NHP lungs with lesions on day 7 after challenge are shown. IM, intramuscular vaccination; IN, intranasal vaccination. Lesions are circled. Download FIG S4, PDF file, 0.3 MB (317.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Virus load in NHP tissue samples on day 7. (A) Total SARS-CoV-2-specific RNA (left) and subgenomic RNA (sgRNA) (right) in lung samples collected from NHPs. RLU, right lobe upper; RLM, right lobe middle; RLL, right lobe lower; LLU, left lobe upper; LLM, left lobe middle; LLL, left lobe lower. (B) Total SARS-CoV-2-specific RNA in tissue samples (right) from NHPs. LN, lymph node; R, right; L, left. Download FIG S5, PDF file, 0.06 MB (69.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
EBOV GP-specific antibodies in NHPs after vaccination and challenge. (A) Serum samples collected throughout the study and (B) bronchoaveolar lavage (BAL) samples collected on day 3 were analyzed by ELISA for EBOV GP-specific IgG. Statistical significance is indicated. Download FIG S6, PDF file, 0.04 MB (46.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Serum cytokine levels in NHPs after vaccination and challenge. (A and B) Serum samples collected throughout the study (A) and BAL samples collected on day 3 (B) were analyzed. Statistical significance is indicated. Download FIG S7, PDF file, 0.1 MB (113.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
BAL fluid RNA sequencing. (A) Principal-component analysis of BAL samples from uninfected animals and vaccinated animals dpc 3 (control, intramuscular vaccination, or intranasal vaccination). (B) Down- and up-regulated differentially expressed genes (DEGs). (C to E) Heatmaps representing DEGs shared by all infected groups and associated with the gene ontology (GO) terms “myeloid cell activation” and “neutrophil degranulation” for upregulated DEGs (C) and downregulated DEGs (D) and those associated with “autophagy” and “positive regulation of cytokine production” for upregulated DEGs (E). Each column represents the median number of rpkm of the given group. The range of colors is based on a scale and centered rpkm values of the represented DEGs. Red represents upregulated DEGs; blue represents downregulated DEGs. (F) Digital cell quantitation using the ImmQuant IRIS database comparing challenged groups to uninfected controls. Red represents upregulation; blue represents downregulation. Each column represents the average relative predicted frequency of the given cell type. P values are calculated relative to values for the uninfected animals. Statistical significance is indicated. For all heatmaps, the range of colors is based on a scale and centered rpkm values of the represented DEGs. Download FIG S8, PDF file, 0.3 MB (310.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Lung RNA sequencing. (A) Principal-component analysis of lower left lung (LLL) samples from uninfected animals and vaccinated animals dpc 7 (control, intramuscular vaccination, or intranasal vaccination). (B) Down- and up-regulated DEGs. (C to E) Heatmaps representing DEGs shared by all infected groups and associated with the GO terms “coagulation,” “response not decreased oxygen levels,” and “wound healing” for downregulated DEGs (C), “chemotaxis”, “cell projection morphogenesis,” and “extracellular structure organization” for upregulated DEGs (D), and “angiogenesis” for upregulated DEGs (E). Each column represents the median number of rpkm of the given group. The range of colors is based on a scale and centered rpkm values of the represented DEGs. Red represents upregulated DEGs; blue represents downregulated DEGs. (F) Digital cell quantitation using the ImmQuant IRIS database comparing challenged groups to uninfected controls. Red represents upregulation; blue represents downregulation. Each column represents the average relative predicted frequency of the given cell type. P values are calculated relative to values for the uninfected animals. Statistical significance is indicated. For all heatmaps, the range of colors is based on a scale and on centered rpkm values of the represented DEGs. Download FIG S9, PDF file, 0.3 MB (296.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
RNA sequencing data are accessible at BioProject accession number PRJNA750507.