Abstract
Introduction
The inflammatory potential of SARS-CoV-2 Spike S1 (Spike) has never been tested in human primary macrophages (MΦ). Different recombinant Spikes might display different effects in vitro, according to protein length and glycosylation, and endotoxin (lipopolysaccharide, LPS) contamination.
Objectives
To assess (1) the effects of different Spikes on human primary MΦ inflammation; (2) whether LPS contamination of recombinant Spike is (con)cause in vitro of increased MΦ inflammation.
Methods
Human primary MΦ were incubated in the presence/absence of several different Spikes (10 nM) or graded concentrations of LPS. Pro-inflammatory marker expression (qPCR and ELISA) and supernatant endotoxin contamination (LAL test) were the main readouts.
Results
LPS-free, glycosylated Spike (the form expressed in infected humans) caused no inflammation in human primary MΦ. Two (out of five) Spikes were contaminated with endotoxins ≥ 3 EU/ml and triggered inflammation. A non-contaminated non-glycosylated Spike produced in E. coli induced MΦ inflammation.
Conclusions
Glycosylated Spike per se is not pro-inflammatory for human MΦ, a feature which may be crucial to evade the host innate immunity. In vitro studies with commercially available Spike should be conducted with excruciating attention to potential LPS contamination.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s10565-021-09693-y.
Keywords: SARS-CoV-2 infection, Human macrophages, Inflammation, Lipopolysaccharide, Spike protein
Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a single-stranded RNA virus of the genus Betacoronavirus in the Coronaviridae family, which also includes SARS-CoV-1 and Middle-East Respiratory Syndrome coronavirus (MERS-CoV) (Zhu et al. 2020).
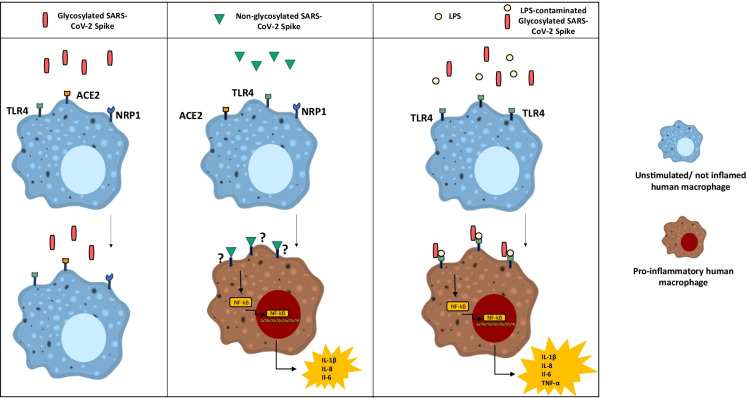
In most cases, SARS-CoV-2 engages, via the viral surface glycosylated Spike protein (Spike), the angiotensin-converting enzyme 2 (ACE2), thereby gaining access to the cytoplasm of host cells (Lan et al. 2020).
The Spike S1 domain contains the receptor-binding sequence responsible for ACE2 binding, whereas the S2 domain is crucial for cell membrane fusion (Hoffmann et al. 2020). High levels of Spike protein glycosylation are instrumental to immune escape, serving as a safety shield from the host’s innate immune system (Bagdonaite and Wandall 2018; Watanabe et al. 2020). Recent evidence demonstrated that Spike binds to additional receptors, including neuropilin-1 (NRP1) (Cantuti-Castelvetri et al. 2020; Daly et al. 2020), a multifunctional receptor for several extracellular ligands playing a crucial role in the regulation of myeloid cell function (Roy et al. 2017), and toll-like receptor (TLR)-4 (Shirato and Kizaki 2021; Choudhury and Mukherjee 2020), essential for the innate immune response to bacteria, mycobacteria, spirochetes, and viruses (Beutler 2009; Lester and Li 2014).
Macrophages (MΦ)—key cells in the innate immune response—play a pivotal role in body’s defense against viral infections mostly by producing inflammatory mediators to kill pathogens and by repairing injured tissues (Sica and Mantovani 2012; Tarique et al. 2015).
Severe COVID-19 is characterized by an aggressive inflammatory response known as cytokine storm (Ye et al. 2020), in which MΦ are implicated through an exaggerated release of inflammatory cytokines/chemokines, which fuel tissue hyperinflammation, further leukocyte recruitment, and release of inflammatory mediators, resulting into a feed-forward, pathogenic vicious cycle (Schultze and Aschenbrenner 2021). The cytokine storm, therefore, plays a major role in tissue, especially lung, damage and in the onset of the acute respiratory distress syndrome (ARDS) (Gustine and Jones 2021).
Recent studies, aimed at unveiling the molecular bases responsible for this MΦ-mediated hypercytokinemia in COVID-19, reported that Spike S1 is a key viral component in triggering MΦ inflammation, independently of virus infection or replication (Chiok et al. 2021). Specifically, SARS-CoV-2 Spike directly induces pro-inflammatory cytokine production in mouse MΦ (Shirato and Kizaki 2021; Chiok et al. 2021), MΦ derived from the human monocytic leukemia cell line THP-1 (Shirato and Kizaki 2021; Chiok et al. 2021; Pantazi et al. 2021; Khan et al. 2021), and human mononuclear cells (MNCs) (Pantazi et al. 2021), via nuclear factor-kappa B (NF-κB) activation through TLR(s) signaling. To the best of our knowledge, thus far, no studies have assessed the effects of Spike glycoprotein on inflammation in human primary MΦ.
The TLR family is associated with a strong pro-inflammatory cytokine and chemokine production—especially TLR4 which can be activated in response to LPS, derived from Gram-negative bacteria, and to few viral proteins (El-Zayat et al., 2019, Lu et al. 2008).
Accordingly, LPS is commonly used to induce in vitro MΦ polarization to classically activated M1 (Mantovani et al. 2002), which represent one extreme (alternatively activated M2 are the other extreme) of a spectrum of macrophage functional phenotypes in response to different microenvironmental cues. M1 macrophages—obtained with LPS and/or IFNγ—are responsible for killing intracellular pathogens and for releasing pro-inflammatory mediators, through TLR4 signaling cascade, which includes the recruitment of IL-1R-associated kinase 1 (IRAK-1) and the downstream activation of MAP kinases and NF-κB. These pathways eventually end up in enhancing the transcription, among others, of tumor necrosis factor alpha (TNFα), monocyte chemoattractant protein-1 (MCP1), interleukin (IL)-6, and IL-1β genes (Guzmán-Beltrán et al. 2017).
It is worth noticing that a recent study showed a specific molecular interaction between SARS-CoV-2 Spike and LPS. In that study, Spike, when combined with low levels of LPS, boosted NF-κB activation via TLR4 in monocytes, and cytokine responses in human MNCs (Petruk et al. 2020).
These findings suggest that LPS and Spike are synergistic in triggering and amplifying the inflammatory response of innate immune cells, with a potential dual relevance: (a) a subclinical Gram bacterial infection or low levels of LPS derived from gut microbiome, e.g., in the obese subject (Cani et al. 2012), may synergistically act with Spike during SARS-CoV-2 infection and significantly affect the clinical course of the disease; (b) at a less relevant level, since LPS is a quite frequent impurity of recombinant proteins derived from E. coli, but also from mammalian cells (e.g., CHO, HEK293) (Wakelin et al. 2006), undetected LPS contamination in recombinant Spike proteins might be a major confounder in experimental studies of Spike effects on innate immune cells and/or endothelium. Based on these premises, we hypothesized that LPS contamination of some commercially available recombinant SARS-CoV-2 Spike proteins might be—at least in part—responsible for the increased MΦ inflammation reported in the literature (Shirato and Kizaki 2021; Chiok et al. 2021; Pantazi et al. 2021; Khan et al. 2021).
The present investigation, therefore, was undertaken with a dual goal: (1) to investigate the in vitro effect of Spike in primary human MΦ inflammation, and, consequently to some of the evidence provided by this study, and (2) to quantify LPS contamination of some commercially available recombinant SARS-CoV-2 Spike proteins, among those reported to be pro-inflammatory in previous papers.
Material and methods
Cell isolation and culture
Monocyte-derived MΦ were isolated from healthy donors’ buffy coats and cultured as previously reported (Derlindati et al. 2015). Briefly, MNCs were isolated by density gradient centrifugation on Lymphoprep (Euroclone, Milano, Italy) and 2 × 107 cells/ml were seeded in multiwell plates. Monocytes were selected from MNCs by plastic adherence for 1 h and cultured for 6 days in RPMI 1640 medium (Euroclone) with 10% fetal bovine serum (FBS), 1% L-glutamine, 1% pen/strep, 1% amphotericin B, and 70 ng/ml macrophage colony-stimulating factor (M-CSF) (Miltenyi Biotec, Bergisch Gladbach, Germany) at 37 °C and 5% CO2.
On day 6, MΦ were stimulated for 16 h with SARS-CoV-2 Spike S1 (Spike A, B, C, D, E, F purchased from the random listed companies Bio-Techne, Minneapolis, MN, USA; Arigo Biolaboratories, Hsinchu, Taiwan; Abcam, Cambridge, UK; BPS Bioscience, San Diego, CA, USA) at the concentration of 10 nM (Table 1). This concentration of 10 nM (corresponding to ~ 1 µg/ml) of Spike was selected on the basis of the literature (Shirato and Kizaki 2021; Zhao et al. 2021; Karwaciak et al. 2021).
Table 1.
Main characteristics of the SARS-CoV-2 Spike proteins herein used
| Spike protein | Amino acid sequence a | Expression system | Glycosylated |
|---|---|---|---|
| Spike A | Val16-Arg685 | CHO cells | Yes |
| Spike B | Val16-Arg685 | CHO cells | Yes |
| Spike C | Val16-Arg685 | CHO cells | Yes |
| Spike D | Val16-Arg685 | HEK293 cells | Yes |
| Spike E | Met15-Cys671 | E. coli | No |
| Spike F | Val16-Pro681 | HEK293 cells | Yes |
aNone of the Spike proteins contains the cleavage site of furin (residues 685–686)
Val valine, Arg arginine, Met methionine, Cys cysteine, Pro proline, CHO Chinese hamster ovary, HEK human embryonic kidney
Where indicated, to neutralize any potential LPS interference (Tsubery et al. 2000), polymyxin B (Poly B) (2 µg/ml; Merck Life Science S.r.l., Milan, Italy) was added in culture.
To define a LPS dose–response curve, MΦ were incubated with increasing LPS concentrations (0.01—0.05—0.1—0.5—1—10—100 ng/ml; Merck Life Science) for 16 h and the effect on cell inflammation was recorded.
Endotoxin quantification
Endotoxin contamination of recombinant spike proteins was assessed in supernatants of MΦ in the different culture conditions by Pierce Chromogenic Endotoxin Quant Kit (Thermo Fisher Scientific, Waltham, MA, USA), strictly following manufacturer’s instructions. Endotoxin concentration in diluted (from 1:10 to 1:100 depending on the degree of contamination) cell supernatants was calculated in duplicate by using the “high” standard curve provided by the kit. The reaction product was photometrically measured at 405 nm (Varioskan Lux, Thermo Scientific), and results were expressed as EU/ml.
Of note, as shown in Fig. 1, Spikes A and B were highly contaminated, while Spikes C and Spike D were uncontaminated compared to control. Spike E showed very low endotoxin level (< 0.4 EU/ml cell supernatant) (Fig. 1).
Fig. 1.
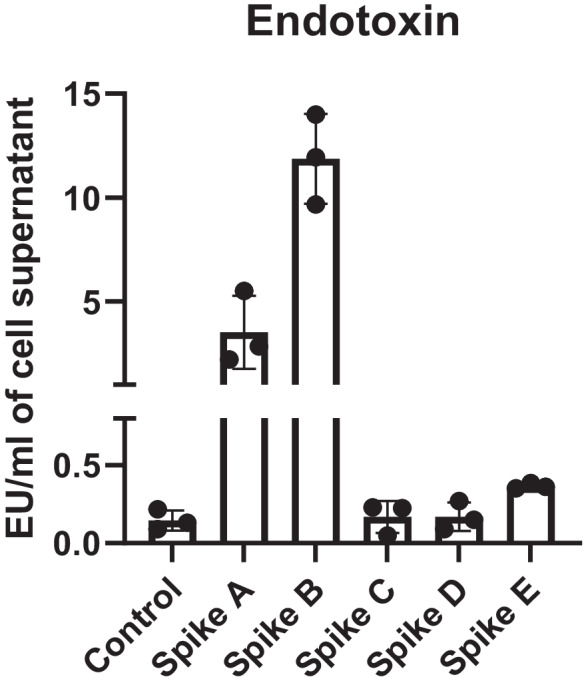
Endotoxin contamination of SARS-CoV-2 recombinant Spike proteins. Endotoxins were quantified in cell supernatants of Spike-treated MΦ. Supernatants from untreated MΦ were used as control. Data expressed as mean ± SEM from 3 independent experiments have been reported in the graph
Inflammatory marker gene expression
To quantify MΦ pro-inflammatory response, quantitative PCR (qPCR) assays were performed on candidate genes. Briefly, cells were lysed by Qiazol and total RNA was isolated by miRNeasy mini kit (Qiagen Ltd, West Sussex, UK) followed by NanoDrop (Thermo Scientific) quantification.
A total of 250 ng of RNA was retro-transcribed by using High-Capacity RNA-to-cDNA Kit (Applied Biosystem, Life Technologies, Foster City, California, USA), following manufacturer’s instructions.
Interleukin (IL)-6 (Hs00174131_m1), IL-8 (Hs00174103_m1), IL-1β (Hs01555410_m1), tumor necrosis factor (TNF)-α (Hs00174128_m1) gene expression was assessed using TaqMan Universal Master Mix (Applied Biosystems) with TaqMan primers and probes (Thermo Scientific) on a CFX Connect Real-Time (Bio-Rad, Hercules, CA, USA), as previously reported (Spigoni et al. 2020). Thermal cycling conditions were as follows: 98° for 30 s, followed by 40 amplification cycles (95 °C for 15 s; 60 °C for 1 min).
Gene expression values were calculated based on the ΔΔCt method (Schmittgen et al. 2000) and normalized to the geometric mean of RPS18 (ribosomal protein S18) (Hs01375212_g1), GAPDH (glyceraldehyde 3-phosphodehydrogenase) (Hs99999905_m1), ACTB (β-actin) (Hs99999903_m1), and B2M (β-2-Microglobulin) (Hs00187842_m1) housekeeping genes. Each sample was analyzed in triplicate and the mean values were used for calculations.
Inflammatory molecule secretion
IL-6, IL-1β, and TNFα levels in cell culture supernatants (diluted 1:2) were quantified in duplicate by a multiparameter kit based on magnetic beads (Luminex Assay, R&D Systems) and analyzed on MagPix instrument (Luminex Corporation, Austin, TX, USA) according to kit instructions, as previously reported (Spigoni et al. 2017). IL-8 quantification was performed by Human IL-8/CXCL8 Quantikine ELISA Kit (R&D Systems) following manufacturer’s instruction. Inter- and intra-assay coefficients of variation were 6.7% and 4.6%, respectively. IL-8 concentration in diluted (1:200) cell supernatants was calculated in duplicate by using a standard curve generated by serially diluting reconstituted standards and by measuring the absorbance at 450 nm in a microplate reader (Multiskan™ FC Microplate Photometer, Thermo Scientific).
Statistical analysis
Normally distributed data are reported as mean ± SE, while skewed data are expressed as median ± interquartile range (IQR). Differences were identified using Kruskal–Wallis with Dunn’s multiple comparison test. Paired t-test was performed to compare protein quantification data. Statistical significance was set at p < 0.05 (two-sided). Data were analyzed using GraphPad PRISM version 5.0 (GraphPad Software Inc., California, USA).
Results
Effects of SARS-CoV-2 Spike proteins on human MΦ inflammation
To test the effect of Spike on MΦ inflammation, we assessed pro-inflammatory marker (IL-1β, IL-6, IL-8, TNFα) gene expression in human primary MΦ treated with Spike (A, B, C, D, E) 10 nM for 16 h.
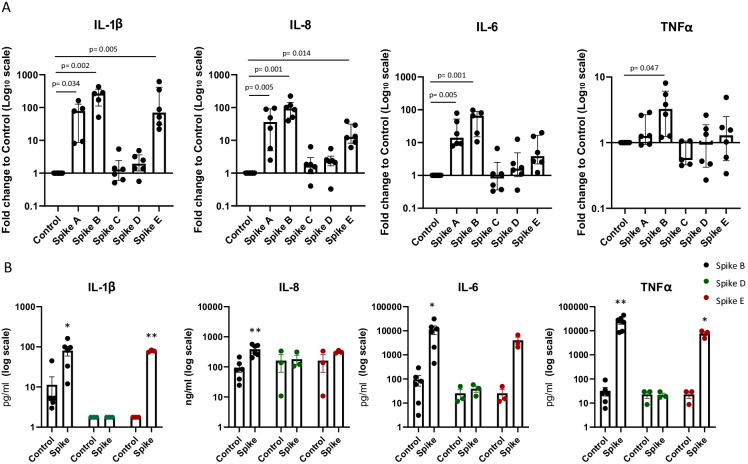
LPS-free Spikes C and D did not increase the gene expression of inflammatory biomarkers in MΦ vs control (Fig. 2A). Conversely—and in line with data on endotoxin contamination (Fig. 1)—Spikes A and B caused a significant increase of IL-1β, IL-8, TNFα (only Spike B), and IL-6 gene expression compared to control. Unexpectedly, Spike E—which was only slightly contaminated (0.33 EU/ml of endotoxins)—significantly boosted IL-1β and IL-8 (but not IL-6 and TNF α) gene expression compared to untreated cells (Fig. 2A).
Fig. 2.
Effects of Spike proteins on pro-inflammatory cytokine/chemokine expression in human MΦ. Inflammatory marker gene (A) and protein (B) expressions were assessed in human MΦ treated/untreated with Spike (purchased from different companies) 10 nM for 16 h. Individual data points representing repeated experiments on at least 3 individual donors are shown. A Data are represented as median ± IQR, and differences were evaluated with Kruskal–Wallis corrected with Dunn’s multiple comparison test. B Data are expressed as mean ± SEM, and differences were evaluated with paired t-test (IL = interleukin; TNF = tumor necrosis factor) (*p < 0.05; **p < 0.01 vs control)
In some selected culture conditions (Control, Spike B, D and E), cytokine release in the medium secretion was assessed to possibly confirm qPCR data (Fig. 2B). In concordance with gene expression data, LPS-free Spike D did not affect cytokine secretion vs control, while Spike B significantly increased the secretion of all tested biomarkers, compared to untreated control. IL-1β and TNFα secretion was also augmented by Spike E.
Thus, we inferred that Spike per se does not affect pro-inflammatory cytokine secretion in primary human MΦ.
LPS concentration–response curve
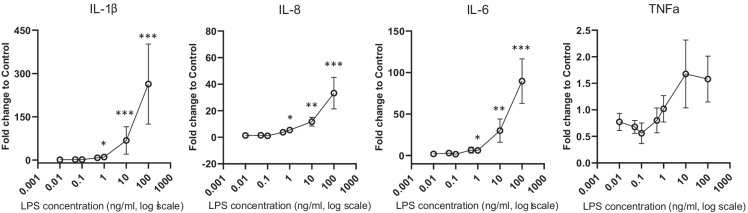
We then performed a dose–response curve of human MΦ inflammation to scalar LPS concentrations to ascertain whether endotoxins are the only triggering factor responsible for MΦ inflammation following stimulation with LPS-contaminated Spike.
As shown in Fig. 3, LPS at concentrations ≥ 1 ng/ml activated MΦ, inducing an increase in pro-inflammatory marker (IL-1β, IL-6 and IL-8, but not TNFα) gene expression (Fig. 3). Lower LPS concentrations were devoid of any effect.
Fig. 3.
LPS dose–response curve on pro-inflammatory marker gene expression in human primary MΦ. MΦ were stimulated with increasing (0—0.01—0.05—0.1—0.5 – 1 – 10—100 ng/ml) LPS concentrations for 16 h and pro-inflammatory marker (IL-1β, IL-8, IL-6, TNFα) gene expression were assessed. Data are represented as mean ± SEM from at least 3 independent experiments and differences evaluated with Kruskal–Wallis followed by Dunn’s multiple comparison test (IL = interleukin; TNF = tumor necrosis factor) *p < 0.05; **p < 0. 005: *** p < 0.001 vs control (LPS = 0.0 ng/ml)
Furthermore, these data demonstrate that the endotoxin contamination (13 EU/ml corresponding to 1.3 ng/ml of LPS) of Spike B is sufficient to drive inflammation in cultured human MΦ.
Since LPS alone is effective only at ≥ 1 ng/ml, Spike A (showing a contamination of only 3 EU/ml of endotoxin corresponding to 0.3 ng/ml of LPS) strongly stimulates inflammation; the data are consistent with the idea that LPS and Spike are synergistic with each other, even at concentrations which are individually ineffective.
The pro-inflammatory effects associated to Spike E, however, were unexpectedly evident also at a negligible (0.03 ng/ml) LPS contamination. This prompted us to run further experiments with Spike E.
Spike E
Spike E displayed a very low endotoxin contamination (0.3 EU/ml), but, when compared to Spikes A–D used in this study, other factors may be implicated: (i) it is shorter than the other Spikes, because it lacks the NRP1-binding domain at the C-terminal; (ii) it is produced in a prokaryotic expression system (E. coli), whereas Spikes A–D are expressed in mammalian cells, and all share the same amino acid sequence.
We then hypothesized that inflammation stimulated by Spike E might be attributable to:
Spike interaction with residual (0.03 ng/ml) LPS concentrations
The lack of the NRP1-binding domain
The absence of post-translational modifications (glycosylation) owing to E. coli expression system
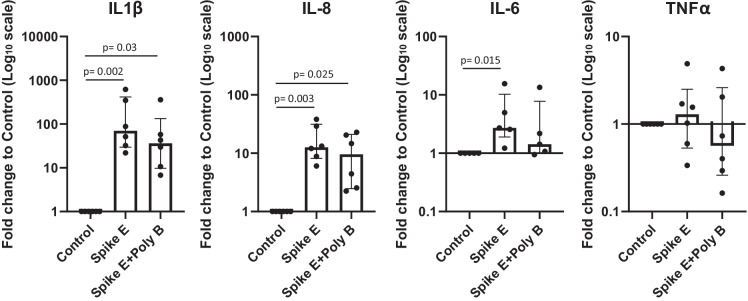
To explore the potential interaction between Spike and LPS (hypothesis a), we premixed Spike E with polymyxin B—which is an antibiotic known for its capacity to bind to and neutralize LPS (Tsubery et al. 2000)—and we tested their combined effect on MΦ inflammation. As shown in Fig. 4, polymyxin B did not affect Spike E–induced inflammation, showing that its pro-inflammatory effect was not due to its minimal LPS contamination.
Fig. 4.
Pro-inflammatory effects of Spike E in the presence/absence of Poly B. Inflammatory marker (IL-1β, IL-8, IL-6, TNFα) gene expression was evaluated in human MΦ treated/untreated with Spike E 10 nM and Poly B 2 µg/ml for 16 h. Data are expressed as median ± IQR from at least 3 independent experiments and differences analyzed by Kruskal–Wallis followed by Dunn’s multiple comparison test (IL = interleukin; TNF = tumor necrosis factor; Poly B = polymyxin B)
Next, to investigate whether the lack of NRP1-binding domain in Spike E affects inflammation (hypothesis b), we first investigated whether NRP1 was present in our cells, and we found that it is highly expressed in human MΦ (Supplementary Fig.S1).
We then tested another recombinant Spike (Spike F) which we found to be completely endotoxin-free and which also misses the NRP1-binding sequence but, at variance with Spike E, is fully glycosylated. Spike F had no effects on MΦ pro-inflammatory response compared to control (data not shown), thereby effectively ruling out hypothesis b. Hence, our data suggest that the inflammatory response induced by Spike E should be attributed to its being unglycosylated, a feature specific of the prokaryotic expression system used to produce it.
Discussion
In the present study, we demonstrate that LPS-free recombinant glycosylated SARS-CoV-2 Spike (i.e., the Spike which is generated in infected humans) exerts no stimulatory effect on inflammation in human primary MΦ. Furthermore, we also show that the huge cytokine/chemokine production, observed in human MΦ tested with several commercially available recombinant SARS-CoV-2 Spikes, is due primarily to endotoxin (i.e., LPS) contamination of the recombinant peptides, and importantly that Spike boosts LPS-mediated pro-inflammatory action in human MΦ. We also report that a recombinant non-glycosylated Spike from E. coli triggers inflammation, even in the absence of endotoxin contamination, unlike glycosylated Spike produced in mammalian cells. A putative explanation of these latter findings is the lack of protein glycosylation, which is a well-known viral mechanism to elude detection by the host innate immune system (macrophages) (Watanabe et al. 2020). Thus, our results shed light on the potential confounding factors which might have affected the flourishing literature about the in vitro effects of SARS-CoV-2 Spike protein on macrophage inflammation (Shirato and Kizaki 2021; Chiok et al. 2021; Khan et al. 2021; Pantazi et al. 2021).
To the best of our knowledge, this is the first study assessing the effects of Spike glycoprotein in monocyte-derived human MΦ, and the absence of any Spike-mediated pro-inflammatory effects is in line with a very recent study showing that the treatment with Spike alone (from 0.1 to 10 µg/ml) had no effect on IL-1β secretion in MΦ from SARS-CoV-2-naïve individuals (Theobald et al. 2021).
Nevertheless, several very recent in vitro studies demonstrated a pro-inflammatory action of SARS-CoV-2 spike protein (Olajide et al. 2021; Theobald et al. 2021) in mouse (Shirato and Kizaki 2021; Khan et al. 2021) or THP-1 (Shirato and Kizaki 2021; Khan et al. 2021; Pantazi et al. 2021; Chiok et al. 2021) derived MΦ, by TLR(s) signaling activation. Importantly, none of these studies investigated a possible endotoxin contamination of the recombinant protein tested. In our work, the endotoxin contamination found in commercially available Spike casts some doubts on the significance of the aforementioned studies regarding the role of Spike in immune cell inflammation through TLR(s) activation. Indeed, in our hands, glycosylated truly LPS-free Spike has no inflammatory effects in human primary MΦ.
Our results militate against a relevant role of Spike per se in the cytokine storm of human infection played through a direct pro-inflammatory effect on MΦ. Apparently, Spike goes undetected by human MΦ only when it is glycosylated. On the other hand, the lack of any response at all to glycosylated Spike may imply that SARS-CoV-2 can evade detection by the innate immune system in the very early phases of infection (Tian et al. 2020; Kasuga et al. 2021) and this presumably is an important component of its pathogenic potential.
Endotoxin contamination of recombinant protein is a very common problem when immune cells are involved, as they can be activated by minimal amounts of LPS, equivalent to the levels of endotoxin contamination detected in some commercially available proteins (Schwarz et al. 2014). Accordingly, here we show that also human MΦ are sensitive to low (1 ng/ml) LPS concentration, which correspond to the levels of endotoxins (10 EU/ml) detected in one of the Spike tested (Spike B).
Moreover, it has been reported that recombinant peptides could be contaminated even if labelled as endotoxin-free or expressed in eukaryotic systems (Wakelin et al. 2006). We confirmed that a broad range of endotoxin contamination is found also in recombinant Spike produced in mammalian cells.
Of note, despite 1 ng/ml of LPS was identified as the concentration threshold for MΦ response, we observed that even lower endotoxin levels (i.e., 3 EU/ml) of Spike were associated to inflammation, suggesting that LPS at very low concentrations did not induce inflammation per se, but only when combined to Spike, i.e. Spike can work as a cofactor of the inflammatory action of LPS. This is in accordance with a recent paper showing that SARS-CoV-2 Spike binds to bacterial LPS, leading to changes in LPS biophysical state, thus boosting its pro-inflammatory activity in monocytes and human MNCs through TLR4-NF-κB activation (Petruk et al. 2020). The proven synergy between LPS and Spike is crucial in an attempt to explain the increased risk of severe COVID-19 in conditions characterized by subclinically increased circulating levels of LPS generated by the host gut microbiome, such as metabolic syndrome, obesity, and type 2 diabetes (Cani et al. 2012; Drucker 2021), as well as provide new therapeutic targets. In line with this observation, evidence from porcine animal models demonstrated that infection with the highly prevalent porcine respiratory coronavirus increases the lung sensibility to LPS (Van Gucht et al. 2006).We also explored the effect of a recombinant Spike (Spike E), which lacks the binding domain of NRP-1 (C-terminal peptide 682Arg-Arg-Ala-Arg685 of the S1 subunit), which is a host cell receptor reportedly able to bind Spike and to promote virus entry and infectivity (Cantuti-Castelvetri et al. 2020; Daly et al. 2020). Although our preliminary data showed a high expression of NRP-1 (and NRP-2) genes in human MΦ (Supplementary Fig. S1), we observed that the presence/absence of NRP-1-binding domain did not affect MΦ susceptibility to Spike, indicating that NRP-1 is not involved in the Spike-mediated activation of MΦ pro-inflammatory pathway.
Commercially available Spikes are produced in different expression systems, mainly E. coli, human HEK293, and Chinese hamster ovary (CHO) epithelial cell lines. The different expression system implies a different degree of glycosylation (null or very low in prokaryotic vs extensive in eukaryotic cells) of the final product (Dell et al. 2010; Brooks 2004). We therefore tested a Spike (Spike E) expressed in E. coli, with low or absent protein glycosylation, and found that it was able to elicit a robust inflammatory response in human primary MΦ.
SARS-CoV-2 Spike exhibits both N and O glycosylations (Shajahan et al. 2020; Reis et al., 2021). In particular, two N-glycosylation sites—specifically glycosylated by the machinery of the host (mammalian) cells (Kornfeld and Kornfeld 1985)—have been identified in the receptor-binding domain and are recognized as important mediators of SARS-CoV-2 binding to host cells via the ACE2 receptor (Reis et al., 2021). The different degree of glycosylation may explain the increased inflammatory profile induced by Spike E vs Spike F, which share the same amino acid sequence but are produced in different protein expression system (E. coli vs HEK293, respectively).
Of note, the presence of glycans—arranging in a shield around the RBD—is a common strategy to escape immune surveillance for coronaviruses and other viruses with heavy glycosylated spike proteins (like HIV-1 Env) (Reis et al., 2021). In this framework, our data strongly suggest that prokaryotic-expressed recombinant Spike, naturally lacking post-translational modification at specific glycosylation sites, can be recognized by host immune cells, in primis MΦ, which build a first-line antiviral response.
In line with our hypothesis, some recent works reported a pro-inflammatory effect of prokaryotic-expressed Spike proteins in immune cells (Shirato and Kizaki 2021; Rotoli et al. 2021).
Thus, speculations based on in vitro studies of immune effects of non-glycosylated Spike should be avoided, as it does not correspond to that found in virions, which is highly glycosylated.
Our studies were limited to MΦ; hence, we cannot rule out that the behavior of non-immune cells, like epithelial and endothelial cells, which also are very sensitive to LPS stimulation (Menden et al. 2013), is different. Future experiments are needed to clarify this crucial issue.
Some study limitations must be acknowledged: (1) it is uncertain whether the concentration of Spike used in the present work (10 nM) is comparable to those occurring in infected subjects—of note, the viral load of an infected individual varies from 102 to 1013 copies/ml (Costa et al. 2021) and that the trimeric spike copy number per virion is 26 ± 15 (Yao et al. 2020): the highest figures should generate in vivo a Spike concentration around 2–7 nM; (2) although our data are compatible with a role of Spike glycosylation in macrophage inflammation, future ad hoc experiments should be performed to prove a causal relationship; (3) the extrapolation of our in vitro data to the complex in vivo setting should be made with caution.
Conclusions
In conclusion, this study demonstrates that (a) LPS-free, glycosylated SARS-CoV-2 Spike proteins do not cause inflammation but, rather, Spike protein advances LPS-mediated pro-inflammatory action in primary human MΦ; and (b) the in vitro high cytokine release, induced by recombinant Spike in human MΦ, has to be attributed primarily to LPS contamination of the recombinant peptides. Moreover, we observed that non-glycosylated Spike—which does not represent the protein expressed on SARS-CoV-2 virions in infected mammals—is pro-inflammatory, thereby highlighting the potential role of glycosylation in SARS-CoV-2 pathogenicity.
In vitro studies with commercially available Spike should be conducted with excruciating attention to potential LPS contamination.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. S1. Gene expression of Spike receptors on human macrophages. Gene expression of ACE2, TMPRSS2, NRP1 and NRP2 was tested in cultured macrophages and in A549 (adenocarcinomic human alveolar basal epithelial cells) and Calu-3 (human lung cancer cell line). Gene expression data, normalized to the RPL15 (Ribosomal like protein 15) housekeeping gene, are represented as mean ± SEM from at least 3 independent experiments (PDF 86 KB)
Acknowledgements
We thank Amelia Barilli, Bianca Maria Rotoli, and Valeria Dall’Asta at the University of Parma, Italy, for useful discussions on the results, for suggesting the use of polymyxin B, and for quantifying macrophage gene expression of ACE2, TMPRSS2, NRP1, and NRP2.
Author contribution
GC, VS, ADC, and RCB designed the study, analyzed the data, and wrote the manuscript. NTI, VP, and GC collected and analyzed the data and contributed to the revision of the document. VS, ADC, and RCB critically revised the manuscript for important intellectual content. RCB is the guarantor of this work and is responsible for its integrity. All authors approved the version to be published.
Funding
This work was supported by the University of Parma through “Bando straordinario di Ateneo per progetti di ricerca biomedica in ambito SARS-CoV-2 e COVID-19” to RCB and through “FIL” funds for research to ADC and RCB.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Declarations
Ethics approval
This study protocol was approved by the Institutional Review Board (Comitato Etico Area Vasta Emilia Nord Prot. 44136) and carried out in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Consent to participate
No informed consent was needed, as blood donor material was completely anonymized.
Competing interests
RCB is a member of the following board/advisory panel: MSD, Eli Lilly Ltd, Amgen, Sanofi and invited speaker for Sanofi, MSD, Eli Lilly Ltd, Astra Zeneca, and Janssen. ADC is an invited speaker for MSD, Astra Zeneca, Sanofi, Eli Lilly Ltd, Doc Generici, and Servier. All other authors declare no competing interests.
Footnotes
Riccardo C. Bonadonna and Alessandra Dei Cas jointly supervised this work.
Gloria Cinquegrani and Valentina Spigoni equally contributed to this work
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gloria Cinquegrani, Email: gloria.cinquegrani@unipr.it.
Valentina Spigoni, Email: valentina.spigoni@unipr.it.
Nicolas Thomas Iannozzi, Email: nicolasthomas.iannozzi@studenti.unipr.it.
Vanessa Parello, Email: vanessa.parello@studenti.unipr.it.
Riccardo C. Bonadonna, Email: riccardo.bonadonna@unipr.it
Alessandra Dei Cas, Email: alessandra.deicas@unipr.it.
References
- Bagdonaite I, Wandall HH. Global aspects of viral glycosylation. Glycobiology. 2018;28(7):443–467. doi: 10.1093/GLYCOB/CWY021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler BA. TLRs and innate immunity. Blood. 2009;113(7):1399–1407. doi: 10.1182/BLOOD-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA. Appropriate glycosylation of recombinant proteins for human use: implications of choice of expression system. Appl Biochem Biotechnol Part B Mol Biotechnol. 2004;28(3):241–255. doi: 10.1385/MB:28:3:241. [DOI] [PubMed] [Google Scholar]
- Cani, PD, Osto M, Geurts L, and Everard A. 2012. “Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity.” Gut Microbes 3 (4). 10.4161/GMIC.19625. [DOI] [PMC free article] [PubMed]
- Cantuti-Castelvetri, L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, et al. 2020. “Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity.” Science (New York, N.Y.) 370 (6518). 10.1126/SCIENCE.ABD2985. [DOI] [PMC free article] [PubMed]
- Chiok Kim, Hutchison K, Miller LG, Bose S, Miura TA. Proinflammatory responses in SARS-CoV-2 infected and soluble spike glycoprotein S1 subunit activated human macrophages. Biorxiv. 2021 doi: 10.1101/2021.06.14.448426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol. 2020;92(10):2105–2113. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R, Bueno F, Albert E, Torres I, Carbonell-Sahuquillo S, Barrés-Fernández A, Sánchez D, et al. Upper respiratory tract SARS-CoV-2 RNA loads in symptomatic and asymptomatic children and adults. Clin Microbiol Infect. 2021 doi: 10.1016/J.CMI.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C, Shoemark DK, et al. “Neuropilin-1 is a host factor for SARS-CoV-2 infection”. Science (New York, N.Y.) 2020;370(6518):861–65. doi: 10.1126/SCIENCE.ABD3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell, Anne, Alaa Galadari, Federico Sastre, and Paul Hitchen. 2010. “Similarities and differences in the glycosylation mechanisms in Prokaryotes and Eukaryotes Int J Microbiol 2010. 10.1155/2010/148178 [DOI] [PMC free article] [PubMed]
- Derlindati E, Dei Cas A, Montanini B, Spigoni V, Curella V, Aldigeri R, Ardigò D, Zavaroni I, and Bonadonna RC. Transcriptomic Analysis of Human Polarized Macrophages: More than One Role of Alternative Activation? PLoS ONE. 2015;10(3). 10.1371/journal.pone.0119751. [DOI] [PMC free article] [PubMed]
- Drucker DJ. Diabetes, obesity, metabolism, and SARS-CoV-2 Infection: the end of the beginning. Cell Metab. 2021;33(3):479–498. doi: 10.1016/J.CMET.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Zayat Salwa Refat, Hiba Sibaii, and Fathia A. Mannaa. 2019. “Toll-like receptors activation, signaling, and targeting: an overview.” Bull Natl Res Cent 2019 43:1 43 (1): 1–12. 10.1186/S42269-019-0227-2.
- Van Gucht S, Atanasova K, Barbé F, Cox E, Pensaert M, Van Reeth K. Effect of Porcine respiratory coronavirus infection on lipopolysaccharide recognition proteins and haptoglobin levels in the lungs. Microbes Infect. 2006;8(6):1492. doi: 10.1016/J.MICINF.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustine JN, Jones D. Immunopathology of hyperinflammation in COVID-19. Am J Pathol. 2021;191(1):4–17. doi: 10.1016/J.AJPATH.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán-Beltrán S, Torres M, Arellano M, Juárez E. Human macrophages chronically exposed to lps can be reactivated by stimulation with mdp to acquire an antimicrobial phenotype. Cell Immunol. 2017;315(May):45–55. doi: 10.1016/J.CELLIMM.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, et al. SARS-CoV-2 cell entry depends on ace2 and tmprss2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/J.CELL.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwaciak I, Sałkowska A, Karaś K, Dastych J, Ratajewski M. Nucleocapsid and Spike proteins of the coronavirus SARS-CoV-2 induce IL6 in Monocytes and macrophages-potential implications for cytokine storm syndrome. Vaccines. 2021;9(1):1–10. doi: 10.3390/VACCINES9010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga Y, Zhu B, Jang KJ, Yoo JS. Innate immune sensing of coronavirus and viral evasion strategies. Exp Mol Med. 2021;53(5):723–736. doi: 10.1038/S12276-021-00602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, Shahanshah, Mahnoush S Shafiei, Christopher Longoria, John Schoggins, Rashmin C Savani, and Hasan Zaki. 2021. SARS-CoV-2 Spike Protein Induces Inflammation via TLR2-Dependent Activation of the NF-ΚB Pathway. BioRxiv. http://biorxiv.org/content/early/2021/03/17/2021.03.16.435700.abstract. [DOI] [PMC free article] [PubMed]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/ANNUREV.BI.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, et al. Structure of the SARS-CoV-2 Spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/S41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lester SN, Li K. Toll-like Receptors in antiviral innate immunity. J Mol Biol. 2014;426(6):1246–1264. doi: 10.1016/J.JMB.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42(2):145–151. doi: 10.1016/J.CYTO.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–555. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- Menden, H, Tate E, Hogg N, and Sampath V. 2013. “LPS-mediated endothelial activation in pulmonary endothelial cells: role of Nox2-dependent IKK-β phosphorylation.” Am J Physiol Lung Cell Mol Physiol. 304 (6). 10.1152/AJPLUNG.00261.2012. [DOI] [PMC free article] [PubMed]
- Olajide, Olumayokun A, Victoria U Iwuanyanwu, Izabela Lepiarz-Raba, and Alaa A Al-Hindawi. 2021. “Exaggerated cytokine production in human peripheral blood mononuclear cells by recombinant SARS-CoV-2 Spike glycoprotein S1 and its inhibition by dexamethasone.” [DOI] [PMC free article] [PubMed]
- Pantazi, I, Al-Qahtani AA, Alhamlan FS, Alothaid H, Matou-Nasri S, Sourvinos G, Vergadi E, and Tsatsanis C. 2021. “SARS-CoV-2/ACE2 interaction suppresses IRAK-M expression and promotes pro-inflammatory cytokine production in macrophages.” Front Immunol 12 (June). 10.3389/FIMMU.2021.683800. [DOI] [PMC free article] [PubMed]
- Petruk G, Puthia M, Petrlova J, Samsudin F, Strömdahl AC, Cerps S, Uller L, Kjellström S, Bond PJ, Schmidtchen A. SARS-CoV-2 spike protein binds to bacterial lipopolysaccharide and boosts proinflammatory activity. J Mol Cell Biol. 2020;12(12):916–932. doi: 10.1093/jmcb/mjaa067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, Celso A., Rudolf Tauber, and Véronique Blanchard. 2021. “Glycosylation is a key in SARS-CoV-2 infection.” Journal of Molecular Medicine. Springer Science and Business Media Deutschland GmbH. 10.1007/s00109-021-02092-0. [DOI] [PMC free article] [PubMed]
- Rotoli, BM, Barilli A, Visigalli R, Ferrari F, and Dall’Asta V. 2021. “Endothelial cell activation by SARS-CoV-2 Spike S1 protein: a crosstalk between endothelium and innate immune cells.” Biomedicines 9 (9). 10.3390/BIOMEDICINES9091220. [DOI] [PMC free article] [PubMed]
- Roy, S, Bag AK, Singh RK, Talmadge JE, Batra SK, and Datta K. 2017. “Multifaceted Role of neuropilins in the immune system: potential targets for immunotherapy.” Front Immunol. 8 (OCT). 10.3389/FIMMU.2017.01228. [DOI] [PMC free article] [PubMed]
- Schmittgen TD, Zakrajsek MAG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study MRNA decay: comparison of endpoint and real-time methods. Anal Biochem. 2000;285(2):194–204. doi: 10.1006/ABIO.2000.4753. [DOI] [PubMed] [Google Scholar]
- Schultze JL, Aschenbrenner AC. COVID-19 and the human innate immune system. Cell. 2021;184(7):1671–1692. doi: 10.1016/J.CELL.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, Harald, Maria Schmittner, Albert Duschl, and Jutta Horejs-Hoeck. 2014. “Residual endotoxin contaminations in recombinant proteins are sufficient to activate human CD1c+ dendritic cells.” PLoS One. 9 (12). 10.1371/journal.pone.0113840. [DOI] [PMC free article] [PubMed]
- Shajahan A, Supekar NT, Gleinich AS, Azadi P. Deducing the N- and o-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato, Ken, and Takako Kizaki. 2021. “SARS-CoV-2 Spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages.” Heliyon. 7 (2). 10.1016/j.heliyon.2021.e06187. [DOI] [PMC free article] [PubMed]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Investig. 2012;122(3):787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigoni, Valentina, Federica Fantuzzi, Cecilia Carubbi, Giulia Pozzi, Elena Masselli, Giuliana Gobbi, Anna Solini, Riccardo C. Bonadonna, and Alessandra Dei Cas. 2020. “sodium-glucose cotransporter 2 inhibitors antagonize lipotoxicity in human myeloid angiogenic cells and ADP-dependent activation in human platelets: potential relevance to prevention of cardiovascular events.” Cardiovasc Diabetol. 19 (1). 10.1186/s12933-020-01016-5. [DOI] [PMC free article] [PubMed]
- Spigoni V, Fantuzzi F, Fontana A, Cito M, Derlindati E, Zavaroni I, Cnop M, Bonadonna RC, Cas AD. Stearic acid at physiologic concentrations induces in vitro lipotoxicity in circulating angiogenic cells. Atherosclerosis. 2017;265(October):162–171. doi: 10.1016/j.atherosclerosis.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Tarique, Abdullah A., Jayden Logan, Emma Thomas, Patrick G. Holt, Peter D. Sly, and Emmanuelle Fantino. 2015. “Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages.” 10.1165/Rcmb.2015-0012OC 53 (5): 676–88. 10.1165/RCMB.2015-0012OC. [DOI] [PubMed]
- Theobald, Sebastian J, Alexander Simonis, Theodoros Georgomanolis, Christoph Kreer, Matthias Zehner, Hannah S Eisfeld, Marie-Christine Albert, et al. 2021. “Long-lived macrophage reprogramming drives spike protein-mediated inflammasome activation in COVID-19.” EMBO Mol Med. e14150 10.15252/EMMM.202114150. [DOI] [PMC free article] [PubMed]
- Tian, W, Zhang N, Jin R, Feng Y, Wang S, Gao S, Gao R, et al. 2020. “Immune suppression in the early stage of COVID-19 disease.” Nat Commun. 11 (1). 10.1038/S41467-020-19706-9. [DOI] [PMC free article] [PubMed]
- Tsubery H, Ofek I, Cohen S, Fridkin M. The functional association of polymyxin B with bacterial lipopolysaccharide is stereospecific: studies on polymyxin B nonapeptide. Biochemistry. 2000;39(39):11837–11844. doi: 10.1021/BI000386Q. [DOI] [PubMed] [Google Scholar]
- Wakelin SJ, Sabroe I, Gregory CD, Poxton IR, Forsythe JL, Garden OJ, Howie SE. ‘Dirty Little secrets’–endotoxin contamination of recombinant proteins. Immunol Lett. 2006;106(1):1–7. doi: 10.1016/J.IMLET.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Watanabe Yasunori, Berndsen Zachary T, Raghwani Jayna, Seabright Gemma E, Allen Joel D, Pybus Oliver G, McLellan Jason S, et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, Zhang J, et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183(3):730–738.e13. doi: 10.1016/J.CELL.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Wang B, Mao J. The pathogenesis and treatment of the `cytokine storm’ in COVID-19. J Infect. 2020;80(6):607–613. doi: 10.1016/J.JINF.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kuang M, Li J, Zhu L, Jia Z, Guo X, Hu Y, et al. SARS-CoV-2 Spike Protein Interacts with and Activates TLR41. Cell Res. 2021. 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMOA2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Gene expression of Spike receptors on human macrophages. Gene expression of ACE2, TMPRSS2, NRP1 and NRP2 was tested in cultured macrophages and in A549 (adenocarcinomic human alveolar basal epithelial cells) and Calu-3 (human lung cancer cell line). Gene expression data, normalized to the RPL15 (Ribosomal like protein 15) housekeeping gene, are represented as mean ± SEM from at least 3 independent experiments (PDF 86 KB)
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Not applicable.