Abstract
Bone marrow-derived endothelial progenitor cells (EPCs) play a fundamental role in postnatal angiogenesis. Currently, EPCs are defined as early and late EPCs based on their biological properties and their time of appearance during in vitro culture. Reports have shown that early EPCs share common properties and surface markers with adherent blood cells, especially CD14+ monocytes. Distinguishing early EPCs from circulating monocytes or monocyte-derived macrophages (MDMs) is therefore crucial to obtaining pure endothelial populations before they can be applied as part of clinical therapies. We compared the gene expression profiles of early EPCs, blood cells (including peripheral blood mononuclear cells, monocytes, and MDMs), and various endothelial lineage cells (including mature endothelial cells, late EPCs, and CD133+ stem cells). We found that early EPCs expressed an mRNA profile that showed the greatest similarity to MDMs than any other cell type tested. The functional significance of this molecular profiling data was explored by Gene Ontology database search. Novel plasma membrane genes that might potentially be novel isolation biomarkers were also pinpointed. Specifically, expression of CLEC5A was high in MDMs, whereas early EPCs expressed abundant SIGLEC8 and KCNE1. These detailed mRNA expression profiles and the identified functional modules will help to develop novel cell isolation approaches that will allow EPCs to be purified; these can then be used to target cardiovascular disease, tumor angiogenesis, and various ischemia-related diseases.
Keywords: Early endothelial progenitor cell (EPCs), Monocyte, Macrophage, Gene expression profile, Surface marker
INTRODUCTION
Defects in angiogenesis (blood vessel growth) or vessel repair are two of the major complications associated with cardiovascular disorders. Postnatal neoangiogenesis and vasculogenesis rely on circulating endothelial progenitor cells (EPCs), which were first identified by Asahara et al. in 1997 (1). EPCs are thought to derive from CD133+ stem cells that reside in bone marrow (2), and a reduced level of circulating EPCs has been found to serve as a biomarker that correlates with an increased frequency of cardiovascular events as well as more frequent death from cardiovascular causes (3–5). It has also become clear that the activation state of these cells is critical to their ability to carry out the vessel repair process.
Given their involvement in pathological and physiological angiogenesis, there has been growing interest in manipulating EPCs for therapeutic purposes. However, recently there has been much controversy in the field over the exact definition and functionality of an EPC. Current EPC definitions are based predominantly on cell phenotype and the biological properties of the cell. Early EPCs (eEPCs; also referred to as circulating angiogenic cells) appear early (<1 week) in culture dishes, whereas late EPCs (also referred to as late outgrowth EPCs or endothelial colony-forming cells) appear late (2–4 weeks) and have a cobblestone-like morphology (1). Strikingly different angiogenic functions have been found to be associated with these two EPC subpopulations. Specifically, late outgrowth EPCs, but not eEPCs, form vascular networks de novo and can be incorporated into vascular networks; these cells can therefore be regarded as true EPCs (6). In contrast, eEPCs indirectly augment tubulogenesis even when physically separated from their target by a Transwell membrane, which implies that their effectiveness occurs via a cytokine-based paracrine mechanism (6,7). We have recently shown that eEPCs can abundantly express various inflammatory cytokines and paracrine angiogenic factors, including HGF, CCL3, CCL4, CCL18, CXCL16, and IL-10; these can promote angiogenesis in a paracrine manner (8).
Early EPCs are believed to represent a cell population that is enriched in monocytes, and this population exerts its angiogenic effects via paracrine and signaling mechanisms (9,10). It has been found that eEPCs can function as antigen-presenting cells (11). The antigen-presenting ability of eEPCs is weaker than that of monocyte-derived dendritic cells, but stronger than that of peripheral blood monocytes (11). EPCs carrying monocyte markers were recently shown to be selectively abnormal in type 1 diabetic patients with early retinopathy (12). Differences in CD14+ expression have been proposed as a means of distinguishing mononuclear cells (MNCs) that represent a source of either early or late EPCs (13). The majority of eEPCs have been suggested to arise from CD14+ subpopulations of peripheral blood mononuclear cells (PBMCs), whereas late outgrowth ones, which have the potential to differentiate toward endothelial cells, have been reported to be derived from the CD14− MNC fractions (13,14). Gene expression microarray and proteomics analysis have shown that eEPCs are hematopoietic cells with a molecular phenotype linked to monocytes; this means that they should be used with caution because of their immunomodulatory nature.
As it has been shown that lineage and functional heterogeneity are present in the population of circulating cells capable of assuming an endothelial phenotype, it is critical to be able to separate out a pure population of eEPCs from immune cells such as CD14+ monocytes and monocyte-derived macrophages (MDMs) (14). Currently, circulating EPCs are enumerated by flow cytometry (15). Few reports exist in the literature concerning the characteristics of eEPCs, especially in relation to their surface antigen expression, other than endothelial markers. In this investigation, our aim is to better define the gene expression patterns of eEPCs and hematological cells that closely resemble them, especially monocytes and MDMs. Novel surface proteins differentially expressed between eEPCs and MDMs are identified in this study and hold the potential to be new biomarkers that will allow a more accurate isolation and/or quantification of eEPCs.
MATERIALS AND METHODS
Isolation and Cultivation of EPCs From Cord Blood
All patients gave informed consent, and the study was approved by the local research ethics committee. The protocols of this study were consistent with ethical guidelines provided in the 1975 Helsinki Declaration. EPC isolation and characterization were done as described previously with minor modifications (16,17). In brief, cord blood MNCs were isolated by Histopaque-1077 (1.077 g/ml; Sigma, St. Louis, MO, USA) density-gradient centrifugation. MNCs (1 × 107) were plated in 2 ml of endothelial growth medium-2 (Lonza Ltd., Basel, Switzerland) with supplementation (hydrocortisone, IGF-1, human EGF, human VEGF, human FGF-B, ascorbic acid, GA-1000, heparin, and 2% fetal bovine serum) on fibronectin-coated six-well plates at 37°C in a 5% CO2 incubator. After 3 days of culturing, attached eEPCs appeared, and medium and nonadherent cells were removed. Thereafter, the medium was replaced every 2 days, and a certain number of eEPCs continued to grow into the late EPC colonies, which emerged 2–4 weeks after the start of MNC culture. The late EPCs exhibited “cobblestone” morphology and a monolayer growth pattern that is typical of mature endothelial cells at confluence.
Characterization of Early and Late EPCs
The early and late EPCs were also assessed for endothelial and progenitor markers by indirect immunostaining with 1,19-dioctadecyl-3,3,39,39-tetramethylindocarbocyanine perchlorate-acetylated low-density lipoprotein (DiI-acLDL; Molecular Probes, Invitrogen, Carlsbad, CA, USA) and costaining with Bandeiraea simplicifolia lectin I (Sigma). Briefly, the adherent cells were first incubated with DiI-acLDL (2.4 µg/ml) for 1 h and then fixed in 4% paraformaldehyde and counterstained with FITC-labeled lectin (10 µg/ml) from Ulex europaeus (UEA-1; Sigma). The fluorescent images were recorded under a laser scanning confocal microscope.
The antibodies used in FACS to characterize the adherent cell population were kinase insert domain receptor (KDR)/VEGF receptor 2 (R&D Systems, Minneapolis, MN, USA), AC133 (CD133), platelet endothelial cell adhesion molecule-1 (CD31; Miltenyi Biotec GmBH, Bergisch Gladbach, Germany), and CD45 (Biolegend, San Diego, CA, USA). Flow cytometry was performed using a FACSCanto flow cytometer (BD Pharmingen, Franklin Lakes, NJ, USA).
Culture of CD14+ MDMs
Macrophage preparation was done as described (18). In brief, peripheral MNCs were isolated from the blood of healthy donors by standard density gradient centrifugation with Ficoll-Paque (Amersham Biosciences, Piscataway, NJ, USA). CD14+ cells were subsequently purified from peripheral MNCs by high-gradient magnetic sorting using the VARIOMACS technique with anti-CD14 microbeads (Miltenyi Biotec). CD14+ monocytes were cultured in complete RPMI-1640 medium (Life Technologies, Gaithersburg, MD, USA) supplemented with hM-CSF (10 ng/ml) for 6 days to obtain MDMs. Fresh medium supplemented with hM-CSF (10 ng/ml) was added on day 3.
Gene Expression Microarray
Array data of CD133+ stem cells, CD34+ precursors, endothelial cells, PBMC, eEPCs, and late EPCs were from our previous publication (GSE39763 and GSE10856) (8,19,20). Public GEO microarray data sets (http://www.ncbi.nlm.nih.gov/geo/) included in this study were also GSE12891 (another batch of early and late EPCs) (20) and GSE11430 (monocyte).
The array data for primary MDMs were obtained by our group using Affymetrix™ HG-U133 Plus 2.0 whole genome chips. RMA log expression units were calculated from Affymetrix GeneChip array data using the “affy” package of the Bioconductor (http://www.bioconductor.org) suite of software for the R statistical programming language (http://www.r-project.org). The default RMA settings were used to background correct, normalize, and summarize all expression values. Significant differences between the sample groups was identified using the “limma” (Linear Models for Microarray Analysis) package of the Bioconductor suite, and an empirical Bayesian moderated t-statistic hypothesis test between the two specified phenotypic groups was performed (21). To control for multiple testing errors, we then applied a false discovery rate algorithm to these p values in order to calculate a set of q values, thresholds of the expected proportion of false positives, or false rejections of the null hypothesis (22).
Heat maps were created by the dChip software (http://www.dchip.org/). Principal components analysis (PCA) was performed by the Partek Genomics Suite (http://www.partek.com/) to provide a visual impression of how the various sample groups are related. The Euclidean distance between two groups of samples is calculated by the average linkage measure [the mean of all pairwise distances (linkages) between members of the two groups concerned] (23). The standard error of the average linkage distance between two groups (the standard deviation of pairwise linkages divided by the square root of the number of linkages) is quoted when intergroup distances are compared in the text. Gene enrichment analysis was performed by the Gene Ontology (GO) database (http://www.geneontology.org/) using the DAVID (Database for Annotation, Visualization and Integrated Discovery) bioinformatics resources v6.7 at the National Institute of Allergy and Infectious Diseases (NIAID), NIH (http://david.abcc.ncifcrf.gov/).
RESULTS
Isolation and Characterization of Human Endothelial Precursor Cells
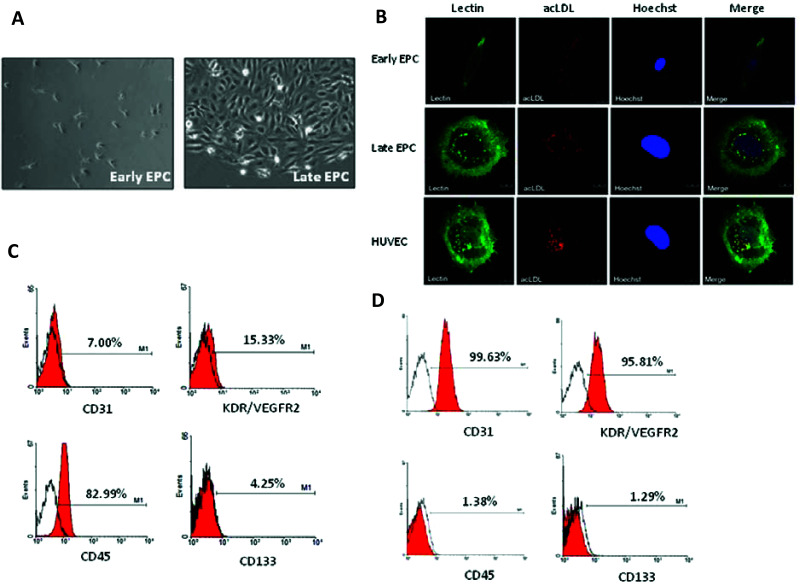
EPCs were obtained from the cord blood of healthy subjects as described (17). The peripheral blood MNCs that were initially seeded on fibronectin-coated wells were round. After the medium was changed on day 4, attached eEPCs with an elongated morphology appeared; then, late EPCs with a cobblestone-like morphology similar to mature endothelial cells grew to confluence at days 14–21 (Fig. 1A). Both EPCs were confirmed as having an endothelial lineage because both precursors were double positive for acLDL uptake and lectin (UEA-1) binding affinity (Fig. 1B).
Figure 1.
Cultivation and characterization of early and late EPCs. (A) Cord blood MNCs were isolated and plated on fibronectin-coated culture dishes for 4 days. Adherent eEPCs are shown in the left panel. Twenty-one days after plating, late EPCs with a cobblestone-like morphology were selected, reseeded, and grown to confluence (right). (B) Both early and late EPCs endocytose DiI-acLDL (acetylated low density lipoprotein; red) and bind fluorescein isothiocyanate UEA-1 (lectin) (green). Cells were counterstained with Hoechst 33258 to show the nucleus (blue). (C, D) Expression of indicated progenitor, endothelial, and hematopoietic markers in early (C) and late (D) EPCs by flow cytometric analysis.
The expression of cell lineage markers on the two EPCs was further validated and quantified by flow cytometry analysis. The majority of eEPCs expressed the hematological marker CD45, whereas the endothelial markers CD31 and KDR were present on only part of the isolated eEPC population (Fig. 1C). In contrast, the majority of late EPCs expressed the endothelial markers CD31 and KDR (Fig. 1D). Both early and late EPC populations expressed the stem cell marker AC133/CD133 to only a limited extent (Fig. 1C, D).
Microarray Analysis Reveals Close Relationships Between eEPCs and Blood Cells, Especially MDMs
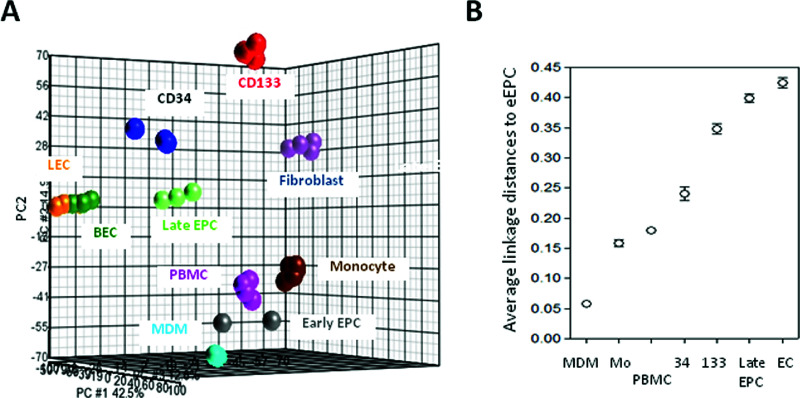
Early EPCs are believed to show greater similarity to hematopoietic cells with a molecular phenotype linked to monocytes (9,10). To evaluate the relationships between eEPCs and blood cells, especially monocytes and MDMs, we analyzed the transcriptomes of eEPC and of blood cell using whole-genome chips and then compared these to those of other endothelial lineage cells obtained by our group (19). A PCA plot using genes differentially expressed between CD133+ ancestor stem cells and mature endothelial progeny cells [n = 8,880, positive false discovery rate (pFDR) q < 10−4] represents the differentiation hierarchical relationship (Fig. 2A). Instead of being close to endothelial lineage cells, eEPCs were found to cluster with PBMCs, monocytes, and MDMs (Fig. 2A).
Figure 2.
Similar yet distinct gene expression patterns between eEPCs and MDMs. (A) A PCA plot using genes differentially expressed between CD133+ stem cells and mature endothelial cells (8,880 probe sets, q < 10−4). LEC, lymphatic endothelial cell; BEC, blood vessel endothelial cell; CD133, CD133+ stem cells; CD34, CD34+ progenitor cells. (B) Transcriptome distance analysis for eEPCs and various blood cell or endothelial cell types. Average linkage distances between transcriptomes were calculated as described using the aforementioned 8,880 probe sets. Mo, monocyte.
To provide more quantitative evidence, we calculated the average linkage distances between eEPCs and various cell types in order to assess the similarity between pairs of gene expression profiles, as described previously (19). As shown in Figure 2B, the distance between eEPCs and MDMs was smaller than the distances between eEPC and any other cell type examined, indicating that the genetic profile of eEPCs is closer to that of MDMs.
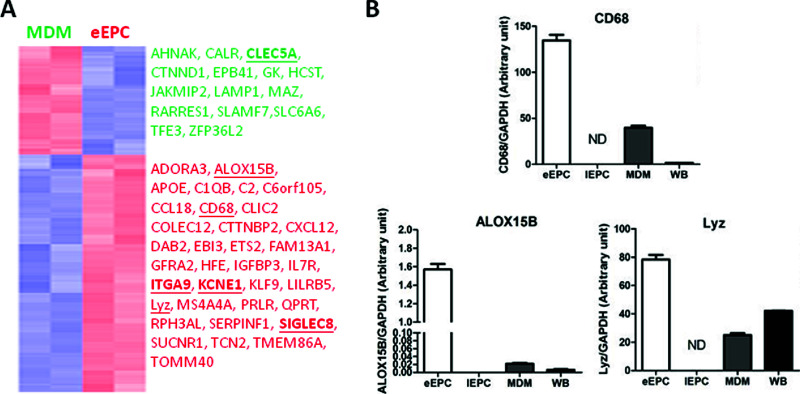
We compared gene expression profiles between eEPCs and MDMs, the two most similar cell types. A total of 290 signature probe sets were found to be differentially expressed between MDMs and eEPCs, with 199 probe sets being unique to eEPCs (fold change > 2, top 50 eEPC genes in Table 1) (full gene lists can be viewed in Supplementary Table 1, available at https://docs.google.com/file/d/0B3m2MizcxKoDcEsxaVlOSzlGc1k/edit?usp=sharing). A gene expression heat map for these genes presents the unique expression patterns of each cell type (Fig. 3A). The differential expression pattern of CD68, ALOX15B (arachidonate 15-lipoxygenase, type B), and Lyz (lysozyme) by eEPCs was verified by qPCR (Fig. 3B). Among the eEPC-enriched genes, CXCL12 [chemokine (C-X-C motif) ligand 12, also known as stromal cell-derived factor 1 (SDF-1)] (Table 1, indicated by an asterisk) plays an important role in angiogenesis by recruiting EPCs from the bone marrow through a CXCR4-dependent mechanism (24). The abundant expression of CXCL12 by eEPCs, but not MDMs, reflects the concept that eEPCs exert their angiogenic effects via a paracrine mechanism rather than by being incorporated into blood vessels directly (9,10). Consistently, CCL8, CCL18, and MMP1 (matrix metallopeptidase 1/interstitial collagenase) were also found to be abundantly expressed in eEPCs (Table 1) (also Supplementary Table 1 available at https://docs.google.com/file/d/0B3m2MizcxKoDcEsxaVlOSzlGc1k/edit?usp=sharing).
Table 1.
Top 50 Known Genes in Early EPCs (Compared With MDMs)
| Probe Set ID | UniGene ID | Gene Title | Gene Symbol | Location |
|---|---|---|---|---|
| 206171_at | Hs.281342 | Adenosine A3 receptor | ADORA3 | chr1p13.2 |
| 222416_at | Hs.500645 | Aldehyde dehydrogenase 18 family, member A1 | ALDH18A1 | chr10q24.3 |
| 1555416_a_at | Hs.111256 | Arachidonate 15-lipoxygenase, type B | ALOX15B* | chr17p13.1 |
| 203381_s_at | Hs.654439 | Apolipoprotein E | APOE | chr19q13.2 |
| 202686_s_at | Hs.590970 | AXL receptor tyrosine kinase | AXL | chr19q13.1 |
| 202953_at | Hs.8986 | Complement component 1, q subcomponent, B chain | C1QB | chr1p36.12 |
| 229070_at | Hs.126409 | Chromosome 6 open reading frame 105 | C6orf105 | chr6p24.1 |
| 214038_at | Hs.271387 | Chemokine (C-C motif) ligand 8 | CCL8* | chr17q11.2 |
| 221019_s_at | Hs.464422 | Collectin subfamily member 12 | COLEC12 | chr18pter-p11.3 |
| 232136_s_at | Hs.592285 | Ccortactin binding protein 2 | CTTNBP2 | chr7q31 |
| 209687_at | Hs.522891 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | CXCL12* | chr10q11.1 |
| 202887_s_at | Hs.523012 | DNA-damage-inducible transcript 4 | DDIT4 | chr10pter-q26.12 |
| 228057_at | Hs.480378 | DNA-damage-inducible transcript 4-like | DDIT4L | chr4q23 |
| 219424_at | Hs.501452 | Epstein-Barr virus induced gene 3 | EBI3 | chr19p13.3 |
| 241981_at | Hs.268874 | Family with sequence similarity 20, member A | FAM20A | chr17q24.2 |
| 224840_at | Hs.407190 | FK506 binding protein 5 | FKBP5 | chr6p21.3-p21.2 |
| 206674_at | Hs.507590 | Fms-related tyrosine kinase 3 | FLT3 | chr13q12 |
| 218706_s_at | Hs.363558 | GRAM domain containing 3 | GRAMD3 | chr5q23.2 |
| 204018_x_at | Hs.449630 | Hemoglobin, alpha 1 /// hemoglobin, alpha 2 | HBA1 /// HBA2 | chr16p13.3 |
| 204419_x_at | Hs.712539 | Hemoglobin, gamma A /// hemoglobin, gamma G | HBG1 /// HBG2 | chr11p15.5 |
| 203819_s_at | Hs.700696 | Insulin-like growth factor 2 mRNA binding protein 3 | IGF2BP3 | chr7p11 |
| 227297_at | Hs.113157 | Integrin, alpha 9 | ITGA9* | chr3p21.3 |
| 236407_at | Hs.121495 | Potassium voltage-gated channel, Isk-related family, member 1 | KCNE1* | chr21q22.1-q22.2|21q22.12 |
| 221583_s_at | Hs.144795 | Potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | KCNMA1 | chr10q22.3 |
| 228977_at | Hs.130652 | Hypothetical protein LOC729680 | LOC729680 | chr13q12.11 |
| 203414_at | Hs.463483 | Monocyte to macrophage differentiation-associated | MMD | chr17q |
| 204475_at | Hs.83169 | Matrix metallopeptidase 1 (interstitial collagenase) | MMP1 | chr11q22.3 |
| 225520_at | Hs.591343 | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | MTHFD1L | chr6q25.1 |
| 228056_s_at | Hs.636624 | Napsin B aspartic peptidase pseudogene | NAPSB | chr19q13.33 |
| 33767_at | Hs.198760 | Neurofilament, heavy polypeptide 200 kDa | NEFH | chr22q12.2 |
| 203708_at | Hs.198072 | Phosphodiesterase 4B, cAMP-specific | PDE4B | chr1p31 |
| 225207_at | Hs.8364 | Pyruvate dehydrogenase kinase, isozyme 4 | PDK4 | chr7q21.3 |
| 220952_s_at | Hs.188614 | Pleckstrin homology domain containing, family A member 5 | PLEKHA5 | chr12p12 |
| 204285_s_at | Hs.96 | Phorbol-12-myristate-13-acetate-induced protein 1 | PMAIP1 | chr18q21.32 |
| 201876_at | Hs.530077 | Paraoxonase 2 | PON2 | chr7q21.3 |
| 204044_at | Hs.513484 | Quinolinate phosphoribosyltransferase (nicotinate-nucleotide pyrophosphorylase) | QPRT | chr16p11.2 |
| 212651_at | Hs.148670 | Rho-related BTB domain containing 1 | RHOBTB1 | chr10q21.2 |
| 225202_at | Hs.445030 | Rho-related BTB domain containing 3 | RHOBTB3 | chr5q15 |
| 205578_at | Hs.98255 | Receptor tyrosine kinase-like orphan receptor 2 | ROR2 | chr9q22 |
| 204900_x_at | Hs.591715 | Sin3A-associated protein, 30 kDa | SAP30 | chr4q34.1 |
| 208253_at | Hs.447899 | Sialic acid binding Ig-like lectin 8 | SIGLEC8* | chr19q13.33-q13.41 |
| 223044_at | Hs.643005 | Solute carrier family 40 (iron-regulated transporter), member 1 | SLC40A1 | chr2q32 |
| 204955_at | Hs.15154 | Sushi-repeat-containing protein, X-linked | SRPX | chrXp21.1 |
| 223939_at | Hs.279575 | Succinate receptor 1 | SUCNR1 | chr3q24-q25.1 |
| 222116_s_at | Hs.369819 | TBC1 domain family, member 16 | TBC1D16 | chr17q25.3 |
| 204043_at | Hs.417948 | Transcobalamin II; macrocytic anemia | TCN2 | chr22q12.2 |
| 209676_at | Hs.516578 | Tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | TFPI | chr2q32 |
| 223594_at | Hs.444668 | Transmembrane protein 117 | TMEM117 | chr12q12 |
| 219410_at | Hs.658956 | Transmembrane protein 45A | TMEM45A | chr3q12.2 |
| 205194_at | Hs.512656 | Phosphoserine phosphatase | PSPH | chr7p15.2-p15.1 |
Verified by RT-qPCR or discussed in the text.
Figure 3.
Genes unique in eEPCs and MDMs. (A) Heat map showing differentially expressed genes in eEPCs or MDMs. Columns represent human tissue and stem cell samples, and rows represent probe sets. Genes in red: increased expression; in blue: decreased expression. Genes picked for RT-qPCR in (B) (underlined) and Figure 4 (underlined and in bold) are also shown. (B) Validation of array data by RT-qPCR. Mean gene expression levels of eEPC proteins (compared to GAPDH control) are shown. Results are expressed as the mean ± SD.
Functional Module Analysis as a Framework for the Interpretation of Early EPC Biology
The gene list outlined above gave us preliminary insights into the functional consequences of differential gene expression between the two types of EPCs. To understand more about how the gene expression profiles might be correlated functionally as well as to provide quantitative evidence, the signature mRNAs were subjected to a GO database search in order to pinpoint statistically overrepresented functional groups within the gene lists. The DAVID bioinformatics web tool (see Materials and Methods for details) was applied to provide the statistical analysis and to visually present the results. The GO biological process categories that were statistically overrepresented (p < 0.05) among genes that formed the eEPC group (compared with those from MDMs) are shown in Table 2. Genes CTTNBP2, APOE, and KCNMA1, which are involved in vasodilation, were significantly overexpressed in eEPCs (p = 0.0197) (Table 2, indicated by an asterisk). Another significant biological process associated with eEPCs was found to be cell proliferation (nine genes including CXCL12, CSGALNACT1, CCND3, DAB2, IGFBP4, PSPH, IL7R, SERPINF1, and TXN) (p = 0.0296) (Table 2, indicated by an asterisk). Other predominant processes found to be associated with the eEPC group include genes involved in cation homeostasis and/or transportation (ATP6V0E1, CXCL12, HFE, APOE, KCNMA1, SLC30A4, SLC40A1, and HFE) and oxidation reduction (BDH2, ALDH18A1, ALDH18A1, CYB5A, ALOX15B, DHRS3, ENOX2, MAOA, TXN, and UQCRB) (Table 2). In contrast, the principal functions associated with MDMs (compared with eEPCs) included those related to cortical actin cytoskeleton organization [calreticulin (CALR), formin-like 1 (FMNL1), and erythrocyte membrane protein band 4.1 (EPB41)] (p = 0.0122) (Table 3, indicated by an asterisk), which reflect the active cell migratory ability of macrophages.
Table 2.
Altered Functional Modules in Early EPCs (Compared With MDMs)
| Biological Process | Count | % | p | Genes |
|---|---|---|---|---|
| Vasodilation* | 3 | 1.91 | 0.0197 | CTTNBP2, APOE, KCNMA1 |
| T-cell activation | 5 | 3.18 | 0.0212 | CXCL12, CCND3, FLT3, IL7R, PRLR |
| Transition metal ion transport* | 4 | 2.55 | 0.0215 | HFE, SLC30A4, SLC40A1, TCN2 |
| Cell proliferation* | 9 | 5.73 | 0.0296 | CXCL12, CSGALNACT1, CCND3, DAB2, IGFBP4, PSPH, IL7R, SERPINF1, TXN |
| Cation homeostasis* | 7 | 4.46 | 0.0327 | ATP6V0E1, CXCL12, HFE, APOE, KCNMA1, SLC30A4, SLC40A1 |
| Oxidation reduction* | 11 | 7.01 | 0.0410 | BDH2, ALDH18A1, ALDH18A1, CYB5A, ALOX15B, DHRS3, ENOX2, MAOA, TXN, UQCRB |
| Cellular homeostasis | 9 | 5.73 | 0.0413 | ATP6V0E1, CXCL12, HFE, APOE, PRDX4, KCNMA1, KCNE1, SLC40A1, TXN |
| Lymphocyte proliferation | 3 | 1.91 | 0.0479 | CXCL12, CCND3, IL7R |
| Homeostatic process | 12 | 7.64 | 0.0491 | ATP6V0E1, CXCL12, HFE, APOE, IL7R, PRDX4, RPH3AL, SLC30A4, SLC40A1, TXN, KCNMA1, KCNE1 |
Discussed in the text.
Table 3.
Altered Functional Modules in MDMs (Compared With Early EPCs)
| Biological Process | Count | % | p |
|---|---|---|---|
| Negative regulation of gene expression | 8 | 10.96 | 0.0033 |
| Response to organic substance | 9 | 12.33 | 0.0066 |
| Negative regulation of macromolecule metabolic process | 9 | 12.33 | 0.0073 |
| Negative regulation of transcription | 7 | 9.590 | 0.0088 |
| Cortical actin cytoskeleton organization* | 3 | 3.371 | 0.0122 |
| Negative regulation of nucleobase and nucleic acid metabolic process | 7 | 9.590 | 0.0145 |
| Negative regulation of nitrogen compound metabolic process | 7 | 9.589 | 0.0154 |
| Negative regulation of macromolecule biosynthetic process | 7 | 9.589 | 0.0195 |
| Negative regulation of cellular biosynthetic process | 7 | 9.589 | 0.0218 |
| Negative regulation of biosynthetic process | 7 | 9.589 | 0.0239 |
| Cell proliferation | 6 | 8.219 | 0.0275 |
| Blood circulation | 4 | 5.479 | 0.0363 |
| Steroid metabolic process | 4 | 5.479 | 0.0446 |
| Response to inorganic substance | 4 | 5.479 | 0.0462 |
| Posttranscriptional regulation of gene expression | 4 | 5.479 | 0.0496 |
| Positive regulation of TGFB receptor signaling pathway | 2 | 2.741 | 0.0498 |
Discussed in the text.
Novel Surface Biomarkers That Distinguish eEPCs From Monocytes/MDMs
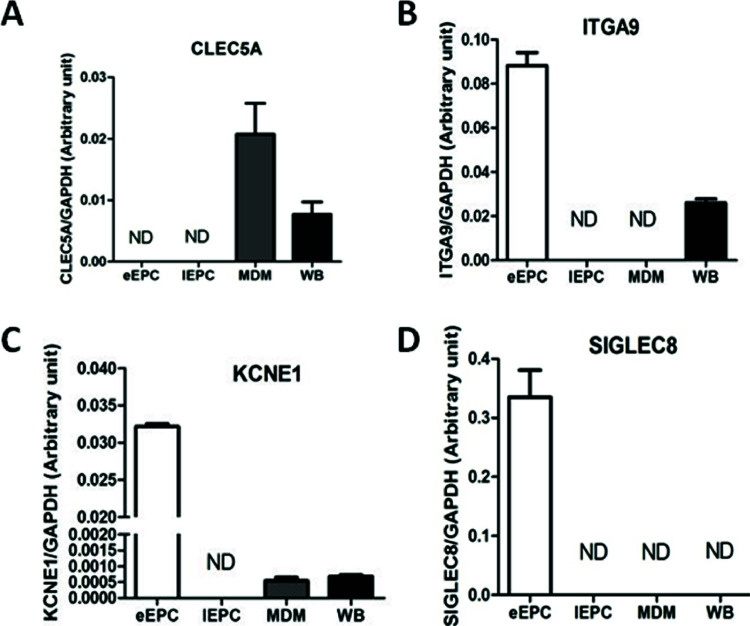
One critical and practical question after gene pattern analysis is how we can make it easier to purify eEPCs without monocyte and/or MDM contamination. In order to pinpoint novel surface antigens that are able to distinguish eEPCs from monocyte/MDMs, we searched for target membrane proteins that might act as novel surface markers when separating eEPCs and immune cells during their purification and isolation. The expression of CLEC5A [C-type lectin domain family 5, member A; also known as myeloid DAP12-associating lectin (MDL-1)], which contains a C-type lectin-like fold similar to the natural killer T-cell C-type lectin domains and associates with a 12-kDa DNAX-activating protein (DAP12) on myeloid cells (25), was found to be unique to MDM and whole-blood cells (Fig. 4A). On the other hand, 3 of the top 50 eEPC genes, including ITGA9 (integrin, alpha 9), KCNE1 (potassium voltage-gated channel, Isk-related family, member 1), and in particular SIGLEC8 (sialic acid binding Ig-like lectin 8), were found to be uniquely expressed in eEPCs (Fig. 4B, C).
Figure 4.
Cell membrane proteins specifically expressed in eEPCs or MDMs. These four genes were selected according to the “Cellular Component” ontology in the GO database. Mean gene expression levels of eEPC proteins (compared to GAPDH control) by RT-qPCR are shown. Results are expressed as the mean ± SD.
DISCUSSION
EPCs play an important role in postnatal vascular repair and the maintenance of vascular homeostasis because they are involved in reendothelialization and neovascularization. CD34, KDR, and CD133+ are considered to be the critical markers associated with isolating circulating EPCs, which need to be different from those of hematopoietic progenitors or leukocytes. Combinations of markers, including CD133+CD34+KDR+, CD34+KDR+, or CD14+CD34low, have been widely used to define or select cells that express the properties attributed to EPCs (26,27). These approaches, however, are unable to distinguish eEPCs from the late EPCs. The lack of known surface biomarkers for the differentiation of the two categories of EPCs and the absence of a standardized protocol in terms of reagents and gating strategy may account for the widespread interlaboratory variations when quantifying EPCs. In our previous study, we have identified, also by mRNA profiling of various novel surface markers, how to distinguish early and late EPCs (8). Among endothelial lineage cells, CD204, CD169, GPNMB, and many other membrane proteins are uniquely expressed on eEPCs, whereas genes such as CXADR, OSAP, and CD106 are uniquely expressed on late EPCs (8). In the present study, new findings provide another group of surface genes that should help to separate circulating eEPCs from other blood cells that closely resemble them. It should now be possible to combine these studies and use these new biomarkers in parallel with CD34 and KDR for the direct isolation and counting of early and late EPCs from cord blood and peripheral blood. The latter point is of clinical importance because the number of EPCs in peripheral blood has been found to correlate with disease prognosis in a number of studies (3–5).
Nevertheless, there are still challenges to the harnessing of EPCs from blood for direct cell therapy. One of these is their rarity (0.01–0.02 per 106 MNCs), which makes the isolation of EPCs challenging. Optimization of the cultivation and amplification of EPCs is therefore required before these cells may be appropriately investigated to determine their use in clinical therapies. It has been shown that eEPCs do not proliferate significantly in vitro (28). The use of these cells is also handicapped by the difficulties associated with separating eEPCs, which are normally regarded as the cells that appear early (<1 week) in culture dishes, from adherent blood cells such as monocytes and macrophages. Finding surface markers distinguishing eEPCs and adherent blood cells (especially monocytes and MDMs) (Fig. 4) should help with the purification of cultured eEPC for use in cell-based therapy. Contamination due to monocytes and macrophages in transplanted eEPCs may cause graft-versus-host disease (GVHD), a common complication of allogeneic cell transplantation. The presence of these immune cells in the cell graft may recognize the recipient (the host) as “foreign” and then attack the host’s body cells.
Early EPCs have long been recognized to have molecular phenotypes and surface markers linked to hematopoietic cells (10,11,13), yet most comparative studies have focused on the relationships between eEPCs and monocytes. Specifically, proteomic analysis has revealed that 90% of spots identified by 2D gel analysis are common between OECs and endothelial cells, whereas eEPCs share 77% spot identity with monocytes (9). In this study, we show that MDMs rather than monocytes share the closest relationship to eEPCs (Fig. 2B). Owing to the inflammatory nature of macrophages, eEPCs should be used clinically with caution and care taken with respect to the disease they are used to treat. If injected into a proinflammatory tissue microenvironment such as ischemic cardiovascular disease, it is possible they might intensify the preexisting pathology because of their inflammatory potential. Late EPCs, on the other hand, should be a better source for cell-based transplantation and therapy that involves neoangiogenesis because these cells have greater similarity to mature endothelial cells and distant in terms of gene expression from immune cells or fibroblasts (Fig. 2A).
GO analysis and qPCR validation work revealed the functional significance of our gene list analysis and in the process identified the functional variations between eEPCs and MDMs. For example, CLEC5A, which is critical to inflammation and autoimmune diseases such as arthritis as well as being involved in the activation of myeloid cells (25,29), is unique to MDM and whole-blood cells (Fig. 4A). CLEC5A/MDL-1 also serves as a receptor for several viruses that infect macrophages and induce lethal inflammation responses. Specifically, in human macrophages, CLEC5A interacts with the dengue virion (DV) directly and is responsible for dengue virus-induced lethal disease and inflammasome activation (30,31). Blockade of CLEC5A-DV’s interactions suppresses the secretion of proinflammatory cytokines. Moreover, anti-CLEC5A monoclonal antibodies inhibit DV-induced plasma leakage as well as subcutaneous and vital organ hemorrhaging; these result in a reduction in the mortality of DV-infected mice (30). CLEC5A also regulates Japanese encephalitis virus (JEV)-induced neuroinflammation and lethality (32). The unique expression of CLEC5A in MDMs (Fig. 4A) indicates that macrophages, but not eEPCs, serve as a natural host for DV or JEV and are involved in viral pathogenesis. This emphasizes that eEPCs and MDMs are actually two distinct cell populations with different physiological and pathological roles. eEPCs seem to be able to contribute more to the area of angiogenesis because they express more genes related to vasodilation, including CTTNBP2, APOE, and KCNMA1 (Table 2). MDMs, on the other hand, are more involved in inflammatory responses. In addition, macrophages may possess better motility than eEPCs because genes involved in cortical actin cytoskeleton organization are more abundant MDMs (Table 3); this may reflect the host defense nature of this immune cell population.
In summary, our results, which combine mRNA profiling and gene set analysis, help to decipher the RNA expression situation in eEPCs compared to various types of blood cells that closely resemble them, especially monocytes and MDMs. Although new research directions and hypotheses can be formed based on the gene expression data provided by this study, careful functional studies of the genes in context using in vitro and in vivo models of angiogenesis are still necessary in order to further support the clinical relevance of the data set. We envision that our report will serve as a resource for future studies that are aimed at improving our understanding of the various regulatory mechanisms that ultimately modulate EPC and EC activities.
ACKNOWLEDGMENTS
This work is supported by Tri-Service General Hospital (TSGH-C102-027).
Footnotes
The authors declare no conflicts of interests.
REFERENCES
- 1. Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science 1997; 275(5302):964–967. [DOI] [PubMed] [Google Scholar]
- 2. Real C, Caiado F, Dias S. Endothelial progenitors in vascular repair and angiogenesis: How many are needed and what to do? Cardiovasc Hematol Disord Drug Targets 2008; 8(3):185–193. [DOI] [PubMed] [Google Scholar]
- 3. Urbich C, Aicher A, Heeschen C, Dernbach E, Hofmann WK, Zeiher AM, et al. Soluble factors released by endothelial progenitor cells promote migration of endothelial cells and cardiac resident progenitor cells. J Mol Cell Cardiol 2005; 39(5):733–742. [DOI] [PubMed] [Google Scholar]
- 4. Fan Y, Shen F, Frenzel T, Zhu W, Ye J, Liu J, et al. Endothelial progenitor cell transplantation improves long-term stroke outcome in mice. Ann Neurol 2010; 67(4):488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Werner N, Kosiol S, Schiegl T, Ahlers P, Walenta K, Link A, et al. Circulating endothelial progenitor cells and cardiovascular outcomes. N Engl J Med 2005; 353(10):999–1007. [DOI] [PubMed] [Google Scholar]
- 6. Sieveking DP, Buckle A, Celermajer DS, Ng MK. Strikingly different angiogenic properties of endothelial progenitor cell subpopulations: Insights from a novel human angiogenesis assay. J Am Coll Cardiol 2008; 51(6):660–668. [DOI] [PubMed] [Google Scholar]
- 7. Mukai N, Akahori T, Komaki M, Li Q, Kanayasu-Toyoda T, Ishii-Watabe A, et al. A comparison of the tube forming potentials of early and late endothelial progenitor cells. Exp Cell Res 2008; 314(3):430–440. [DOI] [PubMed] [Google Scholar]
- 8. James V, Zhang Y, Foxler DE, de Moor CH, Kong YW, Webb TM, et al. LIM-domain proteins, LIMD1, Ajuba, and WTIP are required for microRNA-mediated gene silencing. Proc Natl Acad Sci USA 2010; 107(28):12499–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Medina RJ, O’Neill CL, Sweeney M, Guduric-Fuchs J, Gardiner TA, Simpson DA, et al. Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities. BMC Med Genomics 2010; 3(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bagley RG, Walter-Yohrling J, Cao X, Weber W, Simons B, Cook BP, et al. Endothelial precursor cells as a model of tumor endothelium: Characterization and comparison with mature endothelial cells. Cancer Res 2003; 63(18):5866–5873. [PubMed] [Google Scholar]
- 11. Huang PH, Chen JW, Lin CP, Chen YH, Wang CH, Leu HB, et al. Far infra-red therapy promotes ischemia-induced angiogenesis in diabetic mice and restores high glucose-suppressed endothelial progenitor cell functions. Cardiovasc Diabetol 2012; 11:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X, Gocek E, Liu CG, Studzinski GP. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle 2009; 8(5):736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pekny M, Nilsson M. Astrocyte activation and reactive gliosis. Glia 2005; 50(4):427–434. [DOI] [PubMed] [Google Scholar]
- 14. Yan T, Liu Y, Cui K, Hu B, Wang F, Zou L. MiR-126 regulates EPCs function: Implications for a role of miR-126 in preeclampsia. J Cell Biochem 2013; 114:2148–2159. [DOI] [PubMed] [Google Scholar]
- 15. Qiang L, Hong L, Ningfu W, Huaihong C, Jing W. Expression of miR-126 and miR-508-5p in endothelial progenitor cells is associated with the prognosis of chronic heart failure patients. Int J Cardiol. In press. [DOI] [PubMed] [Google Scholar]
- 16. Huang PH, Chen YH, Chen YL, Wu TC, Chen JW, Lin SJ. Vascular endothelial function and circulating endothelial progenitor cells in patients with cardiac syndrome X. Heart 2007; 93(9):1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen YH, Lin SJ, Lin FY, Wu TC, Tsao CR, Huang PH, et al. High glucose impairs early and late endothelial progenitor cells by modifying nitric oxide-related but not oxidative stress-mediated mechanisms. Diabetes 2007; 56(6):1559–1568. [DOI] [PubMed] [Google Scholar]
- 18. Chang YC, Hsu TL, Lin HH, Chio CC, Chiu AW, Chen NJ, et al. Modulation of macrophage differentiation and activation by decoy receptor 3. J Leukoc Biol 2004; 75(3):486–494. [DOI] [PubMed] [Google Scholar]
- 19. Huang TS, Hsieh JY, Wu YH, Jen CH, Tsuang YH, Chiou SH, et al. Functional network reconstruction reveals somatic stemness genetic maps and dedifferentiation-like transcriptome reprogramming induced by GATA2. Stem Cells 2008; 26(5):1186–1201. [DOI] [PubMed] [Google Scholar]
- 20. Maeng YS, Choi HJ, Kwon JY, Park YW, Choi KS, Min JK, et al. Endothelial progenitor cell homing: Prominent role of the IGF2-IGF2R-PLCbeta2 axis. Blood 2009; 113(1):233–243. [DOI] [PubMed] [Google Scholar]
- 21. Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3(1):Article 3. [DOI] [PubMed] [Google Scholar]
- 22. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003; 100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, et al. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet 2004; 36(7):687–693. [DOI] [PubMed] [Google Scholar]
- 24. Kane NM, Howard L, Descamps B, Meloni M, McClure J, Lu R, et al. Role of microRNAs 99b, 181a, and 181b in the differentiation of human embryonic stem cells to vascular endothelial cells. Stem Cells 2012; 30(4):643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicoli S, Knyphausen CP, Zhu LJ, Lakshmanan A, Lawson ND. miR-221 is required for endothelial tip cell behaviors during vascular development. Dev Cell 2012; 22(2):418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clarke LA, Shah V, Arrigoni F, Eleftheriou D, Hong Y, Halcox J, et al. Quantitative detection of circulating endothelial cells in vasculitis: Comparison of flow cytometry and immunomagnetic bead extraction. J Thromb Haemost 2008; 6(6):1025–1032. [DOI] [PubMed] [Google Scholar]
- 27. Schmidt-Lucke C, Fichtlscherer S, Aicher A, Tschope C, Schultheiss HP, Zeiher AM, et al. Quantification of circulating endothelial progenitor cells using the modified ISHAGE protocol. PLoS ONE 2010; 5(11):e13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rehman J, Li J, Orschell CM, March KL. Peripheral blood “endothelial progenitor cells” are derived from monocyte/macrophages and secrete angiogenic growth factors. Circulation 2003; 107(8):1164–1169. [DOI] [PubMed] [Google Scholar]
- 29. Benndorf RA, Schwedhelm E, Gnann A, Taheri R, Kom G, Didie M, et al. Isoprostanes inhibit vascular endothelial growth factor-induced endothelial cell migration, tube formation, and cardiac vessel sprouting in vitro, as well as angiogenesis in vivo via activation of the thromboxane A (2) receptor: A potential link between oxidative stress and impaired angiogenesis. Circ Res 2008; 103(9):1037–1046. [DOI] [PubMed] [Google Scholar]
- 30. Zhang X, Mao H, Chen JY, Wen S, Li D, Ye M, et al. Increased expression of microRNA-221 inhibits PAK1 in endothelial progenitor cells and impairs its function via c-Raf/MEK/ERK pathway. Biochem Biophys Res Commun 2013; 431(3):404–408. [DOI] [PubMed] [Google Scholar]
- 31. Zhu S, Deng S, Ma Q, Zhang T, Jia C, Zhuo D, et al. MicroRNA-10A* and microRNA-21 modulate endothelial progenitor cell senescence via suppressing high-mobility group A2. Circ Res 2013; 112(1):152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA 2008; 105(5):1516–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]