Abstract
Dual typing (VP4 and VP7) of rotavirus obtained from 257 Mexican children during three epidemiological seasons was performed by reverse transcription-PCR. The P1G1 genotype was the most prevalent (40%), followed by P1G3 (19%) and P2G2 (16%). Thirty-one specimens (12%) presented mixed infections, while some genotypes were not found. This is the first dual typing of isolates from diarrhea cases in Mexico.
Rotavirus is a worldwide cause of severe diarrhea in young children. Genetic or antigenic differences in VP4 (P) and VP7 (G) capsid proteins have been detected in viral isolates (11). Since VP4 and VP7 genes segregate independently, a binary system is used to classify rotavirus (12). There are certain genotypes that circulate with higher frequencies: P1 to P4 for VP4 and G1 to G4 for VP7 (5, 10, 15, 21). The combinations P1G1, P1G3, P2G2, and P1G4 are the most frequent in Brazil, Israel, Japan, South Africa, and the United States (16, 17, 18, 20). However, relative rates of these combinations are different in countries like India and Egypt, where unusual combinations of P and G alleles are also present (2, 8, 14). Rotavirus strains with different combinations may occur simultaneously during seasonal outbreaks, giving rise to the possibility of mixed infections and genetic rearrangements (3, 6, 9, 22).
In this study we performed dual typing of rotavirus strains gathered from eight laboratories included in the Mexican National Network for Rotavirus Diagnosis. The study encompassed three consecutive rotavirus epidemic seasons from July 1994 to June 1997 in several states of Mexico. Stool specimens from 257 children less than 5 years old were received by the reference center (National Institute of Diagnostics and Epidemiological Reference). VP4 (P1 to P4) and VP7 (G1 to G4) genotyping was carried out independently by heminested reverse transcription-PCRs as previously described (4, 7).
From all 257 samples a 1,062-bp product of the VP7 gene was observed, which corresponds to rotavirus group A (Fig. 1). For VP4, the expected 876-bp amplicon was obtained (not shown). The second amplification products of both genes can be seen in Fig. 1 and 2, in which some examples of mixed infections are also shown. One hundred two samples (39.7%) were of the P1G1 genotype, 49 (19.0%) were P1G3, and 42 (16.3%) were P2G2. Genotypes P4 and G4 were not found in this study. The distribution and the rates of these combinations are summarized in Table 1.
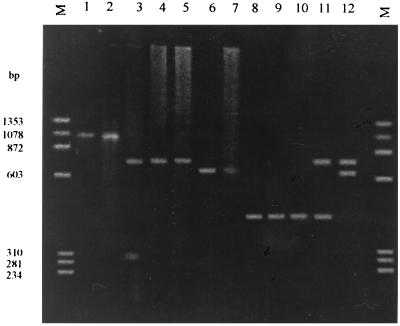
FIG. 1.
Amplification products of the VP7 gene. Lanes 1 and 2, first amplification products (1,062 bp); lanes 3 through 5, genotype G1 (749 bp); lanes 6 and 7, genotype G2 (652 bp); lanes 8 through 10, genotype G3 (347 bp); lanes 11 and 12, mixed infections with the G1G3 and G1G2 genotypes, respectively. φX174 digested with HaeIII (Roche Molecular Biochemicals) was used as the molecular size (M) standard.
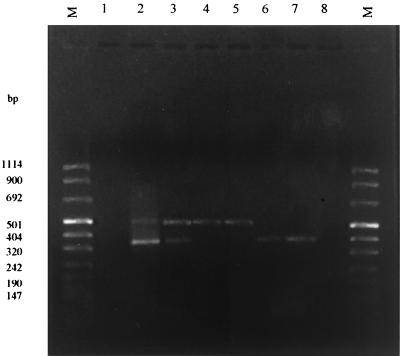
FIG. 2.
VP4 genotype amplicons. Lanes 2 and 3, dual infection with genotype P1P2; lanes 4 and 5, genotype P2 (483 bp); lanes 6 and 7, genotype P1 (345 bp). Marker VIII (Roche Molecular Biochemicals) was used as the molecular size (M) standard.
TABLE 1.
Distribution of VP4 and VP7 genotypes of human rotaviruses in different States of Mexico
| State and yr studied | No. of samples | % of each genotype founda
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| P1G1 | P1G2 | P1G3 | P2G1 | P2G2 | P2G3 | MIb | NTc | ||
| Colima, 1995–1997 | 19 | 47.4 | 10.5 | 5.3 | 10.5 | 10.5 | 0 | 5.3 | 10.5 |
| Mexico City, 1995–1997 | 99 | 52.5 | 0 | 22.2 | 1.1 | 5.5 | 0 | 11.1 | 8.0 |
| Michoacan, 1994–1996 | 25 | 68.0 | 0 | 20.0 | 0 | 4.0 | 4.0 | 0 | 4.0 |
| Nuevo Leon, 1996 | 24 | 8.3 | 0 | 8.3 | 4.2 | 66.7 | 8.3 | 0 | 4.2 |
| Puebla, 1994–1996 | 28 | 17.9 | 0 | 35.7 | 3.6 | 10.7 | 0 | 32.1 | 0 |
| Quintana Roo, 1997 | 18 | 61.1 | 0 | 5.5 | 0 | 0 | 0 | 22.2 | 11.1 |
| Veracruz, 1995–1996 | 21 | 23.8 | 0 | 38.0 | 0 | 4.8 | 0 | 24.8 | 9.5 |
| Yucatan, 1997 | 23 | 4.3 | 13.0 | 0 | 13.0 | 60.8 | 4.3 | 4.3 | 0 |
| Total | 257 | 39.6 | 1.9 | 19.0 | 3.1 | 16.3 | 1.5 | 12.0 | 6.2 |
Numbers in bold are the highest values for that state.
MI, mixed infection.
NT, not typed.
P1G1 was the most widely distributed combination. It was found in southeastern Mexico (Quintana Roo and Yucatan states), on the western coast (Colima and Michoacan), in the central region (Mexico City and Puebla), on the eastern coast (Veracruz), and in northern Mexico (in the Mexico-U.S. border state of Nuevo Leon). The other most frequent genotypes were P1G3 and P2G2, which were found in all studied states except Yucatan and Quintana Roo. The high frequency of P1G1 in Mexico was similar to that found in other countries, like the United States, Japan, South Africa, and Israel, and was different from that reported in India and Brazil (15, 16, 17, 21). The frequency observed for P1G3 was similar to that found in the United States, whereas the high frequency observed for P2G2 has been found solely in South Africa (17).
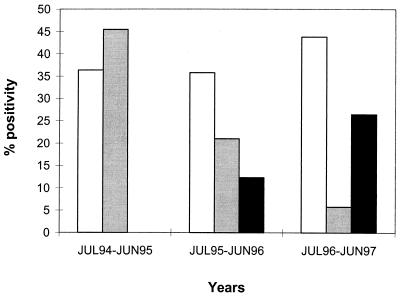
In Mexico, rotavirus infection peaks occurred in winter (November to March) like in other countries (11). Genotype P1G1 occurred during the three seasonal peaks, predominating during the last two, in contrast with P1G3, which predominated during the first peak and decreased in the last two. In this regard, it is worth mentioning that Padilla-Noriega et al. (13) found genotype G3 in high prevalence in Yucatan during the 1994-1995 outbreak, but it was absent in the next peak (1995-1996). Genotype P2G2 was not found in the first period, but an increasing prevalence was observed beginning with the second period (Fig. 3). The seasonal shifts in genotype frequencies might be due to different factors, such as genetic variation of rotavirus strains, host range modifications, host immunity, and climate changes (1, 6, 9, 11, 22).
FIG. 3.
Frequency of rotavirus genotypes by yearly distribution. White bars, P1G1; gray bars, P1G3; black bars, P2G2.
The genetic variation may also be a consequence of simultaneous infection of a single host with different rotavirus strains (3, 6, 19, 22). In this work, 31 samples showed more than one P or G allele, meaning that more than one rotavirus strain was present. The most frequent mixed infection was genotype P1G1G3, which suggests the simultaneous presence of P1G1 and P1G3 rotavirus strains. This result correlates with the high prevalence of those genotypes (Table 1). Mixed infections have also been reported in Brazil (30%), India (6%), the United States (3%), South Africa (3%), and Japan (1%). In this study, Mexico had the second highest percentage (12%) of dual infections reported so far, increasing the likelihood of genetic reassortment, which may yield new gene combinations (2, 3, 9, 15, 20).
Genotypes of rotavirus strains with unusual combinations of P and G alleles were found in lower frequencies as follows: P1G2, 1.9%; P2G1, 3.1%; and P2G3, 1.5% (Table 2). Genotype P2G1 occurred in Yucatan, Colima, Nuevo Leon, and Puebla, while P1G2 was restricted to Colima and Yucatan and P2G3 was present only in Michoacan, Nuevo Leon, and Yucatan. These combinations could have been generated from genetic rearrangements of the most frequent rotavirus genotypes, since they circulated simultaneously in those regions. It is worth mentioning that genotype P2G1 has also been reported at low frequencies (1%) in Brazil and Japan, while P1G2 and P2G3 have not been reported in other countries (8, 10, 19, 20, 21).
TABLE 2.
Mixed infections and untyped samples found in Mexico between 1994 and 1997
| Genotype of infecting strain | No. of samplesa |
|---|---|
| Mixed infection | |
| P1P2G1 | 5 |
| P1P2G2 | 2 |
| P1P2G3 | 7 |
| P1P2G1G2 | 1 |
| P1P2G1G3 | 1 |
| P1G1G3 | 11 |
| P1G1G2 | 2 |
| P2G1G2 | 1 |
| P2G1G3 | 1 |
| Total | 31 (12.0) |
| Untyped strainsb | |
| P1Gnt | 2 |
| P2Gnt | 1 |
| G1Pnt | 9 |
| G3Pnt | 1 |
| G1G2Pnt | 1 |
| G1G3Pnt | 2 |
| Total | 16 (6.2) |
Numbers in parentheses are percentages of the total samples studied (n = 257).
nt, not typed.
Partially untyped samples were also found in this study. In three samples (1.2%), two P1 and one P2 G allele could not be typed, while the P allele was not identified in 13 samples (5%) with the following G alleles: nine G1, one G3, one G1G2 (mixed infection), and two G1G3. This suggests that P alleles different from P1 to P4 segregated with G1 and G3. Thus, there is a larger heterogeneity of P alleles than of G alleles in Mexican field strains. This was also found with monoclonal antibody-based typing (13) and could be due to a higher selective pressure exerted over VP4, since it is the most external viral protein (9, 11). G alleles seemed to be more conserved, because mainly G1, G2, and G3 were found (10, 14, 15, 16, 17, 20, 21). These data support the need for including a larger set of primers to identify other P and G alleles.
In conclusion, our results confirm that there is an important variability among rotavirus strains in Mexico. This is the first report of dual typing, which allowed for identifying one of the main sources of this variability, i.e., the presence of mixed infections.
Acknowledgments
This work was totally supported by INDRE.
We thank Ana Flisser, Dolores Correa, and Alejandro Escobar Gutierrez for encouragement and discussion of the paper, Lizbeth Teran, Angel Barney, Juan Rojas, and Misael Mondragón for technical assistance, and M. del Pilar Bada (Veracruz), Teresa Escobar (Quintana Roo), Sandra Suarez (Michoacan), Socorro Sambrano (Monterrey), Zita Gutierrez (Puebla), Mario Gaytan (Colima), Genny Mendez and Salha (Yucatan), and Felipe Mota and Maricela Echeverria (Mexico City) for collecting and sending samples.
Araceli Rodríguez Castillo and Andrés Velasco Villa contributed equally to this work.
REFERENCES
- 1.Contreras J F, Menchaca G E, Padilla-Noriega L, Tamez R S, Greenberg H B, López S, Arias C F. Heterogeneity of VP4 neutralization epitopes among serotype P1A human rotavirus strains. Clin Diagn Lab Immunol. 1995;2:506–508. doi: 10.1128/cdli.2.4.506-508.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das B K, Gentsch J R, Cicirello H G, Woods P A, Gupta A, Ramachandran M, Kumar R, Bhan M K, Glass R I. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Espejo T R, Calderon E, González N, Salomon A, Martuscelli A, Romero P. Presence of two distinct types of rotavirus in infants and young children hospitalized with acute gastroenteritis in México City, 1977. J Infect Dis. 1979;139:474–477. doi: 10.1093/infdis/139.4.474. [DOI] [PubMed] [Google Scholar]
- 4.Gentsch J R, Glass R I, Woods P, Gouvea V, Gorziglia M, Flores J, Das B K, Bhan M K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorziglia M, Larralde G, Li B, Kapikian A Z, Chanock R M. Extent and distribution of antigenic polymorphism of human rotavirus outer capsid protein VP4. Vaccines. 1991;91:65–68. [Google Scholar]
- 6.Gouvea V, Brantly M. Is rotavirus a population of reassortants? Trends Microbiol. 1995;3:159–162. doi: 10.1016/s0966-842x(00)88908-8. [DOI] [PubMed] [Google Scholar]
- 7.Gouvea V, Glass R I, Woods P, Taniguchi K, Clark H F, Forrester B, Fang Z-Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouvea V, de Castro L, Timenetsky M D C, Greenberg H, Santos N. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J Clin Microbiol. 1994;32:1408–1409. doi: 10.1128/jcm.32.5.1408-1409.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham A, Kudesia G, Allen A M, Desselberger U. Reassortment of human rotavirus possessing genome rearrangements with bovine rotavirus: evidence for host cell selection. J Gen Virol. 1987;68:115–122. doi: 10.1099/0022-1317-68-1-115. [DOI] [PubMed] [Google Scholar]
- 10.Gunasena S, Nakagomi O, Isegawa Y, Kaga E, Nakagomi T, Steele A D, Flores J, Ueda S. Relative frequency of VP4 gene alleles among human rotaviruses recovered over a 10-year period (1982–1991) from Japanese children with diarrhea. J Clin Microbiol. 1993;31:2195–2197. doi: 10.1128/jcm.31.8.2195-2197.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshino Y, Kapikian A Z. Rotavirus vaccine development for the prevention of severe diarrhea in infants and young children. Trends Microbiol. 1994;2:242–249. doi: 10.1016/0966-842x(94)90629-7. [DOI] [PubMed] [Google Scholar]
- 12.Hoshino Y, Sereno M M, Midthun K, Flores J, Kapikian A, Chanock R M. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc Natl Acad Sci USA. 1985;82:8701–8704. doi: 10.1073/pnas.82.24.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padilla-Noriega L, Méndez-Toss M, Menchaca G, Contreras J F, Romero-Guido P, Puerto F I, Guiscafré H, Mota F, Herrera I, Cedillo R, Muñoz O, Calva J, de Lourdes Guerrero M, Coulson B S, Greenberg H B, López S, Arias C F. Antigenic and genomic diversity of human rotavirus VP4 in two consecutive epidemic seasons in Mexico. J Clin Microbiol. 1998;36:1688–1692. doi: 10.1128/jcm.36.6.1688-1692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramachandran M, Das B K, Vij A, Kumar R, Bhambal S S, Kesari N, Rawat H, Bahl L, Thakur S, Woods P A, Glass R I, Bhan M K, Gentsch J R. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos N, Riepenhoff-Talty M, Clark H F, Offit P, Gouvea V. VP4 genotyping of human rotavirus in the United States. J Clin Microbiol. 1994;32:205–208. doi: 10.1128/jcm.32.1.205-208.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silberstein I, Shulman L M, Mendelson E, Shif I. Distribution of both rotavirus VP4 genotypes and VP7 serotypes among hospitalized and nonhospitalized Israeli children. J Clin Microbiol. 1995;33:1421–1422. doi: 10.1128/jcm.33.5.1421-1422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steele A D, van Niekerk M C, Mphahlele M J. Geographic distribution of human rotavirus VP4 genotypes and VP7 serotypes in five South African regions. J Clin Microbiol. 1995;33:1516–1519. doi: 10.1128/jcm.33.6.1516-1519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki Y, Sanekata T, Sato M, Tajima K, Matsuda Y, Nakagomi O. Relative frequencies of G (VP7) and P (VP4) serotypes determined by polymerase chain reaction assays among Japanese bovine rotaviruses isolated in cell culture. J Clin Microbiol. 1993;31:3046–3049. doi: 10.1128/jcm.31.11.3046-3049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timenetsky M C S T, Gouvea V, Santos N, Carmona R C, Hoshino Y. A novel human rotavirus serotype with dual G5-G11 specificity. J Gen Virol. 1997;78:1373–1378. doi: 10.1099/0022-1317-78-6-1373. [DOI] [PubMed] [Google Scholar]
- 20.Timenetsky M D C S T, Santos N, Gouvea V. Survey of rotavirus G and P types associated with human gastroenteritis in São Paulo, Brazil, from 1986 to 1992. J Clin Microbiol. 1994;32:2622–2624. doi: 10.1128/jcm.32.10.2622-2624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ushijima H, Mukoyama A I, Hasegawa A, Nishimura S, Konishi K, Bosu K. Serotyping of human rotaviruses in the Tokyo area (1990–1993) by enzyme immunoassay with monoclonal antibodies and by reverse transcription and polymerase chain reaction amplification. J Med Virol. 1994;44:162–165. doi: 10.1002/jmv.1890440208. [DOI] [PubMed] [Google Scholar]
- 22.Ward R L, Nakagomi O, Knowlton D R, McNeal M M, Nakagomi T, Clemens J D, Sack D A, Schiff G M. Evidence for natural reassortants of human rotaviruses belonging to different genogroups. J Virol. 1990;64:3219–3225. doi: 10.1128/jvi.64.7.3219-3225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]