ABSTRACT
Systemic cryptococcosis is fatal without treatment. Globally, this disease kills 180,000 of the 225,000 infected people each year, even with the use of antifungal therapies. Currently, there is no vaccine to prevent cryptococcosis. Previously, we discovered that Znf2, a morphogenesis regulator that directs Cryptococcus yeast-to-hyphal transition, profoundly affects cryptococcal interaction with the host—overexpression of ZNF2 drives filamentous growth, attenuates cryptococcal virulence, and elicits protective host immune responses. Importantly, immunization with cryptococcal cells overexpressing ZNF2, either in live or heat-inactivated form, offers significant protection to the host from a subsequent challenge by the otherwise lethal wild-type H99 strain. We hypothesize that cellular components enriched in ZNF2oe cells are immunoprotective. Here, we discovered that serum from protected animals vaccinated with inactivated ZNF2oe cells recognizes cryptococcal antigens that reside within the capsule. Consistently, capsule is required for immunoprotection offered by ZNF2oe cells. Interestingly, the serum from protective animals recognizes antigens in both wild-type yeast cells and ZNF2oe cells, with higher abundance in the latter. Consequently, even the heat-inactivated wild-type cells become immunoprotective with an increased vaccination dose. We also found that disruption of a chromatin remodeling factor Brf1, which is important for initiation of filamentation by Znf2, reduces the antigen level in ZNF2oe cells. Deletion of BRF1 drastically reduces the protective effect of ZNF2oe cells in both live and heat-killed forms even though the ZNF2oebrf1Δ strain itself is avirulent. Collectively, our findings underscore the importance of identifying the subset of cryptococcal surface factors that are beneficial in host protection.
KEYWORDS: Cryptococcus neoformans, antigens, capsule, immunofluorescence, morphogenesis, vaccination
INTRODUCTION
Cryptococcus neoformans is a ubiquitous environmental fungus and an opportunistic human fungal pathogen. This fungus enters the host through inhalation and its dissemination results in cryptococcal meningoencephalitis, a disease that claims hundreds of thousands of lives each year (1, 2). Systemic cryptococcosis is fatal without treatment. Even with current antifungal treatments, the mortality rates of this disease range from 10% to 70% depending on the time of diagnosis, the host underlying conditions, and the antifungal regimen used (2–10). Developing effective vaccines based on cryptococcal factors that mediate host-cryptococcal interactions has been an important research topic.
Morphogenesis profoundly shapes cryptococcal interaction with various hosts (11). This basidiomycete fungal pathogen undergoes three key morphological transitions in its life cycle – spore germination to yeast, yeast to hypha/filament, and hyphae to fruiting bodies/sporulation (12). The virulence potential of each morphotype and their cell envelope composition are drastically different. Basidiospores are small, highly infectious cells covered in a polysaccharide spore coat. Basidiospores can survive harsh conditions such as high temperature, desiccation, oxidative stress, chemical insult, and UV irradiation (12–14). Yeast cells are surrounded by large polysaccharide capsules and can replicate rapidly. Yeast cells are immune elusive and highly virulent to a mammalian host (15). Cryptococcal filaments grow by apical extension at the tip, and they are essential for completion of the cryptococcal bisexual cycle. Filaments are resistant to natural predators like soil ameba (11, 16, 17), but they are rarely found in human or animal hosts and are attenuated in virulence in animal models of cryptococcosis (16, 18, 19).
In our research to understand the molecular mechanisms underlying virulence attenuation of filamentous cells in mammalian hosts, we identified the transcription factor Znf2 that drives filamentation in C. neoformans (20, 21). Deletion of ZNF2 abolishes hyphal growth and enhances virulence, while overexpression of ZNF2 drives filamentation and drastically attenuates virulence (20, 22). Further investigation into virulence attenuation caused by ZNF2 overexpression revealed that ZNF2oe strains elicit protective host immune responses (22). Importantly, ZNF2oe cells, either in a live form or in a heat-inactivated form, elicit strong immunoprotective responses from the mammalian host (22). Notably, many cryptococcal mutants that fail to cause fatal infection (e.g., stress-sensitive, temperature-sensitive, or acapsular mutants), do not offer protection to the host against a subsequent challenge by a lethal wild-type strain (23, 24). So far, only two other cryptococcal mutants are reported to offer significant host protection in an inactivated form: a chitosan deficient mutant cda1-3Δ and a ubiquitination E3 ligase mutant fbp1Δ (25, 26). However, the molecular bases underlying the immunoprotection of these strains are unknown. Here we explore the cellular and molecular bases for ZNF2oe-mediated immunity against cryptococcosis.
RESULTS
ZNF2oe cells present more antigens compared to wild-type cells.
We previously showed that the exposure to ZNF2oe cells steered the host to differentiate protective T helper cells, as evident by the polarization toward Th1/Th17 of CD4+ T cells isolated from lungs at day 7 postinfection (22). There was also differential activation of Cryptococcus-specific responses toward Th1/Th17 in CD4+ T cells purified from the lung-draining mediastinal lymph node at day 7 from ZNF2oe inoculated mice (22). Because most T cell receptors are specific for peptide-MHC complexes (27) and B cells depend on T cell help with shared antigen specificity for the secretion of antibodies (28), we speculated that host antibodies should recognize protein antigens present in ZNF2oe cells. As both live and heat-killed (HK) ZNF2oe cells protected mice from the challenge with the wild-type H99 strain, we hypothesized that serum from protected animals should recognize shared protein antigens present in ZNF2oe cells and wild-type cells, and that the beneficial antigenic components are more abundantly expressed in ZNF2oe cells.
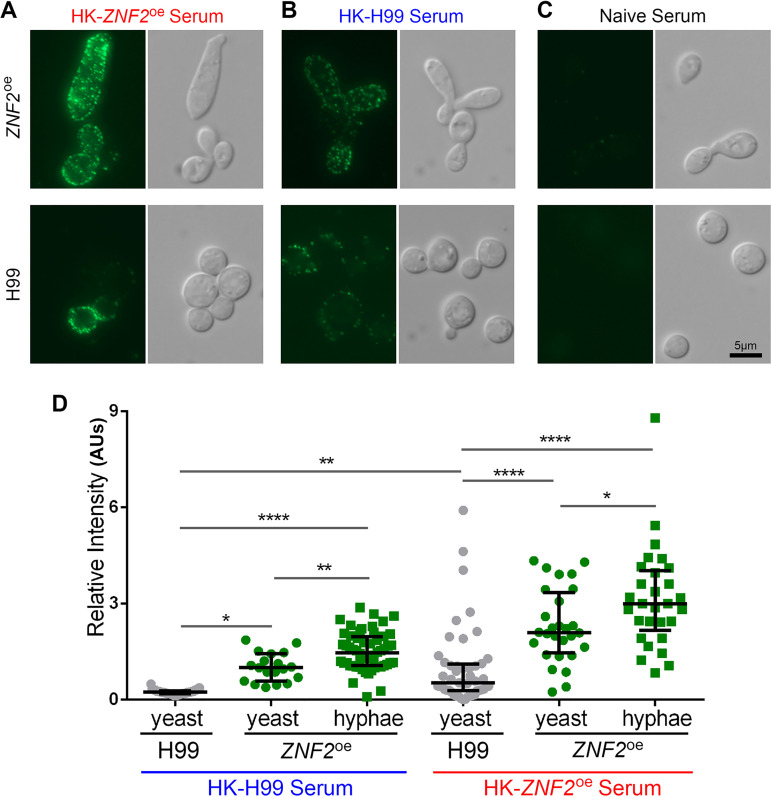
To test our hypothesis, we employed immunofluorescent microscopy using serum collected from naive mice, non-protected mice vaccinated with HK-H99 at the dose of 1 × 107 cells/animal, or protected mice vaccinated with HK-ZNF2oe at the same dose. As expected, naive-serum did not recognize either wild-type H99 or ZNF2oe cryptococcal cells (Fig. 1C). HK-H99-vaccinated-serum weakly recognized antigens present in some wild-type H99 cells, with stronger recognition of ZNF2oe cells (Fig. 1B). In comparison, HK-ZNF2oe-vaccinated-serum reacted strongly with ZNF2oe cells and with some wild-type H99 cells (Fig. 1A). We noticed heterogeneity within cryptococcal populations in terms of antigen expression (Fig. 1A). Nonetheless, these observations indicate that host-recognizable antigens are present in the wild-type cells, but they are much more abundant in ZNF2oe cells.
FIG 1.
ZNF2oe cells are recognized stronger by the host serum based on immunofluorescence. (A–C) Wild-type H99 (bottom images) or ZNF2oe (upper images) cells from an overnight culture in YPD medium were probed using sera collected from mice vaccinated with heat-killed ZNF2oe cells. All images were taken at the same exposure (A), from mice vaccinated with heat-killed H99 cells (B), or from naive mice (C). (D) Quantification of the relative fluorescence intensity of cells (artificial unit based on calculation detailed in the method).
We then quantified the fluorescence intensity of H99 and the ZNF2oe cells recognized by either the HK-H99-vaccinated-serum or the HK-ZNF2oe-vaccinated-serum. Because of the heterogeneity in morphotype in the population of ZNF2oe strain where the ZNF2 gene is controlled by the constitutively active promoter of GPD1, we quantified the yeast form and the hyphal+pseudohyphal form of the ZNF2oe population separately. As shown in Fig. 1D, the average fluorescence intensity of H99 cells immune-stained with the HK-H99-vaccinated-serum was 0.24 AUs (artificial units). It is significantly lower than the fluorescence intensity of ZNF2oe cells either in the yeast form or in the hyphal form immune-stained by the same serum. This result indicates that ZNF2oe cells are better recognized by the host after exposure to heat-inactivated H99. Consistent with our expectations, the HK-ZNF2oe-vaccinated-serum showed much stronger recognition of cryptococcal cells (both wild-type and ZNF2oe). The average fluorescence intensity of H99 cells immune-stained with the HK-ZNF2oe-vaccinated-serum was approximately 4-fold stronger than the fluorescence intensity immune-stained by the HK-H99-vaccinated-serum. Similarly, the signal strength detected from the ZNF2oe strain was strongest, with the fluorescence intensity of the hyphal cells being stronger than that of yeast cells. These results indicate i) that the ZNF2oe strain expresses more host recognizable antigens and ii) that hosts vaccinated with HK-ZNF2oe cells can better recognize both wild-type and ZNF2oe strains. Notably, a higher abundance of the antigens is present in ZNF2oe cells in the yeast form even before hyphal morphogenesis takes place.
Heat-killed wild-type H99 can offer protection if the vaccination dose is increased.
Heat-killed wild-type H99 cells do not provide protection at the vaccination dose of 1 × 107 cells/animal based on the literature published by our lab and others (22, 26, 29). The results presented in Fig. 1, however, indicate that host-recognizable antigens are present in the wild-type cells, albeit at a lower abundance (∼25%) compared with ZNF2oe cells. If increased abundance of host-beneficial antigens of ZNF2oe cells contribute to better host recognition and protection, we would expect that increasing the total amount of these antigens by increasing the vaccination dose of heat-killed wild-type H99 would also offer host protection.
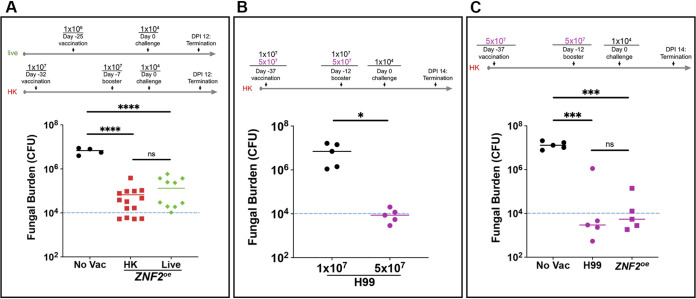
Before testing this hypothesis, we first determined whether or not lung fungal burden within 2 weeks of challenge is reflective of the vaccination effect, as using this index rather than the animal survival rate would reduce the number of animals necessary for the study and shorten the duration of the vaccination experiment. We previously demonstrated in separate animal survival experiments that vaccination with either live or heat-killed ZNF2oe cells protected animals (22). A minority of the vaccinated mice had cleared the infection by the aggressive wild-type H99 at day 60 postchallenge (DPI 60). Here, we directly compared the protective effect of the live and heat-killed ZNF2oe cells as vaccines using the same vaccination regimens as we described previously (22) with the exception that we terminated the experiment at DPI 12 (Fig. 2A). In this experiment, we dissected the lungs and measured the fungal burden based on CFU. As shown in Fig. 2A, the two groups of mice vaccinated with live or heat-killed ZNF2oe cells showed significantly lower fungal burden (1–2 log difference) than the non-vaccinated control group at this time point (Fig. 2A). The data indicate that lung fungal burden within 2 weeks of challenge can in fact be used to measure the protective effect of vaccination.
FIG 2.
Dose-dependent vaccination effect of heat-killed H99 and ZNF2oe cells. (A) Schematic representation of the vaccination regimens using live or heat-killed ZNF2oe cells. The lung fungal burdens at DPI 12 of the (5 mice) control mice without vaccination and of mice vaccinated with (10 mice) live or (10 mice) heat-killed ZNF2oe cells at the indicated doses are shown. (B) Schematic representation of the vaccination regimen using heat-killed H99 cells at two different doses. The lung fungal burdens at DPI 14 of the 5 mice vaccinated with heat-killed H99 cells at the typical dose of 1 × 107 cells/animal or the higher dose of 5 × 107 cells/animal are shown. (C) Schematic representation of the vaccination regimen using heat-killed H99 cells and heat-killed ZNF2oe cells. The lung fungal burden at DPI 14 of the 5 mice vaccinated with these heat-killed cells at the higher dose of 5 × 107 cells/animal are shown. The group without vaccination was included as the control.
To test our hypothesis that increasing the vaccination dose of heat-killed wild-type H99 can also offer protection, we compared the efficacy of HK-H99 cells for protection of animals against cryptococcosis at the typical vaccination dose of 1 × 107 cells/animal to that of a higher dose of 5 × 107 cells/animal. We chose this higher dose based on a recent study of a cryptococcal HK-fbp1Δ mutant (25), where such a high dose did not elicit obvious adverse effects in animals. As expected, animals vaccinated with HK-H99 cells at the typical dose showed an average fungal burden in the lungs approaching 1 × 107 CFU at DPI 14 (Fig. 2B). By stark contrast, animals vaccinated with HK-H99 at the higher dose had only about 1 × 104 CFU in their lungs, a drastic reduction compared to the typical vaccination dose group (>2.5 log difference) (Fig. 2B). This result demonstrates that HK-H99 cells, when used as a vaccine at this higher dose, provide significant protection based on the marked reduction in the lung fungal burden. In addition, when we directly compared the vaccination effect of HK-ZNF2oe cells and HK-H99 cells at this higher dose of 5 × 107 cells/animal, we found no significant difference between the two groups (Fig. 2C), suggesting the observed protection from the higher dose might be at or near peak level. Because the typical vaccination dose of 1 × 107 HK cells/animal can differentiate the more protective strains from the wild-type H99 and has been more frequently employed as the standard vaccination dose in the literature, we decided to use this lower dose in the subsequent experiments unless indicated otherwise.
Antigens are localized within the capsule.
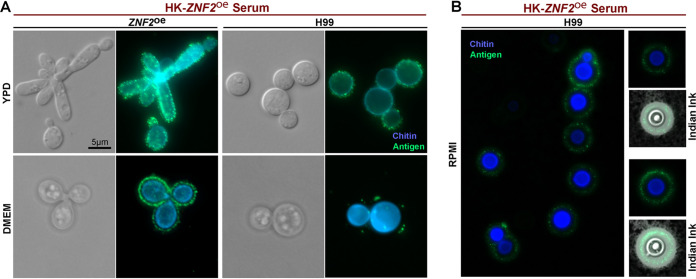
The immunofluorescence signals from cryptococcal cells immune-stained with the vaccinated serum appear to be present at the cryptococcal cell surface (Fig. 1). To determine the exact subcellular location of the recognized antigens, we co-stained H99 and ZNF2oe cells cultured overnight in YPD medium with the HK-ZNF2oe-vaccinated-serum and calcofluor white, a dye that stains chitin in the fungal cell wall. We found that the antigens (green) were located just outside of the chitin layer (blue) (Fig. 3A, upper panel), indicating that these antigens may reside in the capsule surrounding the cell wall. Capsule is a defining feature of Cryptococcus and its size varies widely depending on the culture conditions (30, 31). The capsule size of cryptococcal cells cultured in YPD medium overnight is small (this growth condition does not promote the formation of a large capsule), we cultured the H99 and ZNF2oe strains for 2 days in ambient air at 37°C in DMEM medium, a mammalian cell culture medium that promotes the formation of a larger capsule to further define the localization of these antigens. We then co-stained cells with calcofluor white and performed immunofluorescence microscopy. When compared with the cells cultured in YPD medium, both ZNF2oe cells and H99 cells cultured in DMEM medium showed clearer separation of the antigens (green) from the cell wall (blue) (Fig. 3A, lower panel) as a result of a large capsule. Similarly, a clear spatial separation of antigens from chitin was observed when cells were cultured in RPMI medium (Fig. 3B, left panel), another mammalian cell culture medium that promotes capsule production in Cryptococcus. To reveal the capsule, we negatively stained the cells with Indian ink. The capsule appears as a white halo surrounding the yeast cell due to exclusion of the ink particles (Fig. 3B). In these cells with large capsule, the antigens were clearly enriched within the capsule outside of the cell wall.
FIG 3.
The antigens are localized within the capsule and outside of the cell wall. (A) The ZNF2oe strain and the wild-type H99 were cultured in YPD medium at 30°C overnight (upper panel) or in DMEM medium at 37°C for 2 days in ambient air (lower panel). The cells were then used for immunofluorescence and probed with sera from mice vaccinated with HK-ZNF2oe cells (green). The cells were co-stained with calcofluor white to reveal chitin in the cell wall (blue). All images were taken at the same exposure. (B) The wild-type H99 cells were cultured in RPMI medium for 2 days. The cells were then used for immunofluorescence and labeled with sera from mice vaccinated with HK-ZNF2oe cells (green). The cells were co-stained with calcofluor white to reveal chitin in the cell wall (blue). Then the cells were negatively stained with Indian ink to reveal the capsule (white halo surrounding the yeast cells).
Capsule is required for host-protection offered by ZNF2oe.
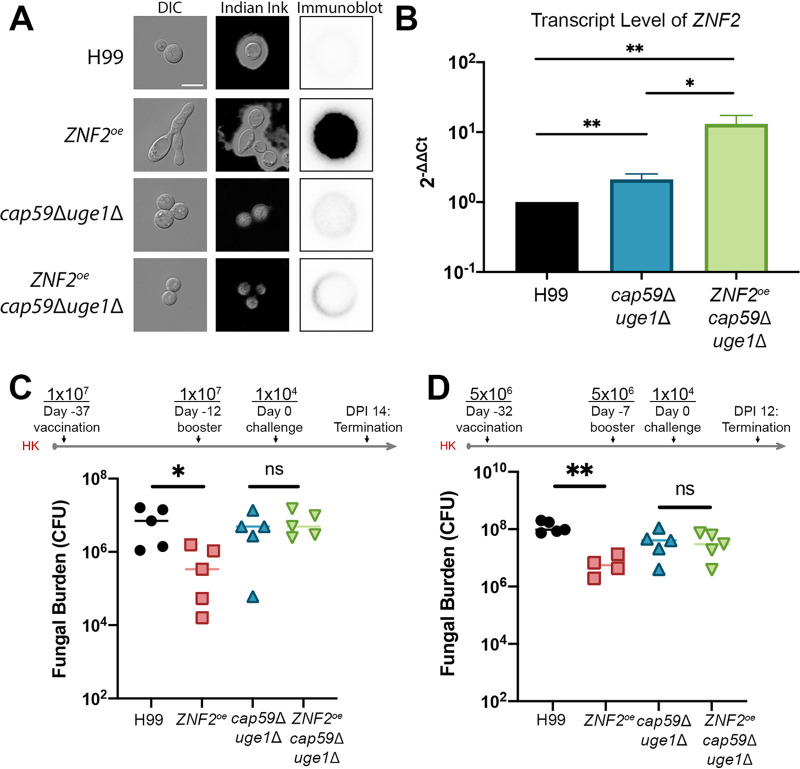
Given that the antigens are present within the capsule, we speculate that removing capsule would reduce the immunoprotection offered by ZNF2 overexpression. To test this hypothesis, we overexpressed ZNF2 in the acapsular mutant cap59Δuge1Δ that is devoid of both glucuronoxylomannan and galactoxylomannan (Fig. 4A)(32). We first confirmed that ZNF2 was indeed overexpressed in the ZNF2oe strain and the ZNF2oecap59Δuge1Δ strain by real-time PCR (Fig. 4B). Because ZNF2oecap59Δuge1Δ cells clumped together, we could not reliably use immunofluorescence to quantify the cell surface antigen level. We therefore resorted to colony immunoblot to detect only antigens released from these cells. Although this latter method is more qualitative than quantitative as growth (and death) rate, capsule shedding, and secretion rate could all influence the signal strength, we found that the antigen level released from ZNF2oecap59Δuge1Δ cells was drastically reduced comparing to ZNF2oe (Fig. 4A). We then compared the vaccination effect of HK-ZNF2oecap59Δuge1Δ and HK-cap59Δuge1Δ at the typical vaccination dose of 1 × 107 cells/animal using the same vaccination regimen as described in Fig. 2B We then measured the fungal burden in the lungs at day 14 postchallenge with live H99. Mice vaccinated with HK-H99 cells and mice vaccinated with HK-ZNF2oe cells were included as the negative control and the positive control, respectively. As expected, mice vaccinated with HK ZNF2oe cells showed a lower lung fungal burden compared to the ones vaccinated with HK H99 cells (Fig. 4C). However, we did not observe any significant difference in lung fungal burden between mice vaccinated with the HK-ZNF2oecap59Δuge1Δ cells and those vaccinated with the HK-cap59Δuge1Δ cells (Fig. 4C). To further test our hypothesis, we also vaccinated animals with HK-ZNF2oecap59Δuge1Δ cells and HK-cap59Δuge1Δ cells using a slightly different regimen. In this experiment, we halved the vaccination dose to 5 × 106 HK cells/animal and used the regimen as diagramed in Fig. 2A. Once again, we included mice vaccinated with HK-H99 cells and mice vaccinated with HK-ZNF2oe cells at the same dose as the negative control and the positive control respectively. As expected, mice vaccinated with HK ZNF2oe cells showed a much lower lung fungal burden compared to the ones vaccinated with HK H99 cells even at this low dose (Fig. 4D). The group vaccinated with HK-cap59Δuge1Δ acapsular mutant cells showed a slightly but not statistically significant reduction in fungal burden compared to the control group vaccinated with HK-H99 cells. Again, no significant difference in fungal burden was observed between mice vaccinated with the HK-cap59Δuge1Δ cells and those vaccinated with HK-ZNF2oecap59Δuge1Δ cells (Fig. 4D). Thus, both vaccination experiments indicate that ZNF2 overexpression in the acapsular mutant background offers no additional benefit to the host, indicating that the immunoprotective effect of ZNF2oe cells requires the presence of the capsule.
FIG 4.
The host protection effect caused by ZNF2 overexpression requires capsule. (A) Images of cells of wild-type H99, LW10 (ZNF2oe), NE644 (cap59Δuge1Δ), and KH35 (ZNF2oecap59Δuge1Δ) when cultured in YPD medium (1st column) and RPMI medium (2nd column with Indian ink staining); and colony immunoblot of these colonies probed with HK-ZNF2oe-vaccinated serum. (B) The relative transcript level of ZNF2 in wild-type H99, NE644 (cap59Δuge1Δ) and KH35 (ZNF2oecap59Δuge1Δ) after overnight culture in YPD. The transcript level of TEF1 of each sample was used as the internal control. (C-D) Two independent animal experiments testing the vaccination effect of heat-killed ZNF2oe cells in the wild-type background or in the acapsular mutant background using two slightly different vaccination regimens. The acapsular mutant cap59Δuge1Δ was included as a control in both experiments. The no vaccination group was included in (C) and the group vaccinated with heat-killed H99 cells was included in (D) as additional controls.
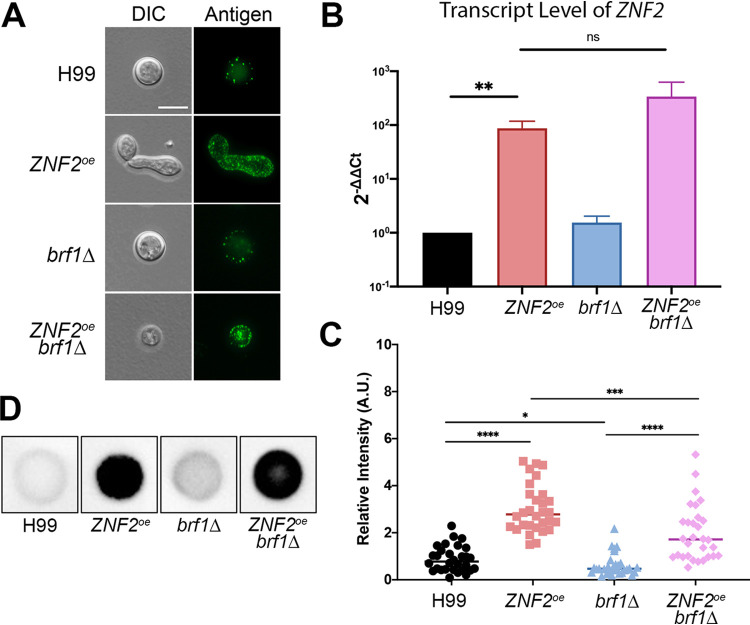
Brf1, a subunit of SWI/SNF chromatin remodeling complex, is critical for Znf2 to execute its function in morphogenesis and is important for the protective effect of ZNF2oe cells.
We showed previously that Znf2 requires the SWI/SNF chromatin-remodeling complex to execute its function in filamentation (33). When the SWI/SNF complex is not functional, for example, by the disruption of the basidiomycete-specific subunit Brf1, Znf2 cannot fully bind to some of its downstream targets important for filamentation (33). Thus, the ZNF2oe strain is filamentous and the ZNF2oebrf1Δ mutant grows mostly in the yeast form (Fig. 5A)(33), even though ZNF2 is highly expressed in both strains (Fig. 5B). To determine if Brf1 is required for the increased antigens observed in ZNF2oe cells, we compared the immunofluorescence signal of the ZNF2oebrf1Δ mutant to that of the ZNF2oe strain using HK-ZNF2oe-vaccinated-serum (Fig. 5A and C). To avoid potential complications caused by different morphotypes, we only quantified the relative fluorescent intensity of ZNF2oe cells in the yeast form for this comparison. We found that the immunofluorescence intensity of the ZNF2oebrf1Δ mutant was much lower than that of the ZNF2oe strain (Fig. 5C). The brf1Δ mutant even showed a slight reduction in immunofluorescence intensity compared to the wild-type H99 control. Qualitative colony immuno-blot analysis indicated more abundant antigens were released from ZNF2oe and ZNF2oebrf1Δ strains compared to the H99 or brf1Δ strains (Fig. 5D), consistent with the pattern of immunofluorescence detected on the cell surface of these strains (Fig. 5C).
FIG 5.
Brf1 is critical for Znf2-controlled filamentation and it contributes to the protection by ZNF2oe. (A) Immunofluorescent images of wild-type H99, ZNF2oe, brf1Δ and ZNF2oebrf1Δ when probed with serum from protected animals vaccinated with HK-ZNF2oe cells. (B) The relative transcript level of ZNF2 in wild-type H99, the ZNF2oe strain, the brf1Δ mutant, and the ZNF2oebrf1Δ strain after overnight culture in liquid YPD medium as measured by RT-PCR. The transcript level of TEF1 in each RNA sample was used as the internal control. The error bar indicates the standard derivations among three biological replicates. n.s.: non-significant. **: P < 0.01. (C) Quantification of the relative fluorescence intensity of cells probed with serum from HK-ZNF2oe cells vaccinated animals. (n =30). *: P < 0.05, ***: P < 0.001, ****: P < 0.0001. (D) Colony immunoblot of H99, ZNF2oe, brf1Δ and ZNF2oebrf1Δ probed with sera collected from mice immunized with heat-killed ZNF2oe cells.
Given that the deletion of BRF1 almost completely blocks filamentation but modestly reduces antigen abundance in the ZNF2oe strain, we choose to determine the contribution of Brf1 to the vaccination effect induced by ZNF2 overexpression. If HK ZNF2oebrf1Δ cells still provide host protection, it suggests that the filamentous morphotype per se may not be critical for eliciting host protective immunoresponses. In that case, focusing on cryptococcal factors that depend on Znf2 but not Brf1 would be a reasonable approach for future investigation. If Brf1 is important for host protection elicited by ZNF2 overexpression (i.e., HK-ZNF2oebrf1Δ vaccination is not protective), it would suggest that cryptococcal factors required for filamentation and host protection depend on both Znf2 and Brf1. As not all antigens will elicit the same magnitude or quality of host responses that can control the fast-replicating fungus (H99 in this case), either of the findings would provide direction for future investigations.
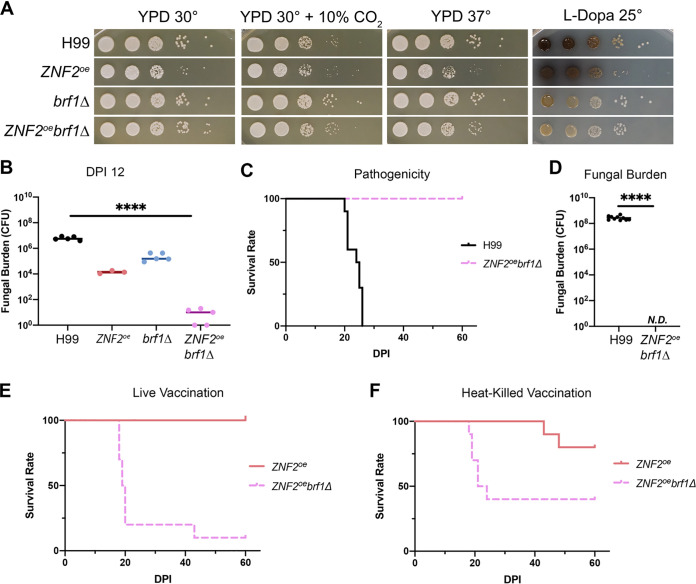
We first examined the virulence level of the ZNF2oebrf1Δ strain. Based on in vitro phenotypical analysis, the ZNF2oebrf1Δ strain and the brf1Δ mutant grew similarly well as H99 on YPD medium at 30°C, 30°C + 10% CO2, and 37°C (Fig. 6A). Lastly, the deletion of BRF1 decreased cryptococcal melanization (Fig. 6A). By contrast, the ZNF2oe strain grew slower, likely because a proportion of cells were in the filamentous form, and they only grew by apical extension and consequently did not replicate as fast as yeast cells. We previously showed that animals can eventually clear ZNF2oe cells, although this strain can persist in animals for many weeks (20, 22). Based on the in vitro growth phenotypes, we expect that the ZNF2oe strain might be the least virulent while the ZNF2oebrf1Δ strain and the brf1Δ mutant would be highly virulent. At day 12 postinoculation, mice infected with the ZNF2oe strain or the brf1Δ mutant showed 2.5 log (ZNF2oe) or 1.5 log (brf1Δ) lower fungal burden in the lungs compared to the control group infected with H99 (Fig. 6B). To our surprise, mice infected by the ZNF2oebrf1Δ strain showed almost 6 log lower fungal burden in the lungs compared to the H99 control group (Fig. 6B). This suggests that the ZNF2oebrf1Δ strain might have been completely cleared by the animals if given more time postinfection. Consistent with this idea, all mice infected by the ZNF2oebrf1Δ strain survived to DPI 60 when we terminated the experiment while all animals infected by H99 succumbed to the infection by DPI 26 (Fig. 6C). Out of all 10 surviving mice inoculated with the ZNF2oebrf1Δ strain, all had cleared the fungus by DPI 60 (Fig. 6D). In comparison, when mice become moribund with H99 infection, their lungs carried over 108 fungal cells.
FIG 6.
Brf1 is critical for the protective effect elicited by ZNF2 overexpression. (A) Phenotypical analyses of H99, ZNF2oe, brf1Δ, and ZNF2oebrf1Δ on thermo- or CO2- tolerance (YPD at 30°C, 30°C + 10% CO2, and 37°C) and melanization (L-Dopa medium at 22°C). (B) The lung fungal burden at day 12 postinoculation with live H99 (5 mice), ZNF2oe (5 mice), brf1Δ (5 mice), or ZNF2oebrf1Δ (5 mice) at the inoculum of 1 × 104 cells/animal. (C) Animal survival was monitored for 60 days after inoculation with live H99 (10 mice) or ZNF2oebrf1Δ (10 mice) at 1 × 104 cells/animal. The survival rate was plotted against the days postinoculation. (D) The lung fungal burden of the surviving mice inoculated with live ZNF2oebrf1Δ (10 mice) at the time of termination (DPI 60) compared the lung fungal burden of mice infected with live H99 (10 mice) at time of euthanization when they reached defined clinical endpoints. (E) Survival of mice vaccinated with live ZNF2oebrf1Δ (10 mice) or ZNF2oe (10 mice) after challenge with live H99. (F) Survival of mice vaccinated with heat-killed ZNF2oebrf1Δ (10 mice) or ZNF2oe (10 mice) after challenge with live H99.
Given that the ZNF2oebrf1Δ strain is avirulent, we decided to test whether this strain can serve as a live attenuated vaccine. We vaccinated the animals with the live ZNF2oebrf1Δ strain and live ZNF2oe strain with the dose of 1 × 106 cells/animal at day -25 and then challenged the mice with live H99 at 1 × 104 cells/animal. We used the same regimen previously to test the ZNF2oe strain as a live attenuated vaccine (22). The median survival of the mice vaccinated with live ZNF2oebrf1Δ strain (19.5 days) was similar to that of the unvaccinated control (Fig. 6C and E). We also vaccinated animals with the HK-ZNF2oebrf1Δ strain with the typical dose of 1 × 107 cells/animal at day −32 and day −7 and challenged the mice with live H99 at 1 × 104 cells/animal. In that case, 60% of animals vaccinated with HK-ZNF2oebrf1Δ cells died before DPI 23 (Fig. 6F). Thus, the deletion of BRF1, which almost abolishes filamentation driven by Znf2, also significantly reduces the ability of the ZNF2oe cells to protect the host.
DISCUSSION
Morphogenesis profoundly shapes Cryptococcus interaction with various hosts (12, 19). Although the inverse relationship between Cryptococcus filamentation and its virulence in mammalian hosts have been observed since the 1960s and 1970s (34–42), the molecular bases for attenuated virulence of the filamentous form are unknown. We have previously discovered that the transcription factor Znf2 controls cryptococcal yeast-to-hypha transition and is a potent anti-virulence factor (20, 21). Unlike a typical avirulent strain (e.g., an acapsular mutant or a temperature-sensitive mutant) that is rapidly cleared by the host, the ZNF2oe cells can persist in the host for many weeks. The virulence attenuation of the ZNF2oe strain and its host protective effect are most likely caused by the ability of ZNF2oe cells to elicit strong protective host responses, which was confirmed in our previous study (22). Consistently, the ZNF2oe cells, either in the live-attenuated form or the inactivated heat-killed form, offer host protection as a vaccine (22). These findings provide a platform to dissect the relationship between the host protection and cryptococcal filamentation.
As morphological changes require dramatic remodeling of the cell envelope, overexpression of ZNF2 not only alters cell shape but also cell surface factors. We have shown here that antigens are more abundantly present in ZNF2oe cells and they reside in the capsule. Cryptococcal capsule is composed of mostly high molecular weight polysaccharides glucuronoxylomannan (90–95%) and galactoxylomannan (5-8%) with some mannoproteins (MPs:1–2%) (18). As Znf2 regulon is highly enriched with secretory proteins and mannoproteins (20, 33, 43) and the pattern of antigen localization detected by immunofluorescence is suggestive of minor components in capsule, we hypothesize that the antigens are likely mannoproteins. However, it is possible that other predicted cytosolic proteins, such as heat shock proteins or abundant housekeeping enzymes like Gpd1, could end up in the cell wall and capsule, as these proteins have been detected in the cell wall fractions or culture supernatants in other studies. Various proteins carried in extracellular cellular vesicles could also potentially end up in the capsule. Nonetheless, mannoproteins could be highly antigenic and are considered to be the main cell surface components recognized by the anti-cryptococcal cell-mediated immune response in mice (45). Although mannoproteins are minor components of the capsule, they are the primary components recognized by the anti-cryptococcal cell-mediated immune response in mice (45, 46). For example, vaccination with recombinant GPI-anchored mannoproteins and chitin deacetylases Cda1, Cda2, and Cda3 together with glucan particles affords a significant survival advantage to mice against cryptococcosis (47). The mannoprotein Krp1 has a strong serological reactivity in human patients with cryptococcosis even though deletion of the gene does not affect cryptococcal pathogenicity in animals (48). We postulate that increased expression and exposure of certain mannoproteins in ZNF2oe cells likely contribute to the strain’s vaccination effect. Consistent with this idea, we found that deletion of BRF1, which is required for Znf2 to drive filamentation (33), reduced antigen level in ZNF2oe cells by 37%. The ZNF2oebrf1Δ strain, although not capable of causing fatal infection itself and cleared by the host relatively quickly, showed much reduced level of protection to the host in the heat-inactivated form when compared to HK-ZNF2oe cells. This information indicates that some of Znf2’s downstream targets that are dependent on Brf1 are important for host protection. Our data also support previous observations that there is no correlation between strains’ virulence potential and their vaccination effect (23, 24).
As mentioned in the introduction, three cryptococcal mutants in the heat-killed inactivated form offer remarkable host protection: the morphology strain ZNF2oe, the chitosan deficient mutant cda1-3Δ, and the ubiquitination E3 ligase mutant fbp1Δ (22, 25, 26). Among these three strains, the fbp1Δ mutant appears to require a higher vaccination dose for host protection (49) although these three strains have never been compared directly for their protective effect. It is also not known if these strains share host protection mechanisms. We found that Znf2 does not regulate FBP1 expression. Znf2 might affect chitin deacetylase activity as the transcript levels of the chitin deacetylase genes CDA2 and CDA3 are slightly reduced but CDA1 transcript level is modestly higher in the ZNF2oe strain (Fig. S1A in the supplemental material). The chitin deacetylases are the enzymes that convert chitin (polymer of N-acetylglucosamine) to chitosan (polymer of glucosamine). Although it is unlikely, it is possible that immunoprotection offered by ZNF2oe cells and chitosan deficient cda1-3Δ cells could occur through a similar mechanism. However, based on Eosin Y staining of chitosan (49), we found no apparent difference between ZNF2oe cells and wild-type H99 cells cultured in YPD medium (Fig. S1B), the growth condition used to prepare heat-inactivated cells for vaccination. This is consistent with Upadhya’s report that chitosan level in three single CDA deletion strains is similar to H99 when cultured in YPD (50). Only the cda1Δ mutant shows 2.5-fold reduction in chitosan level when cultured in a host-physiological condition for 5 days (50). Thus, HK-ZNF2oe cells and HK-cda1-3Δ cells likely elicit protective host responses through distinct mechanisms.
The ZNF2oe strain shows no obvious deficiency in chitosan. (A) The relative transcript level of CDA1, CDA2, and CDA3 in three biological replicates of wild-type H99 and the ZNF2oe strain after overnight culture in liquid YPD medium as measured by RT-PCR. The transcript level of TEF1 in each RNA sample was used as the internal control. The error bars indicate the standard derivations among three technical replicates. Unpaired t-tests were performed. n.s.: non-significant. *: P < 0.05. **: P < 0.01. ***: P = 0.0002. ****: P < 0.0001 (B) The wild-type H99 and the ZNF2oe cells after overnight culture in liquid YPD medium were stained with either calcofluor white or Eosin Y. Download FIG S1, TIF file, 2.4 MB (2.4MB, tif) .
Copyright © 2022 Lin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The effect of capsule on vaccination is likely to be complicated. On one hand, capsule acts as a mask covering the highly antigenic cell wall and helps the fungus evade the immune system (15, 18). On the other hand, capsule contains immunogenic and protective factors such as mannoproteins. Consistently, acapsular mutants, although easily recognized by the host due to exposed cell wall, are not protective (51) (Fig. 4). A recent study showed that immunoprotection elicited by the live sterylglucosidase sgl1Δ mutant, which accumulates sterylglucosides, requires capsule (52). Here we showed that immunoprotection offered by heat-killed ZNF2oe cells also requires capsule. It would be interesting to examine whether capsule is also required for host protection elecited by inactivated cda1-3Δ and fbp1Δ mutants.
Developing effective cryptococcal vaccines remains one of the most important and challenging research areas that could help prevent and/or manage the deadly cryptococcal infections (53). Many questions remain in terms of the molecular bases for the vaccination effect of any of the protective cryptococcal strains. It has been a challenging but critical task for the cryptococal research community to identify and characterize immunogenic factors (47, 54–59). Some immunogenic factors might be beneficial while others could be deleterious for the host. Thus, distinguishing the “good” immunogens from the “bad” would also be critical. Collectively, these efforts can help develop vaccines with multiple epitopes, offer a quality control for whole cell vaccines, and improve our understanding of cryptococcus-host interaction.
MATERIALS AND METHODS
Ethical statements.
This study was performed according to the guidelines of NIH and the University of Georgia Institutional Animal Care and Use Committee (IACUC). The animal models and procedures used have been approved by the IACUC (AUP protocol numbers: A2017 08–023 and A2020 06–015).
Murine model of cryptococcosis.
(i) Virulence. Female A/J mice of 8–10 weeks old were purchased from the Jackson Laboratory (Bar Harbor, Maine). In survival assays, 10 mice were assigned to each group. In fungal burden assays at the specified time point, 5 mice were assigned to each group. Cryptococcal strains were inoculated in 3 ml of YPD medium with the initial inoculum of approximately 106 cells/ml. Cells were cultured at 30°C with shaking at 220 rpm for 15 h. Cells were washed with sterile saline 3 times and adjusted to the final concentration of 2 × 105 cells/ml. Mice were sedated with Ketamine and Xylazine via intraperitoneal injection and then inoculated intranasally with 50 μl fungal cell suspension (1 × 104 cells per animal) as previously described (60–62). After infection, animals were monitored daily for disease progression, including weight loss, gait changes, labored breathing, or fur ruffling. For fungal burden measurement, animals were euthanized on the designated day postinfection. For the survival experiment, mice were euthanized when they reached the clinical endpoint. All the surviving animals were terminated at day 60 postinfection.
(ii) Vaccination. To prepare fungal cells used for vaccination, each strain was inoculated in 3 ml of YPD media with 106 cells/ml. Cells were cultured at 30°C with shaking at 220 rpm for 15 h. The fungal cells were washed with sterile saline 3 times and adjusted to the final concentration of cell suspension with saline to 2 × 107 cell/ml (live vaccination with 1 × 106 cells per animal), 2 × 108 cell/ml (heat-killed/HK vaccination at the typical dose of 1 × 107 cells per animal), or 10 × 108 cell/ml (HK vaccination at the higher dose of 5 × 107 cells per animal). For inactivation of cells for vaccination, the cell suspension was heated at 95°C for 20–25 min. Mice were sedated with Ketamine and Xylazine via intraperitoneal injection and then inoculated intranasally with 50 μl cell suspension using the same procedures as previously described (22). Three vaccination regimens were used in this study. (i) For live cell vaccination, mice were vaccinated once with a live strain at day −25 as we previously described (22). (ii) In one regimen of vaccination with heat-inactivated cells, mice were vaccinated with heat-killed cells twice, at day −32 and at day −7 (22). (iii) In another regimen of vaccination with heat-inactivated cells, mice were vaccinated with heat-killed cells twice, at day -37 and at day −12. For challenge, live H99 cells with the initial inoculum of 105 were cultured in 3 ml of YPD media at 30°C with shaking at 220 rpm for 15 h. Cells were washed with sterile saline 3 times and adjusted to the final concentration of 2 × 105 cell/ml (1 × 104 cells/animal). The infection process was the same as previously described.
(iii) Serum collection. To collect serum, mice were vaccinated and challenged with live H99 using the protocols as described above in the vaccination section. At DPI 12, sera of euthanized mice were collected through cardiovascular puncture. The serum of naive uninfected mice was collected as a control.
(iv) Fungal burden analysis. At the indicated time of euthanization or at the termination of the survival experiments (DPI 60), the lungs, kidneys, and brains of the euthanized mice were dissected. The dissected organs were homogenized in 2 ml cold PBS buffer using an IKA-T18 homogenizer with the same setting for each type of organ as we described previously (22, 60). The tissue suspensions were serially diluted (10×), plated onto YNB agar medium, and incubated at 30°C for 2 days such that the colonies became visible to count CFU.
Strains and culture conditions.
All strains were stocked in 15% glycerol and stored at -80°C. Fresh cultures were used for experiments. Fungal strains were maintained on yeast peptone dextrose (YPD) at 30°C unless indicated otherwise. DEME and RPMI 1640 media buffered with MOPS at pH 7.0 was used for growing Cryptococcus at 37°C in ambient air for some experiments to increase capsule production.
Colony immuno-blot assay.
Strains were grown in YPD at 30°C with shaking at 220 rpm for 16 h. Approximately 1 × 107 cells were washed with sterile ddH2O and resuspended in 1 ml of sterile ddH2O. 5 μl of cells were spotted onto YPD and grown at 30°C for 24 h. After 24 h, nitrocellulose membrane was placed on top of the colonies, and the cells were allowed to grow for an additional 72 h at 30°C. Then the nitrocellulose membranes were removed, washed thoroughly with TBST pH 7.4 to remove adherent cells, and blocked with 1× TBS 1% Casein Blocker (Bio Rad). A 1:1000 dilution of serum from HK-ZNF2oe vaccinated animals in blocking buffer was used as a primary antibody, and goat anti-mouse IgG conjugated to HRP was used as secondary antibody. Blots were developed with SuperSignal West Pico PLUS chemiluminescent substrate (Thermo Scientific). Images were obtained with ChemStudio Imaging system (Analytik Jena).
Generation of mutants.
The capsule mutant strain cap59::NEO uge1::NAT was graciously gifted from Dr. Guilhem Janbon (Institut Pasteur, France)(63). To create the ZNF2oebrf1Δ strain, we deleted the BRF1 ORF from the ZNF2oe strain using a CRISPR-Cas9 transient expression system TRACE (64). Transformants were validated using stability testing, diagnostic PCR screening for the absent of the BRF1 open reading frame. To overexpress ZNF2 in the acapsular mutant, the PGPD1-ZNF2-HYG overexpression construct was transformed into the capsule mutant strain cap59::NEO uge1::NAT using TRACE (64). Transformants were validated using stability testing, diagnostic PCR screening for the presence of the ZNF2oe construct in the intergenic region Safe Haven 2 or SH2 (65) using the same procedures as we described previously (66). The overexpression of ZNF2 was confirmed by quantitative real-time PCR. All primers used to generate mutant strains are listed in the Table S1 in the supplemental material.
Primers used in this study. Download Table S1, DOCX file, 0.02 MB (17.1KB, docx) .
Copyright © 2022 Lin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA extraction and quantitative real-time PCR.
To examine the transcript level of the indicated genes (ZNF2, CDA1, CDA2, CDA3), the wild-type H99, LW10 (ZNF2oe ectopic), NE644 (cap59Δuge1Δ), KH35 (ZNF2oe at SH2 in cap59Δuge1Δ), and JL131 (brf1Δ), and JL392 (ZNF2oebrf1Δ) were grown in triplicates in 3 ml of YPD for 16 h at 30°C shaking at 220 rpm. Cells were collected by centrifugation and were snap-frozen in liquid nitrogen. 1.75 ml of sterile glass beads were added to each 15 ml falcon tube and cells were then freeze-dried in a lyophilizer at −80°C and 0.008 mBar for 48 h (Labconco corp., Kansas City, MO). Freeze-dried cells were then mechanically broken into fine powder by hand shaking for 2 min. RNA was then extracted from these cells using the PureLink RNA minikit according to the instructions from the manufacturer (Life Technologies, Carlsbad, CA). RNA quality was examined by gel electrophoresis. RNA concentrations were quantified using Nanodrop 2000c (Thermo Fisher Scientific, Waltham, MA). Each RNA sample (10 μg) was treated with DNase (TURBO DNA-freeTM) according to the manufacture’s instruction and potential DNA contamination was examined with diagnostic PCR. Clean DNase-treated RNA samples were then used as templates in the synthesis of the first strand cDNA using GoScriptTM Reverse Transcription System (Promega) per the instruction from the manufacture. The resulting cDNA products were diluted to 10 ng/μl and 2 μl was used as template for quantitative real-time PCR (qRT-PCR). qRT-PCR was performed using the SYBR FAST qPCR master mix (KAPA Biosystems, Wilmington, MA) on a RealPlex2 instrument (Eppendorf, Hamburg, Germany). The housekeeping gene TEF1 was used as the internal control for each sample to normalize the gene transcript level as we described previously (67). The relative levels of transcripts were quantified using the ΔΔCt method as we described previously (20, 68). We used one-way ANOVA for multiple comparisons and P values ≤ 0.05 were considered statistically significant. Primers for qRT-PCRs are included in the Table S1 in the supplemental material.
Immuno-fluorescence staining.
Cryptococcal cultures were washed twice with sterile ddH2O and 2 × 107 cells/ml were enumerated using hemocytometer and OD600 analysis. 1 ml of cells were aliquoted into 1.75 ml Eppendorf tubes and cells were collected by centrifugation at 11,363 × g for 1 min. Next, cells were resuspended in 1 ml of 4% formaldehyde and fixed for 5 min at 22°C. Fixed cells were then washed twice with 1 ml 1× PBS pH 7.4. Cells were resuspended and blocked in 1 ml of 1% bovine serum albumin (BSA) in 1×x PBS pH 7.4 at 22°C for 1 h or at 4°C overnight. Cells were then washed twice with 1× PBS pH 7.4 and resuspended in a 1:10 dilution of HK-vaccination animal serum and 1× PBS pH 7.4 for a total volume of 100 μl. Cells were incubated at 22°C for 1 h or at 4°C overnight. Next, cells were washed twice with 1 ml of 1× PBS pH 7.4. Cells were then resuspended in a 1:200 dilution of goat IgG IgM anti-mouse secondary antibody conjugated to Alexa 488 and 1× PBS pH 7.4 for a total volume of 200 μl and incubated at 22°C for 1 h or at 4°C overnight. Following that, cells were washed twice with 1× PBS pH 7.4 and resuspended in 100 μl of 1× PBS pH 7.4 and directly imaged using fluorescence microscopy. Alternatively, these cells were co-stained with calcofluor white or Indian Ink and then imaged.
Calcofluor white co-staining.
Immunostained cells were collected by centrifugation at 11,363 × g for 1 min and then resuspended in 1 ml of 1× PBS pH 7.4 containing 1 μg/ml calcofluor white and incubated for 2 min at 22°C. Cells were then washed twice with 1 ml 1× PBS pH 7.4. Finally, stained cells were resuspended in 100 μl of 1× PBS pH 7.4 and directly imaged for fluorescence microscopy.
Eosin Y staining.
Cells were grown in YPD, pelleted, and washed twice with 1 ml McIlvaine’s buffer (0.2 M Na2HPO4 and 0.1 M citric acid [pH 6.0]). The pellet was resuspended in 500 μl McIlvaine’s buffer and stained with 30 μl eosin Y (5 mg/ml stock; Sigma, St. Louis, MO). Cells were incubated at 22°C in the dark for 10 min. Excess dye was washed twice with McIlvaine’s buffer and cells were resuspended in 500 μl McIlvaine’s buffer.
India ink co-staining.
Immunostained cells were collected by centrifugation at 11,363 × g for 1 min and resuspended in 100 μl of 1× PBS pH 7.4. Ten μl of immunostained cells were then added to a new 1.75 ml Eppendorf microcentrifuge tube. One to two μl of India ink was then added to the 10 μl cell suspension and mixed by pipetting up and down. Finally, 6–10 μl of cells were added to a glass slide and imaged for fluorescence microscopy.
Microscopy.
Regular light microscopy was performed using the Olympus CX41 microscope (Olympus Life Sciences, Tokyo, Japan). Colony images were photographed with a SZX16 stereoscope (Olympus Life Sciences, Tokyo, Japan). Images were acquired with an AxioCam MRm camera and processed with the software Zen pro.
Fluorescent microscopy was performed using a Zeiss Imager M2 microscope (Zeiss, Oberkochen, Germany). Images were acquired with an AxioCam MRm camera and processed with the software Zen pro. The fluorescence intensity of 50 individual cells from each strain imaged at ×63 magnification was quantified using ZEN 2.6 Blue edition software (Zeiss, Oberkochen, Germany). Fluorescence intensity was quantified using the Zen ‘Histo definition’ quantification software application. Each cell and background were selected using the circular selection tool and the average fluorescence intensity within that circle was recorded. The fluorescence intensity of the background around each cell was measured and served as a blank. The fluorescence intensity of each cell was normalized by subtracting the fluorescence intensity of the cell’s associated blank.
Statistical analysis.
Statistical significance of the survival data between different groups was assessed by the Gehan-Breslow Wilconxon test. The one-way ANOVA tests were used in the fungal burden studies. All statistical analyses were performed using the GraphPad Prism version 8.11, with P values lower than 0.05 considered statistically significant.
Data availability statement.
All of the data supporting this study are presented herein, and the reported fungal strains generated for this study are available on request.
ACKNOWLEDGMENTS
We thank Dr. Guilhem Janbon at Institut Pasteur for the kind gift of capsule mutants used in this study. We thank all Lin lab members for their helpful suggestions. This work was supported by National Institutes of Allergy and Infectious Diseases (https://www.niaid.nih.gov/) (R01AI140719 and R21AI150641 to XL). The funder had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Conceived and designed the experiments: J.L., T.P., K.H., N.G., Y.F., and X.L. Performed the experiments: J.L., T.P., K.H., N.G., Y.F., and X.L. Analyzed the data: J.L., T.P., K.H., N.G., Y.F., and X.L. Wrote the manuscript: X.L. Edited the manuscript: J.L., T.P., K.H., and X.L.
We declare no competing interests.
Contributor Information
Xiaorong Lin, Email: xiaorong.lin@uga.edu.
Anuradha Chowdhary, Vallabhbhai Patel Chest Institute.
REFERENCES
- 1.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 3.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC. 2010. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaiwarith R, Vongsanim S, Supparatpinyo K. 2014. Cryptococcal meningitis in HIV-infected patients at Chiang Mai University Hospital: a retrospective study. Southeast Asian J Trop Med Public Health 45:636–646. [PubMed] [Google Scholar]
- 5.Perfect JR, Bicanic T. 2015. Cryptococcosis diagnosis and treatment: What do we know now. Fungal Genet Biol 78:49–54. doi: 10.1016/j.fgb.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong-James D, Meintjes G, Brown GD. 2014. A neglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol 22:120–127. doi: 10.1016/j.tim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Idnurm A, Lin X. 2015. Rising to the challenge of multiple Cryptococcus species and the diseases they cause. Fungal Genet Biol 78:1–6. doi: 10.1016/j.fgb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaskell KM, Rothe C, Gnanadurai R, Goodson P, Jassi C, Heyderman RS, Allain TJ, Harrison TS, Lalloo DG, Sloan DJ, Feasey NA. 2014. A prospective study of mortality from cryptococcal meningitis following treatment induction with 1200 mg oral fluconazole in Blantyre, Malawi. PLoS One 9:e110285. doi: 10.1371/journal.pone.0110285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothe C, Sloan DJ, Goodson P, Chikafa J, Mukaka M, Denis B, Harrison T, van Oosterhout JJ, Heyderman RS, Lalloo DG, Allain T, Feasey NA. 2013. A prospective longitudinal study of the clinical outcomes from cryptococcal meningitis following treatment induction with 800 mg oral fluconazole in Blantyre, Malawi. PLoS One 8:e67311. doi: 10.1371/journal.pone.0067311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin J, Idnurm A, Lin X. 2015. Morphology and its underlying genetic regulation impact the interaction between Cryptococcus neoformans and its hosts. Med Mycol 53:493–504. doi: 10.1093/mmy/myv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Lin J, Fan Y, Lin X. 2019. Life cycle of Cryptococcus neoformans. Annu Rev Microbiol 73:17–42. doi: 10.1146/annurev-micro-020518-120210. [DOI] [PubMed] [Google Scholar]
- 13.Hull CM, Heitman J. 2002. Genetics of Cryptococcus neoformans. Annu Rev Genet 36:557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- 14.Walsh NM, Botts MR, McDermott AJ, Ortiz SC, Wuthrich M, Klein B, Hull CM. 2019. Infectious particle identity determines dissemination and disease outcome for the inhaled human fungal pathogen Cryptococcus. PLoS Pathog 15:e1007777. doi: 10.1371/journal.ppat.1007777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM. 2009. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot Cell 8:595–605. doi: 10.1128/EC.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magditch DA, Liu TB, Xue C, Idnurm A. 2012. DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathog 8:e1002936. doi: 10.1371/journal.ppat.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu MS, Liporagi-Lopes LC, Dos Santos SRJ, Tenor JL, Perfect JR, Cuomo CA, Casadevall A. 2021. Amoeba predation of Cryptococcus neoformans results in pleiotropic changes to traits associated with virulence. mBio 12. doi: 10.1128/mBio.00567-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Meara TR, Alspaugh JA. 2012. The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev 25:387–408. doi: 10.1128/CMR.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin X. 2009. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect Genet Evol 9:401–416. doi: 10.1016/j.meegid.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Zhai B, Lin X. 2012. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog 8:e1002765. doi: 10.1371/journal.ppat.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet 6:e1000953. doi: 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhai B, Wozniak KL, Masso-Silva J, Upadhyay S, Hole C, Rivera A, Wormley FL, Jr, Lin X. 2015. Development of protective inflammation and cell-mediated immunity against Cryptococcus neoformans after exposure to hyphal mutants. mBio 6:e01433-15. doi: 10.1128/mBio.01433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromtling RA, Kaplan AM, Shadomy HJ. 1983. Immunization of mice with stable, acapsular, yeast-like mutants of Cryptococcus neoformans. Sabouraudia 21:113–119. doi: 10.1080/00362178385380181. [DOI] [PubMed] [Google Scholar]
- 24.Wormley FL, Jr, Cox GM, Perfect JR. 2005. Evaluation of host immune responses to pulmonary cryptococcosis using a temperature-sensitive C. neoformans calcineurin A mutant strain. Microb Pathog 38:113–123. doi: 10.1016/j.micpath.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Wang K, Masso-Silva JA, Rivera A, Xue C. 2019. A heat-killed Cryptococcus mutant strain induces host protection against multiple invasive mycoses in a murine vaccine model. mBio 10:02145-19. doi: 10.1128/mBio.02145-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upadhya R, Lam WC, Maybruck B, Specht CA, Levitz SM, Lodge JK. 2016. Induction of protective immunity to cryptococcal infection in mice by a heat-killed, chitosan-deficient strain of Cryptococcus neoformans. mBio 7:00547-16. doi: 10.1128/mBio.00547-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. 2015. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 28.Lanzavecchia A. 1985. Antigen-specific interaction between T and B cells. Nature 314:537–539. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- 29.Wozniak KL, Ravi S, Macias S, Young ML, Olszewski MA, Steele C, Wormley FL. 2009. Insights into the mechanisms of protective immunity against Cryptococcus neoformans infection using a mouse model of pulmonary cryptococcosis. PLoS One 4:e6854. doi: 10.1371/journal.pone.0006854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frases S, Pontes B, Nimrichter L, Rodrigues ML, Viana NB, Casadevall A. 2009. The elastic properties of the Cryptococcus neoformans capsule. Biophys J 97:937–945. doi: 10.1016/j.bpj.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown JCS, Nelson J, VanderSluis B, Deshpande R, Butts A, Kagan S, Polacheck I, Krysan DJ, Myers CL, Madhani HD. 2014. Unraveling the biology of a fungal meningitis pathogen using chemical genetics. Cell 159:1168–1187. doi: 10.1016/j.cell.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moyrand F, Fontaine T, Janbon G. 2007. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol 64:771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- 33.Lin J, Zhao Y, Ferraro AR, Yang E, Lewis ZA, Lin X. 2019. Transcription factor Znf2 coordinates with the chromatin remodeling SWI/SNF complex to regulate cryptococcal cellular differentiation. Commun Biol 2:412. doi: 10.1038/s42003-019-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmer BL, Hempel HO, Goodman NL. 1983. Pathogenicity of the hyphae of Filobasidiella neoformans. Mycopathologia 81:107–110. doi: 10.1007/BF00436987. [DOI] [PubMed] [Google Scholar]
- 35.Shadomy HJ, Utz JP. 1966. Preliminary studies on a hypha-forming mutant of Cryptococcus neoformans. Mycologia 58:383–390. doi: 10.2307/3756912. [DOI] [PubMed] [Google Scholar]
- 36.Shadomy HJ, Lurie HI. 1971. Histopathological observations in experimental cryptococcosis caused by a hypha-producing strain of Cryptococcus neoformans (Coward strain) in mice. Sabouraudia 9:6–9. doi: 10.1080/00362177185190031. [DOI] [PubMed] [Google Scholar]
- 37.Lurie HI, Shadomy HJ. 1971. Morphological variations of a hypha-forming strain of Cryptococcus neoformans (Coward strain) in tissues of mice. Sabouraudia 9:10–14. doi: 10.1080/00362177185190041. [DOI] [PubMed] [Google Scholar]
- 38.Neilson JB, Ivey MH, Bulmer GS. 1978. Cryptococcus neoformans: pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect Immun 20:262–266. doi: 10.1128/iai.20.1.262-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neilson JB, Fromtling RA, Bulmer GS. 1981. Pseudohyphal forms of Cryptococcus neoformans: decreased survival in vivo. Mycopathologia 73:57–59. doi: 10.1007/BF00443015. [DOI] [PubMed] [Google Scholar]
- 40.Fromtling RA, Blackstock R, Hall NK, Bulmer GS. 1979. Kinetics of lymphocyte transformation in mice immunized with viable avirulent forms of Cryptococcus neoformans. Infect Immun 24:449–453. doi: 10.1128/iai.24.2.449-453.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fromtling RA, Blackstock R, Hall NK, Bulmer GS. 1979. Immunization of mice with an avirulent pseudohyphal form of Cryptococcus neoformans. Mycopathologia 68:179–181. doi: 10.1007/BF00578527. [DOI] [PubMed] [Google Scholar]
- 42.Fromtling RA, Blackstock R, Bulmer GS. 1980. Immunization and passive transfer in immunity in murine cryptococcosis, p 122–124. In Kuttin ES, Baum GL (ed), Human and animal mycology: proceedings of the VII Congress of ISHAM. Excerpta Medica; distributors for the USA, Elsevier North-Holland, Amsterdam. [Google Scholar]
- 43.Wang L, Tian X, Gyawali R, Lin X. 2013. Fungal adhesion protein guides community behaviors and autoinduction in a paracrine manner. Proc Natl Acad Sci USA 110:11571–11576. doi: 10.1073/pnas.1308173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reference deleted.
- 45.Murphy JW, Mosley RL, Cherniak R, Reyes GH, Kozel TR, Reiss E. 1988. Serological, electrophoretic, and biological properties of Cryptococcus neoformans antigens. Infect Immun 56:424–431. doi: 10.1128/iai.56.2.424-431.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietrella D, Cherniak R, Strappini C, Perito S, Mosci P, Bistoni F, Vecchiarelli A. 2001. Role of mannoprotein in induction and regulation of immunity to Cryptococcus neoformans. Infect Immun 69:2808–2814. doi: 10.1128/IAI.69.5.2808-2814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Specht CA, Lee CK, Huang H, Hester MM, Liu J, Luckie BA, Torres Santana MA, Mirza Z, Khoshkenar P, Abraham A, Shen ZT, Lodge JK, Akalin A, Homan J, Ostroff GR, Levitz SM. 2017. Vaccination with recombinant Cryptococcus proteins in glucan particles protects mice against cryptococcosis in a manner dependent upon mouse strain and cryptococcal species. mBio 8:eo1872-17. doi: 10.1128/mBio.01872-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reuwsaat JCV, Motta H, Garcia AWA, Vasconcelos CB, Marques BM, Oliveira NK, Rodrigues J, Ferrareze PAG, Frases S, Lopes W, Barcellos VA, Squizani ED, Horta JA, Schrank A, Rodrigues ML, Staats CC, Vainstein MH, Kmetzsch L. 2018. A predicted mannoprotein participates in Cryptococcus gattii capsular structure. mSphere 3:e00023-18. doi: 10.1128/mSphere.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker LG, Specht CA, Donlin MJ, Lodge JK. 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell 6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Upadhya R, Baker LG, Lam WC, Specht CA, Donlin MJ, Lodge JK. 2018. Cryptococcus neoformans Cda1 and its chitin deacetylase activity are required for fungal pathogenesis. mBio 9:e02087-18. doi: 10.1128/mBio.02087-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Decote-Ricardo D, LaRocque-de-Freitas IF, Rocha JDB, Nascimento DO, Nunes MP, Morrot A, Freire-de-Lima L, Previato JO, Mendonca-Previato L, Freire-de-Lima CG. 2019. Immunomodulatory role of capsular polysaccharides constituents of Cryptococcus neoformans. Front Med (Lausanne) 6:129. doi: 10.3389/fmed.2019.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colombo AC, Rella A, Normile T, Joffe LS, Tavares PM, de S Araújo GR, Frases S, Orner EP, Farnoud AM, Fries BC, Sheridan B, Nimrichter L, Rodrigues ML, Del Poeta M. 2019. Cryptococcus neoformans glucuronoxylomannan and sterylglucoside are required for host protection in an animal vaccination model. mBio 10:e02909-18. doi: 10.1128/mBio.02909-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caballero Van Dyke MC, Wormley FL, Jr, 2018. A Call to arms: quest for a cryptococcal vaccine. Trends Microbiol 26:436–446. doi: 10.1016/j.tim.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang H, Ostroff GR, Lee CK, Specht CA, Levitz SM. 2013. Characterization and optimization of the glucan particle-based vaccine platform. Clin Vaccine Immunol 20:1585–1591. doi: 10.1128/CVI.00463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Specht CA, Lee CK, Huang H, Tipper DJ, Shen ZT, Lodge JK, Leszyk J, Ostroff GR, Levitz SM. 2015. Protection against experimental cryptococcosis following vaccination with glucan particles containing Cryptococcus alkaline extracts. mBio 6:01905-15. doi: 10.1128/mBio.01905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaturvedi AK, Weintraub ST, Lopez-Ribot JL, Wormley FL, Jr, 2013. Identification and characterization of Cryptococcus neoformans protein fractions that induce protective immune responses. Proteomics 13:3429–3441. doi: 10.1002/pmic.201300213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen LC, Goldman DL, Doering TL, Pirofski L, Casadevall A. 1999. Antibody response to Cryptococcus neoformans proteins in rodents and humans. Infect Immun 67:2218–2224. doi: 10.1128/IAI.67.5.2218-2224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy JW, Schafer F, Casadevall A, Adesina A. 1998. Antigen-induced protective and nonprotective cell-mediated immune components against Cryptococcus neoformans. Infect Immun 66:2632–2639. doi: 10.1128/IAI.66.6.2632-2639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young M, Macias S, Thomas D, Wormley FL, Jr, 2009. A proteomic-based approach for the identification of immunodominant Cryptococcus neoformans proteins. Proteomics 9:2578–2588. doi: 10.1002/pmic.200800713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhai B, Wu C, Wang L, Sachs MS, Lin X. 2012. The antidepressant sertraline provides a promising therapeutic option for neurotropic cryptococcal infections. Antimicrob Agents Chemother 56:3758–3766. doi: 10.1128/AAC.00212-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhai B, Zhu P, Foyle D, Upadhyay S, Idnurm A, Lin X. 2013. Congenic strains of the filamentous form of Cryptococcus neoformans for studies of fungal morphogenesis and virulence. Infect Immun 81:2626–2637. doi: 10.1128/IAI.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Y, Wang Y, Upadhyay S, Xue C, Lin X. 2020. Activation of Meiotic Genes Mediates Ploidy Reduction during Cryptococcal Infection. Curr Biol 30:1387–1396. doi: 10.1016/j.cub.2020.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moyrand F, Lafontaine I, Fontaine T, Janbon G. 2008. UGE1 and UGE2 regulate the UDP-glucose/UDP-galactose equilibrium in Cryptococcus neoformans. Eukaryot Cell 7:2069–2077. doi: 10.1128/EC.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan Y, Lin X. 2018. Multiple Applications of a Transient CRISPR-Cas9 Coupled with Electroporation (TRACE) System in the Cryptococcus neoformans Species Complex. Genetics 208:1357–1372. doi: 10.1534/genetics.117.300656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Upadhya R, Lam WC, Maybruck BT, Donlin MJ, Chang AL, Kayode S, Ormerod KL, Fraser JA, Doering TL, Lodge JK. 2017. A fluorogenic C. neoformans reporter strain with a robust expression of m-cherry expressed from a safe haven site in the genome. Fungal Genet Biol 108:13–25. doi: 10.1016/j.fgb.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin J, Fan Y, Lin X. 2020. Transformation of Cryptococcus neoformans by electroporation using a transient CRISPR-Cas9 expression (TRACE) system. Fungal Genet Biol 138:103364. doi: 10.1016/j.fgb.2020.103364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chacko N, Zhao Y, Yang E, Wang L, Cai JJ, Lin X. 2015. The lncRNA RZE1 controls cryptococcal morphological transition. PLoS Genet 11:e1005692. doi: 10.1371/journal.pgen.1005692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Tian X, Gyawali R, Upadhyay S, Foyle D, Wang G, Cai JJ, Lin X. 2014. Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Pathog 10:e1004185. doi: 10.1371/journal.ppat.1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ZNF2oe strain shows no obvious deficiency in chitosan. (A) The relative transcript level of CDA1, CDA2, and CDA3 in three biological replicates of wild-type H99 and the ZNF2oe strain after overnight culture in liquid YPD medium as measured by RT-PCR. The transcript level of TEF1 in each RNA sample was used as the internal control. The error bars indicate the standard derivations among three technical replicates. Unpaired t-tests were performed. n.s.: non-significant. *: P < 0.05. **: P < 0.01. ***: P = 0.0002. ****: P < 0.0001 (B) The wild-type H99 and the ZNF2oe cells after overnight culture in liquid YPD medium were stained with either calcofluor white or Eosin Y. Download FIG S1, TIF file, 2.4 MB (2.4MB, tif) .
Copyright © 2022 Lin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download Table S1, DOCX file, 0.02 MB (17.1KB, docx) .
Copyright © 2022 Lin et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All of the data supporting this study are presented herein, and the reported fungal strains generated for this study are available on request.