Abstract
A new quantitative reverse transcription (RT)-PCR assay for human immunodeficiency virus type 1 (HIV-1) RNA (Abbott LCx HIV RNA Quantitative assay) has been compared with the Organon NucliSens assay on 521 retrospective samples obtained from HIV-1-positive patients monitored during highly active antiretroviral therapy, 79 of whom were assayed also by the Chiron Quantiplex 3.0 system and on characterized panels. The LCx system showed a moderate correlation (r = 0.795) and gave higher results than the NucliSens system on 245 of 327 concordant positive samples, with similar sensitivity. Correlation with Quantiplex system results was higher (r = 0.943). LCx reproducibility was very good; the procedure was simple, well controlled, and rapid (up to 48 results in 7 h). The HIV RNA quantitative assay on the LCx system is suitable for routine use.
Several data indicate that the quantitative evaluation of viral-RNA levels is an important prognostic factor in human immunodeficiency virus type 1 (HIV-1) infection. In acute infection and during the symptomless stage (14), high viral-RNA levels are correlated with a faster disease progression, and pregnant women with higher viral-RNA levels are considered at higher risk than those with lower viral-RNA levels for transmission of the infection to their offspring. In patients who are candidates for antiretroviral treatment, it is appropriate to carry out two determinations 1 to 2 weeks apart before starting the therapy (to obtain baseline levels), then one determination within 1 or 2 months after starting therapy, and, if the results show therapy to be effective (i.e., reduction of the viral load of >0.5 log10 or threefold), the patient should be retested every 3 or 4 months thereafter. If the clinical or immune state is modified, evaluations of viral load should be performed at closer intervals than those recommended above (4). Three commercial assays for the measurement of viral load have been developed during the last few years (1, 12, 13, 21), based on three different methods: target amplification by reverse transcription-PCR or by nucleic acid sequence-based amplification (NASBA) or signal amplification by branched-DNA (bDNA) analysis. These assays have ensured more standardization of in-house procedures, but the quantitative measurements of HIV RNA obtained with each method are seldom if ever replicated by the others (11, 15, 19), and this adds up to the variability in HIV-1 RNA results that may exist between laboratories.
In this study we evaluated a new assay for the quantitative determination of HIV-1 RNA in plasma specimens (LCx HIV RNA Quantitative assay; Abbott Laboratories, North Chicago, Ill.) on selected samples obtained from the ca. 10,000 samples routinely processed in our laboratory each year.
Five hundred and twenty-one plasma specimens from patients undergoing highly active antiretroviral therapy and tested for HIV RNA using the NASBA (Organon NucliSens) system were included in this evaluation; 79 of these 521 samples were also tested with a third-generation bDNA method (Chiron Quantiplex 3.0). Samples were processed by the LCx HIV RNA Quantitative assay. Briefly, after addition of an internal standard (IS) that differed slightly from the target sequence, the assay controls and clinical samples were extracted by centrifugation and separation using a QIAAmp column and subjected to amplification with primers targeted at a highly conserved 170-bp sequence within the HIV-1 pol region, which enabled this assay to detect HIV-1 of the M group (A to G subtypes) and the O group. IS and HIV-1 primers were conjugated with two different synthetic capture haptens. Target and IS amplifications were carried out simultaneously in a Perkin-Elmer 4800 thermal cycler in a ready-to-use, closed tube by reverse transcription-PCR (Thermus thermophilus polymerase) that contained all necessary reagents. Two different specific probes, one for the HIV-1 target and the other for the IS-produced target, conjugated with different haptens from the primers (detection haptens), were added during the last step of amplification. The tubes were then transferred unopened into an automatic analyzer (LCx assay) for detection and quantification using the Microparticle Enzyme Immunoassay (MEIA) system (7). Two different conjugates (alkaline phosphatase and beta-galactosidase) are employed; the first one binds to the HIV-1-specific probes, and the second to the IS-specific probes. The two substrates (7-beta galactosidase coumarin-4-acetic [2-hydroxyethylamine] and 4-methylumbelliferyl phosphate) were added in succession, and the resulting fluorescence was read by the MEIA optic system. A calibration was carried out, with each different lot of LCx-assay-determined HIV-1 RNA, by testing, in duplicate, six calibrators in which the HIV RNA and IS concentrations increased and decreased inversely from calibrator 1 through 6. The results were used with the LCx software to draw a curve on which sample and control results were subsequently calculated; the logarithm (log10) ratio of the fluorescent measure of HIV and IS was calculated and related to the ratio values obtained from the calibration curve, in order to extrapolate the HIV-1 RNA concentration for each sample or control. The resulting RNA concentration value in a sample can be expressed either as HIV-1 RNA copies per milliliter or as log10 copies per milliliter. Extraction and amplification procedures and the possible presence of inhibitors were controlled by the IS. The instrument software was used to carry out a second control for a possible inhibition of RNA amplification by checking the fitness of HIV/IS ratios on each sample and control. Three control levels (negative, low positive, and high positive) were required to be tested for each run and had to give valid results for the LCx to express the HIV RNA values for clinical samples.
The assay dynamic range was between 178 copies/ml (2.25 log10 [low limit of detection {LLD}]) and 5 million copies/ml (6.70 log10 copies) with a 0.2-ml sample procedure and between 50 copies/ml (LLD) and 1 million copies/ml (1.70 to 6 log10 copies) when 1 ml of plasma was employed. The 0.2-ml procedure was used for all of the clinical samples and panel members, while the 1.0-ml procedure was used only for 68 selected samples that were below the LLD with the 0.2-ml procedure and on the Boston Biomedica Inc. (BBI) panel.
The Organon NucliSens assay (6, 18) and the Chiron Quantiplex 3.0 bDNA assay (8, 10) were used according to the manufacturer's recommendations. The former uses the NASBA isothermal-target amplification for an HIV-1 gag sequence on 1 ml of human plasma, with a reportable range between 80 and 1 million copies/ml (1.90 to 6.0 log10). The procedure can be partially automated by the use of nucleic acid extraction. The latter is also performed on 1 ml of plasma and has an analytical range between 1.70-log10 and 5.70-log10 HIV RNA copies/ml.
A commercial panel (BBI QRD 701 [one negative and five positive specimens]) and two samples from the National External Assessment (NEQAS) external quality control program were tested to check LCx accuracy. For both panels the reference values and values obtained by means of other HIV-1 RNA-quantitative methods currently on the market were already known. LCx values (0.2-ml and 1.0-ml procedures) were all within the expected range for all samples and also reflected the declared sensitivity. Testing of the two specimens from the NEQAS quality control program yielded a difference between the two samples of 0.62 log10 by LCx, while with the NASBA system the difference was 1.07 log10, closer to the expected difference of 1 log10.
The two LCx assay positive controls were assayed singly over 39 sessions using four different batches of reagents and both the 0.2-ml and the 1.0-ml LCx procedures. All results were valid, and the reproducibility was very good for both the low positive (log10 concentration, 3.573 ± 0.120; coefficient of variation [CV], 3.4%) and the high positive control (log10 concentration, 5.658 ± 0.185; coefficient of variation, 3.3%). Retesting of 17 positive clinical specimens, whose values ranged from 3.56 to 5.32 log10 copies/ml, with a different lot of reagents yielded a mean log10 variation of 0.160 plus or minus a standard error of 0.128, and the difference between replicates ranged from −0.01 to 0.45 log10 copies/ml.
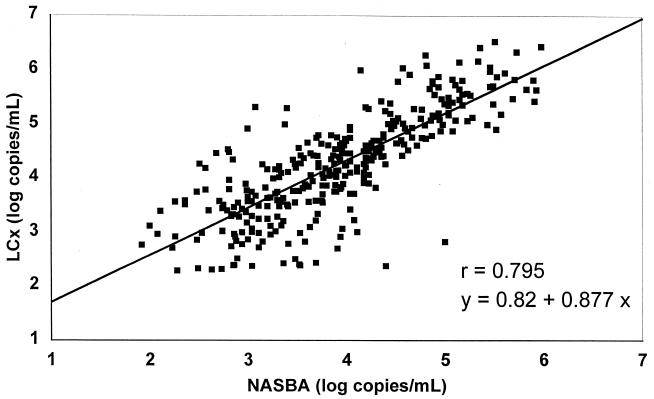
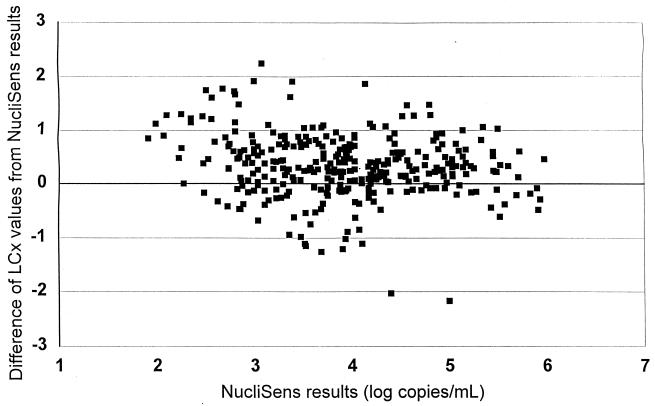
Out of 521 clinical specimens, 382 were above the limit of detection by the NucliSens assay (73.3%) and 360 were above the limit of detection by the LCx assay (69.1%), but the difference was not statistically significant (P > 0.05 by the chi-square test). The qualitative comparison of assay results showed a concordance on 423 samples (81.2%), i.e., 322 positive and 101 negative; the NucliSens assay yielded measurable results on 60 samples (11.5%) that were below the LLD by the LCx assay, and, conversely, 38 samples (7.3%) were HIV-1 positive by the LCx assay but negative by the NucliSens assay. We also compared the rates of negativity with each assay for the HIV RNA log10 values on 523 samples. On 138 samples with a NucliSens result of <2.25 log10, 101 (73.2%) were below the LLD by LCx as well; of the 385 samples that were >2.25 log10 by NucliSens assay, the percentage of negative samples by LCx assay decreased as NucliSens results increased, from 36.8% (28 of 76) in the 2.25 to 3 log10 NucliSens results group to 18.3% (30 of 164) in the 3.01 to 4 log10 group and to 2% (2 of 101) for the 4.01 to 5 log10 class. In the reverse comparison, 62.7% of the samples of <2.25 log10 by LCx assay (101 of 161) were also <2.25 log10 by NucliSens assay, and the NucliSens assay method also showed a decrease in the percentage of negative results as the positive values for LCx increased: 35.8% (19 of 53) for the 2.25 to 3 log10 LCx class, 13.7% (14 of 102) for the 3.01 to 4 log10 LCx class, 3.1% (4 of 130) for the 4.01 to 5 log10 LCx class, and 1.3% (1 of 77) for the 5.01 to 6 log10 LCx class. The quantitative results for the 327 specimens that were >LLD by both LCx and NucliSens assays were compared by a linear regression analysis (Figure 1), which showed a moderate correlation (r = 0.795). A very similar value (r = 0.802) was observed when NucliSens and Quantiplex results on the 79 samples assayed in triplicate were compared, while, on those samples, the correlation between LCx and Quantiplex results was much better (r = 0.943). The regression data suggest also that LCx results were generally higher than NucliSens or Quantiplex results, since positive intercepts of 0.82 and 0.83, respectively, have been observed. Indeed, a difference plot analysis of the log10 values between LCx and NucliSens (Figure 2) showed LCx results to be generally higher compared with the whole range of NucliSens results. On the 327 samples above LLD by both assays, 245 (75%) were higher by LCx assay, 80 (24.5%) were higher by the NASBA method, and 2 (0.5%) gave exactly the same value. The majority of samples (191 [58.4%]) fell within a difference of ≤0.5 log10, while 94 (28.7%) showed a difference between 0.5 and 1 log10. We considered the results to be truly discordant when a difference of 1 log10 or more was observed, and there were 42 such samples (12.8% of samples above LLD on both assays), 34 of which (81% of discordant results) showed higher values by the LCx assay.
FIG. 1.
Linear regression analysis of HIV-1 RNA quantitative assay results (log10 copies/ml) between LCx and NucliSens assay methods on 327 plasma specimens with results >LLD by both assays.
FIG. 2.
Difference plot of LCx HIV RNA Quantitative-assay values according to NucliSens results (log10 copies/ml) on 327 plasma specimens with results >LLD by both assays.
Table 1 shows the results for the 68 samples that were below LLD, as determined by the LCx assay with the 0.2-ml procedure, and were retested with the 1.0-ml LCx procedure, which increased the sensitivity to 50 HIV RNA copies/ml, and the table also reports the final HIV RNA concentrations that were below LLD but comprised between 20 and 49 copies/ml. Eleven out of 14 samples with <80 RNA copies as determined by NucliSens assay were also below the LLD by LCx assay, 2 had levels between 50 and 178 copies, and 1 gave a value of >10,000 copies/ml (this last specimen, however, was highly hemolyzed). Of the 26 samples classified at between 81 and 1,000 copies/ml by NucliSens assay, 13 were classified at <50 copies/ml, 3 at between 50 and 178 copies/ml, and 10 exceeded 178 copies/ml (range, 350 to 3,000 copies/ml). Also, among the 26 samples with NucliSens results between 1,000 and 10,000 copies/ml, the majority (16 samples) were classified at <50 copies by LCx, and only 8 exceeded 178 copies (range, 200 to 2,400 copies/ml). Both samples classified in the >10,000-copies/ml group by NucliSens assay were in the <50-copies/ml group with the LCx assay. Unfortunately, the small amount of plasma available did not allow retesting by NucliSens assay.
TABLE 1.
Retesting, with the LCx 1.0-ml procedure, of 68 samples that yielded <178 copies with the LCx 0.2-ml procedure
| NucliSens results | LCx 1.0 ml results (RNA copies/ml)
|
||||
|---|---|---|---|---|---|
| <20 | 20–50 | 51–178 | >178 | Total | |
| <80 | 9 | 2 | 2 | 1a | 14 |
| 81–1,000 | 11 | 2 | 3 | 10 | 26 |
| 1,000–10,000 | 13 | 3 | 2 | 8 | 26 |
| >10,000 | 2 | 0 | 0 | 0 | 2 |
| Total | 35 | 7 | 7 | 19 | 68 |
Hemolyzed sample.
Of a total of 685 determinations by the LCx method, 25 results (3.6%) were invalid and flagged by the instrument either for sample unsuitability (extraction problems or inhibitory factors, 20 [2.9% of cases]) or for reasons related to the LCx instrument hardware (5 [0.7%]). After a second aliquot of the samples already extracted was retested, only five specimens (0.7%) still gave invalid results, all due to sample unsuitability.
The determination of HIV-1 viral load may be influenced by several factors, either linked to assay methodology (e.g., extraction of RNA from plasma, efficiency of the amplification method, or methods employed for detecting the amplification and reporting results) or to the virus itself. For the latter, it has been demonstrated that HIV variants can influence viral-load determination considerably (15, 16, 17). The development of a quantitative test was based on the B subtype of HIV-1, which is dominant in the United States and in Europe. Over the last 10 years, other subtypes have been found circulating also in Western countries (2, 20) in different percentages; therefore, a periodic evaluation of quantitative methods for HIV-1 RNA is necessary to guarantee a constant and accurate quantification over time, as changes occur in virus spread.
The differences among assay results have been confirmed in our experience, since we found only a moderate correlation between results with LCx and NucliSens procedures (r = 0.795), although a better one was seen between LCx and Quantiplex 3.0 (Chiron) procedures (r = 0.943). Interestingly, the correlation between NucliSens and bDNA assay methods was also poor, and this is at variance with the 0.912 correlation reported by Ginocchio, et al. (9). As a partial explanation of this finding, we must consider that the two procedures determine the presence of HIV RNA by amplifying sequences in the gag region for the NucliSens assay and in the pol region for the LCx assay. This difference is stressed in a recent study that compared the current NucliSens assay and a new version, with primers aimed at the long terminal repeat region (5), and evidenced many differences in HIV-1 RNA results, especially among non-HIV-1B-infected samples. Several papers have compared different methods for HIV RNA quantitation (5, 6, 9, 10, 17, 18, 19, 22); in some instances the statistical correlation between assay results has been good (6, 17) or even excellent (9, 10). In a comparative study of the newest versions of Chiron Quantiplex and Roche Amplicor assays (10), Highbarger et al. found r to be as high as 0.98 and suggested that a mathematical equation could be employed to convert the results from one method to the other. Indeed even in that observation the difference in absolute values was, in quite a few cases, higher than 0.5 log10, i.e., more than threefold the absolute value. In our experience, this occurred in 8% of the specimens assayed by LCx and NucliSens assays. In clinical practice, this difference reflects a high probability of obtaining divergent results on single patients or single results from the same patient, which indicates that results obtained from such tests are not interchangeable (4) and which discourages converting the results obtained with any given assay for viral load into any other assay's results with simple formulas. Brambilla et al. (3) have proposed the adoption of a common external standard to adjust the results obtained by different methods. This standard would be useful when comparing clinical-trial results (22) but could prove difficult in clinical practice, and thus it seems advisable to recalculate baseline HIV RNA levels when switching methods.
The performance characteristics of the LCx HIV assay were quite good. Since the nominal sensitivities of NucliSens and the 0.2-ml LCx procedures are different (80 versus 178 copies/ml, respectively), a higher sensitivity of the former was expected, but the difference between the two assays was indeed not significant, and the analysis of 68 low-level samples demonstrated that in only 6 of them (8.8%) were HIV-1 RNA levels between 80 and 178 copies/ml.
On the basis of our experience, we can say that HIV RNA quantitative determination by the LCx assay is a simple and robust procedure which proved adequate for our routine uses, since we were able to process 40 and once even 60 clinical samples on the same day, and this assay guarantees good control over possible problems inherent in the use of PCR. The reproducibility was very good both on assay controls and on the 17 clinical samples that were retested with a different lot of reagents; if these data are confirmed in other experiments, the LCx assay will achieve the proposed limit for intra-assay standard deviation (22) that has been suggested in order to guarantee a good capability of recognizing changes in RNA concentration. Viral-load measurement has become one of the most important clinical markers; a precise, accurate, and reliable quantitation with commercial assays is indeed possible, but we would recommend using the same method and also possibly the same laboratory to monitor patients (4).
REFERENCES
- 1.Bagnarelli P, Menzo S, Valenza A, Paolucci S, Petroni S, Scalise G, Sampaolesi R, Manzin A, Varaldo P E, Clementi M. Quantitative molecular monitoring of human immunodeficiency virus type 1 activity during therapy with specific antiretroviral compounds. J Clin Microbiol. 1995;33:16–23. doi: 10.1128/jcm.33.1.16-23.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barin F, Courouce A-M, Pillonel J, Buzelay L. Increasing diversity of HIV-1 M serotypes in French blood donors over a 10-year period (1985–95) AIDS. 1997;11:1503–1508. doi: 10.1097/00002030-199712000-00015. [DOI] [PubMed] [Google Scholar]
- 3.Brambilla D, Leung S, Lew J, Todd J, Herman S, Cronin M, Shapiro D E, Bremer J, Hanson C, Hillyer G V, McSherry G D, Sperling R S, Coombs R W, Reichelderfer P S. Absolute copy number and relative change in determination of human immunodeficiency virus type 1 RNA in plasma: effect of an external standard on kit comparison. J Clin Microbiol. 1998;36:311–314. doi: 10.1128/jcm.36.1.311-314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. Morb Mortal Wkly Rep. 1998;47(RR-5):42–82. [PubMed] [Google Scholar]
- 5.de Baar M P, van der Schoot A M, Goudsmith J, Jacobs F, Ehren R, van der Horn K H, Oudshoorn P, de Wolf F, de Ronde A. Design and evaluation of a human immunodeficiency virus type 1 RNA assay using nucleic acid sequence-based amplification technology able to quantify both group M and O viruses by using the long terminal repeat as target. J Clin Microbiol. 1999;37:1813–1818. doi: 10.1128/jcm.37.6.1813-1818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyer J R, Pilcher C D, Shepard R, Schock J, Eron J J, Fiscus S A. Comparison of NucliSens and Roche Monitor assays for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1999;37:447–449. doi: 10.1128/jcm.37.2.447-449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiore M, Mitchell J, Doan T, Nelson R, Winter G, Grandone C, Zeng K, Haraden R, Smith J, Harris K, Leszcynski J, Berry D, Safford S, Barnes G, Scholnick A, Luddington K. The Abbott IMx automated benchtop immunochemistry analyzer system. Clin Chem. 1988;34:1726–1732. [PubMed] [Google Scholar]
- 8.Gale H. Evaluation of the Quantiplex human immunodeficiency virus type 1 RNA 3.0 assay in a tertiary-care centre. J Clin Microbiol. 2000;7:122–124. doi: 10.1128/cdli.7.1.122-124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginocchio C C, Tetali S, Washburn D, Zhang F, Kaplan M H. Comparison of levels of human immunodeficiency virus type 1 RNA in plasma as measured by the NucliSens nucleic acid sequence-based amplification and Quantiplex branched-DNA assays. J Clin Microbiol. 1999;37:1210–1212. doi: 10.1128/jcm.37.4.1210-1212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Highbarger H C, Alvord W G, Jiang M-K, Shah A K, Metcalf J A, Lane H C, Dewar R L. Comparison of the Quantiplex version 3.0 assay and a sensitised Amplicor Monitor assay for measurement of human immunodeficiency virus type 1 RNA levels in plasma samples. J Clin Microbiol. 1999;37:3612–3614. doi: 10.1128/jcm.37.11.3612-3614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holguin A, de Mendoza C, Soriano V. Comparison of three different commercial methods for measuring plasma viraemia in patients infected with non-B HIV-1 subtypes. Eur J Clin Microbiol Infect Dis. 1999;18:256–259. doi: 10.1007/s100960050273. [DOI] [PubMed] [Google Scholar]
- 12.Holodniy M, Katzenstein D A, Sengupta S, Wang A M, Casipit C, Schwartz D H, Konrad M, Groves E, Merigan T C. Detection and quantification of human immunodeficiency virus RNA in patient serum by use of the polymerase chain reaction. J Infect Dis. 1991;163:862–866. doi: 10.1093/infdis/163.4.862. [DOI] [PubMed] [Google Scholar]
- 13.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellors J, Kingsley L, Rinaldo C, Gupta P, White R M, Todd J A, Kingsley L A. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 15.Nolte F S, Boysza J, Thurmond C, Clark W S, Lennox J L. Clinical comparison of an enhanced-sensitivity branched-DNA assay and reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:716–720. doi: 10.1128/jcm.36.3.716-720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parekh B, Phillips S, Granade T C, Baggs J, Hu D J, Respess R. Impact of HIV type 1 subtype variation on viral RNA quantitation. AIDS Res Hum Retrovir. 1999;15:133–142. doi: 10.1089/088922299311556. [DOI] [PubMed] [Google Scholar]
- 17.Pasquier C, Sandres K, Salama G, Puel J, Izopet J. Using RT-PCR and bDNA assays to measure non-clad B HIV-1 subtype RNA. J Virol Methods. 1999;81:123–129. doi: 10.1016/s0166-0934(99)00057-9. [DOI] [PubMed] [Google Scholar]
- 18.Segondy M, Ly T-D, Lapeyre M, Montes B. Evaluation of the NucliSens HIV-1 QT assay for quantitation of human immunodeficiency virus type 1 RNA levels in plasma. J Clin Microbiol. 1998;36:3372–3374. doi: 10.1128/jcm.36.11.3372-3374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segondy M, Izopet J, Pellegrin I, Montes B, Dumon B, Pasquier C, Peeters M, Fleury H J A, Puel J, Reynes J. Comparison of the Quantiplex HIV-1 RNA 2.0 assay with the Amplicor HIV-1 Monitor 1.0 assay for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma of patients receiving stavudine-didanosine combination therapy. J Clin Microbiol. 1998;36:3392–3395. doi: 10.1128/jcm.36.11.3392-3395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnerborg A, Durdevic S, Giesecke J, Salleberg M. Dynamics of HIV-1 subtype distribution in the Swedish HIV-1 epidemic during the period 1980 to 1993. AIDS Res Hum Retrovir. 1997;13:343–346. doi: 10.1089/aid.1997.13.343. [DOI] [PubMed] [Google Scholar]
- 21.Van Gemen B, Kievits T, Schukkink R, van Strijp D, Malek L T, Sookanan R, Huisman H G, Lens P. A one tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J Virol Methods. 1994;49:157–167. doi: 10.1016/0166-0934(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 22.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, King H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS clinical trials group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]