Abstract
Synergistic regulation among microRNAs (miRNAs) is important to understand the mechanisms underlying the complex molecular regulatory networks in goats. Goat milk fat synthesis is driven by a gene network that involves many biological processes in the mammary gland. These biological processes are affected by several miRNAs rather than a single miRNA. Therefore, identifying synergistic miRNAs is necessary to further understand the functions of miRNAs and the metabolism of goat milk fat synthesis. Using qRT-PCR, we assessed the expression of 11 miRNAs that have the potential to regulate milk fat synthesis in the goat mammary gland. Six of these miRNAs exhibited expression during the lactation cycle. Additionally, we also found that prolactin, the key hormone that regulates lactation, promotes the expression of four miRNAs (miR-23a, miR-27b, miR-103, and miR-200a). Further functional analysis showed that overexpression of all four miRNAs by using recombinant adenovirus in goat mammary gland epithelial cells can affect gene mRNA expression associated with milk fat synthesis. Specifically, elevated miR-200a expression suppressed the mRNA expression of genes involved in fat droplet formation. To analyze the synergistic regulation among these four miRNAs (miR-23a, miR-27b, miR-103, and miR-200a), we used the Pearson correlation coefficient to evaluate the correlation between their expression levels in 30 lactating goats. As a result, we found a strong correlation and mutual regulation between three miRNA pairs (miR-23a and miR-27b, miR-103 and miR-200a, miR-27b and miR-200a). This study provides the first experimental evidence that miRNA expression is synergistically regulated in the goat mammary gland and has identified the potential biological role of miRNAs in goat milk fat synthesis. The identification of synergistic miRNAs is a crucial step for further understanding the molecular network of milk fat synthesis at a system-wide level.
Keywords: Synergistic miRNAs, Metabolism of milk fat synthesis, Dairy goat
INTRODUCTION
Milk fat synthesis in the epithelial cells of the mammary gland involves many processes including de novo synthesis of fatty acids, triglyceride synthesis, fat droplet formation, and fatty acid uptake and transport. Goat milk fat contains greater amounts of short-chain, medium-chain, and unsaturated fatty acids than cow milk (1–3), indicating a unique network in goat milk fat synthesis.
MicroRNAs (miRNAs) are short endogenous RNAs known to posttranscriptionally repress gene expression in animals and plants (4–6). They are key regulators in many metabolic processes, including tissue development (7,8), cell differentiation (9,10), and lipid metabolism (11,12). Recent studies have indicated that miRNAs are involved in lactation of the mammary gland. MiR-27 decreased fat accumulation by targeting peroxisome proliferator-activated receptor γ (PPARγ) in 3T3-L1 preadipocytes (13); miR-103 silencing in OB/OB mice showed reduced fat-pad weights (14). Furthermore, 431 miRNAs were identified in the mammary gland during early lactation of the Chinese Laoshan dairy goat by high-throughput sequencing (15). Information from previous studies indicated that miRNAs are important for goat lactation. miRNAs are expressed spatially and temporally to precisely control the patterns of gene expression. A relatively small number of miRNA networks can effectively modulate multiple factors involved in biological metabolic processes (16). In addition, a single biological process can be affected by several miRNAs rather than a single miRNA (16). However, the molecular network (including miRNA network) for lactation in dairy goats has been barely characterized. Therefore, analysis of synergistic regulations among multiple miRNAs in the mammary gland of goats is important to understand the function of miRNAs as well as the mechanisms underlying the molecular network of lactation in dairy goats.
To identify synergistic miRNAs, we screened miRNAs that were differentially expressed during the lactation cycle and positively respond to prolactin concentration. We analyzed the correlation and interaction between the expression levels of four miRNAs and overexpressed them in goat mammary gland epithelial cells. This study has identified miRNAs that synergistically regulate milk fat synthesis of dairy goats and has provided important clues that miRNAs form a regulatory network during lactation.
MATERIALS AND METHODS
Animals, Tissue Sampling, and RNA Extraction
Thirty healthy 3-year-old Xinong Saanen goats of similar weight were selected from the Experimental Farm of Northwest A&F University for this study. The goats were in their second lactation. Mammary gland tissues were surgically collected from these 30 goats at midlactation (120 days after parturition) and immediately frozen in liquid nitrogen. In addition, 10 of the 30 goats were used for mammary gland tissue collection at early lactation (30 days after parturition).
Total RNA was extracted from the mammary gland tissue and mammary gland epithelial cells using a mirVana miRNA Isolation Kit (Ambion, USA) according to the manufacturer’s instructions. The quantity and quality of RNA was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop, USA). Total RNA was stored at −80°C for further use.
Primers, cDNA Synthesis, and qRT-PCR for miRNAs and mRNAs
For miRNA, first-strand cDNA was synthesized using 100 ng of total RNA and a TaqMan® MicroRNA Reverse Transcription Kit (Ambion). The first-strand cDNA was diluted with DNase/RNase-free water. The PCR assay was carried out with a TaqMan MicroR-103 Assay (Ambion) on a Bio-Rad CFX96 real-time PCR detection system (Bio-Rad, USA). The primers for miRNA are listed in Table 1. The relative expression levels of the miRNAs were normalized with the U6 or U2 snRNA expression level, which was calculated using the 2−ΔΔCT method.
Table 1.
miRNA Primers for qPCR Analysis
| miRNA | Primers (5′–3′) |
|---|---|
| miR-9 | AGAAACCAATAGATCGACATACT |
| miR-23a | CATCACATTGCCAGGGATTA |
| miR-27b | TTCACAGTGGCTAAGTTCCG |
| miR-29c | ATCGTGGTAAACTTTAGCCAAGG |
| miR-33a | CACGTAACATCAACGTAACG |
| miR-103 | AGCAGCATTGTACAGGGCTA |
| miR-130a | GTCACGTTACAATTTTCCCTTG |
| miR-143 | ACTCTACTTCGTGACATCGAGG |
| miR-200a | TAACACTGTCTGGTAACGATGT |
| miR-223 | ACAGTCAAACAGTTTATGGGGT |
| miR-335 | TGTTCTCGTTATTGCTTTTACGCG |
| U6 | CACTATTGCGGGTCTGC |
| U2 | CGCTTCTTCGGCTTATTAG |
For mRNA, 1 µg of total RNA was synthesized into cDNA using the PrimeScript® RT Reagent Kit (Perfect Real Time, Takara, Japan). The qPCR assay was performed using SYBR® Premix Ex Taq™ II (Perfect Real Time, Takara) on a Bio-Rad CFX96. Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) was used as an internal control. Data were analyzed using the relative quantification (ΔΔCt) method. The mRNA primers are listed in Table 2. In addition to the primers from published papers, other primers were designed using goat genes that were cloned in our laboratory.
Table 2.
mRNA Primers for qPCR Analysis
| Gene | Accession No. | Primers (5′–3′) | Source |
|---|---|---|---|
| FASN | DQ915966* | Forward: GTCGTTGTCTACAGCACAGCCT | This article |
| Reverse: ATGGCGAGGTTCCACTCAAAC | |||
| ACACA | DQ370054.1* | Forward: CATCTTGTCCGAAACGTCGAT | (26) |
| Reverse: CCCTTCGAACATACACCTCCA | |||
| SCD | GU947654* | Forward: CCATCGCCTGTGGAGTCAC | This article |
| Reverse: GTCGGATAAATCTAGCGTAGCA | |||
| GPAM | AY515690† | Forward: GCAGGTTTATCCAGTATGGCATT | (26) |
| Reverse: GGACTGATATCTTCCTGATCATCTTG | |||
| AGPAT6 | NM_00108366.1† | Forward: AAGCAAGTTGCCCATCCTCA | This article |
| Reverse: AAACTGTGGCTCCAATTTCGA | |||
| LPIN1 | NM_00120615.1 | Forward: TCCCTGCTCGGACGTAATTG | This article |
| Reverse: TGGCCACCAGAATAAAGCATG | |||
| DGAT1 | HM566448* | Forward: CCACTGGGACCTGAGGTGTC | This article |
| Reverse: GCATCACCACACACCAATTCA | |||
| ADFP | HQ846826* | Forward: CCCCAGAAGCCGAGTTACTATGTT | This article |
| Reverse: CACGCAGCCAGGACAGATAGAG | |||
| TIP47 | HQ846826* | Forward: GGTGGAGGGTCAGGAGAAA | This article |
| Reverse: TCACGGAACATGGCGAGT | |||
| gBTN1A1 | EF102891* | Forward: TCACGAGGGAGAGGAGTTTC | This article |
| Reverse: GGAAGAAGGATGCTGGTATG | |||
| CD36 | JF69774.1* | Forward: GTACAGATGCAGCCTCATTTCC | (26) |
| Reverse: TGGACCTGCAAATATCAGAGGA | |||
| SLC27A6 | NM_00110116.1†† | Forward: CCAAGACTCCCAGAAGGT | This article |
| Reverse: GGCTGTTGTTCCAGAAGTAA | |||
| ABCA1 | NM_00102469.1† | Forward: CGGCGGCTTCTCTTGTATAGC | (26) |
| Reverse: TTCAAGCGTGAGCTGAAACG | |||
| ABCG1 | NM_001205528.1† | Forward: CGTCCATAGGTTTCCACTGTGT | (26) |
| Reverse: GCACAGCAGAAGAATCTCCATA | |||
| FABP3 | AY466498* | Forward: CCTTCAAATTGGGCCAGGA | This article |
| Reverse: CAGCACCAGCTTATCATCCAC | |||
| GPR41 | HM013824* | Forward: CGCATTCTTCACCACCATCT | This article |
| Reverse: GCAGGTCCCGTTGATACC | |||
| PPARγ | HQ589347* | Forward: TCCGTGATGGAAGACCACTC | This article |
| Reverse: CCCTTGCATCCTTCACAAGC | |||
| SREBP-1c | JN790254.1* | Forward: CCAGCTGACAGCTCCATTGA | This article |
| Reverse: TGCGCGCCACAAGGA | |||
| LXRα | GU332719* | Forward: CATCAACCCCATCTTCGAGTT | This article |
| Reverse: CAGGGCCTCCACATATGTGT | |||
| GAPDH | AJ431207.1* | Forward: GCAAGTTCCACGGCACAG | This article |
| Reverse: GGTTCACGCCCATCACAA |
Accession number of goat genes.
Accession number of cow genes. Bold symbols mean that this gene was cloned in our laboratory from the goat.
Cell Culture and Hormone Treatments
Mammary gland epithelial cells were cultured in DMEM/F12 medium (Invitrogen Corp., USA) containing insulin (5 µg/ml), hydrocortisone (0.25 µmol/L), penicillin (50 U/ml)/streptomycin (50 U/ml), epidermal growth factor 1 (EGF-1, Gibco; 10 ng/ml), and 10% FBS at 37°C in a humidified atmosphere with 5% CO2 (17). The medium was changed every day. At confluence, the mammary gland epithelial cells were dissociated using trypsin-EDTA solution (0.25% trypsin and 0.05% EDTA). Some of the passage 1 cells were seeded on DMEM/F12 medium in culture plates (Nunc, Denmark) at a density of 5 × 104 cells/cm2, and then human prolactin (Sigma-Aldrich, USA) was added to the medium to obtain a final concentration of 0, 2, 6, or 10 µg/ml. The cells were cultured at 37°C in a humidified atmosphere with 5% CO2. After 48 h, the cells were collected, and the total RNA was extracted. The other passage 1 cells were seeded on DMEM/F12 medium to obtain passage 2 cells for adenovirus infection as described in the following.
Ad-miRNA Generation and Infection
The stem–loop of the miRNAs and about 250 nucleotides of the flanking sequences on the 5′ and 3′ ends of miRNAs were amplified from normal Xinong Saanen dairy goat genomic DNA. The primers for the miRNA were designed using the Oligo software (version 6.0). The primers for miR-23a, miR-27b, miR-103, and miR-200a are listed in Table 3. The adenovirus vectors pAd and pAd-miRNAs were constructed using a commercial system (AdEasy, Stratagene). Mammary gland epithelial cells were infected with Ad-miRNAs or Ad (control). The infected cells were cultured at 37°C for 0, 24, 48, and 72 h and then used for total RNA extraction as described herein. Furthermore, we found that Ad-miR-23a, Ad-miR-27b, Ad-miR-103, and Ad-miR-200a all successfully mediated their respective miRNA overexpression throughout the observation period in epithelial cells.
Table 3.
miRNA Primers for pri-miRNAs
| miRNA | Primers (5′–3′) | Pri-miRNA Length (bp) |
|---|---|---|
| miR-23a | Forward: TTATGAAAGATTTGGTCTGCC | 306 |
| Reverse: ATAACGCTTTCAGTTGACCTT | ||
| miR-27b | Forward: GGAAGAAGGATGCTGGTATG | 312 |
| Reverse: TCCACGAGGGAGAGGAGTTTC | ||
| miR-103 | Forward: CGCTAGAAGCTTTTGGGTTAATACTCCATTGAG | 328 |
| Reverse: GCCCTAGACCATGGATTTGTCATTTTGTAAAACT | ||
| miR-200a | Forward: GCCTAACACAAGTAATGTAAAA | 281 |
| Reverse: AACTGAAGAGACGGTCAATGA |
Statistical Analysis
All infection experiments were repeated six times. The qRT-PCR assays were performed in triplicate, and each experiment was repeated at least three times. The data are presented as the means ± SD of three or more independent experiments. Differences were considered statistically significant at p < 0.05 using Student’s t test.
RESULTS
Screening MiRNAs Whose Expression Correlates With Lactation and Prolactin Concentrations
A previous study has reported 300 miRNA profiles in goat mammary gland during early lactation (15). Among these 300 miRNAs, we found 11 miRNAs that have been reported to directly or indirectly regulate lipid synthesis in mammalian tissue or cells. The functions of the majority of these miRNAs have been reported in adipose tissue, whereas others were validated in the liver, muscle, and pancreas (Table 4). There is no miRNA function reported in goat mammary gland. The mammary gland is a “lipid synthesis machine” during lactation (18). Therefore, we speculated that these 11 miRNAs might be crucial for goat lactation. In addition, we predicted the targets of the miRNAs using MicroCosm and PicTar. We found that some of the predicted targets were related to fat synthesis (Table 4).
Table 4.
miRNAs Involved in Fat Synthesis
| miRNA | Tissue | Function | Known Targets | Reference(s) | Predicted Targets* |
|---|---|---|---|---|---|
| miR-9 | Pancreas | Insulin secretion | SIRT1 | (54) | RXRα, INSIG1, ABCA1 |
| miR-23 | Adipose | Enhancing glutamine metabolism | MuRF1, c-Myc | (22) | SLC4A5, ADRP, STAT5B |
| miR-27 | Adipose | Adipogenesis | PPARγ, C/EBP | (13,55) | LPIN1, ABCA1, LPL 1 |
| miR-29 | Adipose | Glucose transport | INSIG1, CAV2, | (56) | LPL |
| miR-33 | Liver | Lipid homeostasis | ABCA1, CPT1A | (44,57) | MAP3K3, IRS2, PPARGC1A |
| miR-103 | Adipose | Adipogenesis and insulin sensitivity | CDK5R1, CAV1 | (14,23) | ACSL1, GPR41, STAT5A, GLUD1 |
| miR-130 | Adipose | Adipogenesis | PPARγ | (58) | ACSL4, ACSL1, ABCA1 |
| miR-143 | Adipose | Insulin resistance | MAPK7 | (59) | MAP3K7, SLC16A2 |
| miR-200 | Adipose | Regulating insulin pathway | ZEB1/2, BMI1 | (24) | MGF, BTN1A1, GPR43, LPL |
| miR-223 | Muscle | Glucose uptake | GLUT4 | (60) | ACSL3, MAP3K2 |
| miR-335 | Adipose | Fatty acid and triglyceride synthesis | SIRT1, ACSL1 | (61) | MAP3K2, PANK2 |
ABCA1, ATP-binding cassette subfamily A member 1; GLUT, solute carrier; ACSL, acyl-CoA synthetase; CAV2, caveolin 2; CPT1A, carnitine palmitoyltransferase 1A; INSIG1, insulin-induced gene 1; IRS2, insulin receptor substrate 2; PPARγ, peroxisome proliferator-activated receptor γ; SIRT, sirtuin; SLC, solute carrier; MuRF1, muscle ring finger 1; c-Myc, c-myc oncogene; C/EBP, CCAAT/enhancer-binding protein; RXRα, retinoid X receptor α; ADRP, adipose differentiation related protein; STAT5, signal transducer and activator of transcription subfamily 5; LPIN1, Lpin1; LPL, lipoprotein lipase; BTN1A1, butyrophilin, subfamily 1, member A1; MAP3K, mitogen-activated protein kinase kinase kinase 3; CDK5R1, cyclin-dependent kinase 5 regulatory subunit 1; ZEB, zinc-finger E-box binding homeobox; BMI1, polycomb group gene; MAPK7, mitogen-activated protein kinase 7; GPR, orphan G protein-coupled receptor; PANK2, pantothenate kinase 2; SCD, stearoyl-CoA desaturase; MGF, mammary gland factor; PPARGC1A, peroxisome proliferator-activated receptor gamma coactivator 1 α.
Genes are predicted by MicroCosm (Release 8.1, 2012) or PicTar (Release 3.26, 2007).
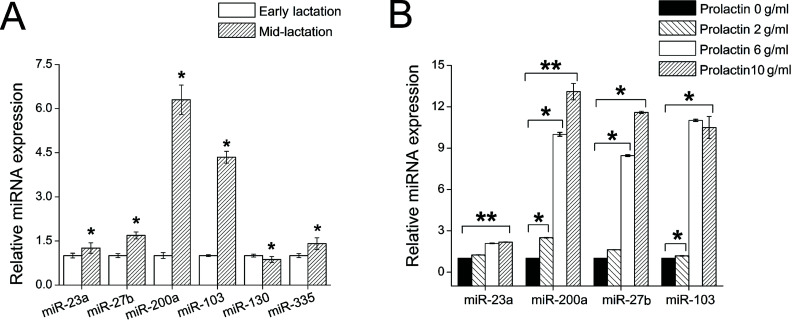
To screen these miRNAs for their involvement in milk fat synthesis, we measured the changes in expression of these 11 miRNAs during the lactation stages. Based on overall milk production, the lactation cycle can be divided into three periods: early, mid-, and late lactation. Milk yield in midlactation is higher than during the other two periods. We chose 10 goats and sampled the mammary gland tissues at midlactation (120 days after parturition) and early lactation (30 days after parturition). These tissues were then pooled in each sample data and subjected to RNA extraction. We compared miRNA expression in these two different stages by quantitative real-time PCR. As a result, we found six differently expressed miRNAs. The expression levels of miR-23a (1.42-fold, p < 0.05), miR-27b (2.2-fold, p < 0.05), miR-103 (4.7-fold, p < 0.05), miR-200a (6.4-fold, p < 0.05), and miR-335 (1.4-fold, p < 0.05) were all higher during midlactation than during early lactation (Fig. 1A). In contrast, miR-130a (0.87-fold, p < 0.05) expression was lower during midlactation than during early lactation (Fig. 1A). The correlation of the expression of these six miRNAs and the lactation stages demonstrates that these miRNAs may regulate milk fat synthesis in goats. The other five miRNAs (miR-9, miR-29c, miR-33a, miR-143, and miR-223) whose expression was nearly unchanged during the lactation cycle may be involved in basic metabolism in the mammary gland.
Figure 1.
miRNA expression correlates with lactation stage and prolactin concentration. (A) MiR-23a, miR-27b, miR-103, and miR-200a in goat mammary gland are differentially regulated during early (30 days after parturition) and midlactation (120 days after parturition). The relative expression levels of miR-23a, miR-27b, miR-103, and miR-200a were calculated by the 2−ΔΔCT method, normalized with U6, and plotted relative to their respective expression in early lactation. Data are presented as the mean ± SD (n = 10). (B) MiR-23a, miR-27b, miR-103, and miR-200a positively respond to prolactin concentration in goat mammary gland epithelial cells. MiRNA expression was assessed by qRT-PCR, normalized to U6 internal control, and plotted relative to the expression at a concentration of 0 µg/ml. All data are presented as the mean ± SD (n = 9). *p < 0.05, **p < 0.01.
To further understand these miRNAs, we investigated the interactions between the miRNAs and prolactin, a primary hormone that regulates lactation in mammals (19). The serum concentration of prolactin paralleled changes in milk yield throughout the lactation (20,21). Prolactin activates the STAT5/JAK2 signaling pathway to increase milk fat synthesis in mammary gland (19). As expected, we observed a dose-dependent effect of prolactin on the expression of four miRNAs (i.e., miR-23a, miR-27b, miR-103, and miR-200a) in goat mammary gland epithelial cells (Fig. 1B). The expression levels of these four miRNAs were upregulated as the prolactin concentration increased from 0 to 10 µg/ml (Fig. 1B). This suggested that miR-23a, miR-27b, miR-103, and miR-200a may be downstream of prolactin and thereby related to milk fat synthesis. In addition, the other two miRNAs (miR-130a and miR-335) that are not sensitive to prolactin may not be correlated with the prolactin or the lactation. Therefore, we chose miR-23a, miR-27b, miR-103, and miR-200a for further study.
Elevated miRNA Expression Affects Gene mRNA Expression Associated With Milk Fat Synthesis in Goat Mammary Epithelial Cells
Previous studies have shown the following: miR-23 enhanced glutamine metabolism in human heart tissue (22); miR-27 decreased fat accumulation in 3T3-L1 preadipocytes (13); overexpressing miR-103 in preadipocyte 3T3-L1 cells increased triglyceride accumulation at an early stage of adipogenesis (23); and miR-200a promoted insulin signaling in human adipose cell (24). In this study, the expression patterns of miR-23a, miR-27b, miR-103, and miR-200a were found to be similar in mammary gland, and these miRNAs were all regulated by prolactin. Furthermore, these miRNAs have a common function in the regulation of lipid metabolism in adipose tissue (Table 4). To investigate the role of these four miRNAs in milk fat synthesis, we generated four recombinant adenoviruses expressing miR-23a, miR-27b, miR-103, and miR-200a in goat mammary gland epithelial cells. We assessed the expression of key genes related to milk fat synthesis in epithelial cells infected with each Ad-miRNA (Fig. 2 and Table 5). Ad-infected cells were used as a control.
Figure 2.
Overexpression of miRNA regulates gene mRNA expression associated with milk fat synthesis in epithelial cells. (A) miRNAs are overexpressed by using their respective Ad-miRNA in mammary gland epithelial cells. miRNA expression was assessed at 0, 24, 48, and 72 h postinfection. The expression levels of miR-23a, miR-27b, miR-103, and miR-200a were measured by qRT-PCR, normalized with internal control U2, and plotted relative to level of Ad-infected cells (control). Data are expressed as the mean ± SD (n = 10). *p < 0.05. (B) Overexpression of miR-23a promotes mRNA expression of FASN, ADRP, CD36, and TIP47. (C) Overexpression of miR-27b increases mRNA levels of SCD, but decreases mRNA levels of FASN, ADRP, and BTN1A1. (D) Overexpression of miR-103 upregulates mRNA levels of ACACA, FASN, ADRP, and LPL. (E) Overexpression of miR-200a promotes mRNA expression of SLC27A6 and CD36 but suppresses mRNA expression of GPR41 and TIP47. mRNA expression (B–E) was assessed 0, 24, 48, and 72 h after goat mammary gland epithelial cells were infected with their respective Ad-miRNA. The mRNA levels were measured by qRT-PCR, normalized with GAPDH, and presented relative to the mRNA amounts of Ad-infected cells (control). All data are expressed as the mean ± SD (n = 18). *p < 0.05.
Table 5.
Relative mRNA Expression in miR-200a Overexpression Background
| Gene Symbol | Gene Description | Ad-miR-200a to Ad |
|---|---|---|
| Do novo fatty acid synthesis | ||
| FASN | Fatty acid synthase | 1.38* |
| ACACA | Acetyl-coenzyme A carboxylase α | 1.47** |
| Triglyceride synthesis | ||
| SCD | Stearoyl-CoA desaturase (delta-9-desaturase) | 1.04 |
| GPAM | Glycerol-3-phosphate acyltransterase | 1.11 |
| AGPAT6 | 1-Acylaglycerol-3-phosphate O-acyltransferase 6 | 0.92 |
| LPIN1 | Lipin 1 | 0.15 |
| DGAT1 | Diacylglycerol acyltransferase 1 | 1.52* |
| Lipid drop formation and secretion | ||
| ADFP | Adipose differentiation-related protein | 0.49* |
| TIP47 | PAT-related proteins family, member 47 | 0.11* |
| BTN1A1 | Goat butyrophilin, subfamily 1, member A1 | 0.97 |
| Fatty acid uptake | ||
| CD36 | CD 36 molecule (thrombospondin receptor) | 1.08* |
| SLC27A6 | Solute carrier family 27 transporter, subfamily A, member 6 | 1.49* |
| FA transport | ||
| ABCA1 | ATP-binding cassette, subfamily A, member 1 | 0.85* |
| ABCG1 | ATP-binding cassette, subfamily G, member 1 | 0.41 |
| FA intracellular transport | ||
| FABP3 | Fatty acid-binding protein 3 | 1.81* |
| Fatty acids receptor | ||
| GPR41 | Orphan G protein-coupled receptor family, member 41 | 0.59* |
| Transcription factor | ||
| SREBP-1c | Sterol regulatory element-binding protein, member 1c | 1.15* |
| PPARγ | Peroxisome proliferator-activated receptor, member γ | 1.28* |
| LXRα | Nuclear oxysterol receptor, member α | 0.81** |
Genes were clustered based on main functions relative to milk fat synthesis. Ad-infected cells were used as control for Ad-miR-200a-infected cells. MRNA expression levels were determined at 72 h postinfection in goat mammary gland epithelial cells. mRNA levels were measured by qRT-PCR, normalized to GAPDH and plotted relative to controls. Data are presented as mean ± SD (n = 18).
*p < 0.05, **p < 0.01.
Milk fat synthesis in mammary gland epithelial cells is a complex biological process involving various enzymatic reactions (25–27). Fatty acids are synthesized by fatty acid synthase (FASN) and acetyl-coenzyme A carboxylase α (ACACA), unsaturated by stearoyl-CoA desaturase (SCD), and then processed into triglyceride by diacylglycerol acyltransferase1 (DGAT1). Lipids outside of cells are hydrolyzed by lipoprotein lipase (LPL) and transported into cells by thrombospondin receptor (CD36) and solute carrier family 27 transporter subfamily A member 6 (SLC27A6). These fatty acids are also processed into triglyceride. All triglycerides coalesce to form fat droplets by adipose differentiation-related protein (ADRP) and PAT-related protein family member 47 (TIP47), and then they are secreted by butyrophilin (BTN1A1). We assessed the expression of key genes related to these processes at 0, 24, 48, and 72 h after infecting mammary gland epithelial cells with Ad-miRNA or Ad (control). Our results showed that miR-23a, miR-27b, and miR-103 selectively regulated mRNA expression associated with milk fat synthesis. Overexpression of miR-23a (Fig. 2A) upregulated the mRNA expression levels of ADRP, FASN, CD36, and TIP47 throughout the observation period (Fig. 2B). CD36 expression gradually increased as miR-23a expression increased (Fig. 2B). At 72 h postinfection, the changes in mRNA expression of CD36 was the greatest among the four genes (Fig. 2B). This suggests that miR-23a positively regulates CD36 expression and thereby may accelerate fatty acid transport in epithelial cells. Elevated miR-27b expression (Fig. 2A) upregulated the mRNA expression level of SCD but downregulated the mRNA expression levels of FASN, ADRP, and BTN1A1 (Fig. 2C). Specifically, the mRNA expression of ADRP in Ad-miR-27b-infected cells was reduced by 60% compared with Ad-infected cells at 72 h postinfection (p < 0.05, Fig. 2C). This indicates a potential role of miR-27b in the negative regulation of fat droplet formation by suppressing ADRP expression in goat. The SCD expression in Ad-miR-27b-infected cells was higher than in Ad-infected cells, suggesting another potential role of miR-27b in regulation of unsaturated fatty acid composition. Furthermore, overexpression of miR-103 (Fig. 2A) gave an increase in the mRNA expression levels of FASN, ADRP, ACACA, and LPL (Fig. 2D). The mRNA expression level of LPL was nearly 10-fold higher in Ad-miR-103-infected cells than in Ad-infected cells at 72 h postinfection (Fig. 2D), indicating a potential role of miR-103 in the positive regulation of lipid hydrolysis, which supplies fatty acids for milk fat synthesis. These results indicate that these genes regulated (either directly or indirectedly) by miRNAs nearly cover the whole milk fat synthesis pathway, suggesting that miR-23a, miR-27b, and miR-103 are crucial for lactation in goat. In addition, we found that miR-23a, miR-27b, and miR-103 all affected FASN expression. FASN is a crucial enzyme with a central role in short- and medium-chain fatty acid synthesis of milk fat (28,29). The change in the mRNA expression of FASN indicates that these miRNAs might regulate fatty acid composition.
miR-200a Has an Extensive Role in the Entire Process of Milk Fat Synthesis
Among miR-23a, miR-27b, miR-103, and miR-200a, the change in miR-200a expression was the greatest in both the mammary gland during different lactation stages and in epithelial cells after alterations in the prolactin concentration. Therefore, we chose miR-200a for further functional validation. We assessed the mRNA expression of key genes associated with the entire fat synthesis process 72 h after infection with Ad-miR-200a or Ad (control) in goat mammary gland epithelial cells (Table 5). Overexpression of miR-200a suppressed the mRNA expression of ADRP and TIP47 associated with fat droplet formation, whereas it increased the mRNA expression of SLC27A6 and CD36 associated with fatty acid uptake, ACACA and FASN involved in fatty acid synthesis, and SCD and DGAT1 related to triglyceride synthesis (Table 5). The expression levels of fatty acid binding protein 3 (FABP3), GPR41, and ABCA1 also changed (Table 5). The gene that exhibits the greatest changes was found to be TIP47. The mRNA expression level of TIP47 in Ad-miR-200a-infected cells was reduced by 89% compared with that in Ad-infected cells (p < 0.05). In addition, the mRNA expression level of ADRP was also markedly decreased (0.49-fold, p < 0.05). ADRP and TIP47 are two crucial proteins for milk fat droplet formation in mammary gland epithelial cells (30). miR-200a strongly suppressed these two gene expression, suggesting that miR-200a may negatively regulate milk fat droplet formation of lactating goats. Furthermore, we assessed the expression of four random genes (CD36, SLC27A6, FASN, and TIP47) at 0, 24, 48, and 72 h postinfection. Elevated miR-200a expression (Fig. 2A) decreased the mRNA expression levels of TIP47, whereas it increased the mRNA expression levels of CD36, SLC27A6, and FASN throughout the observation period (Fig. 2E), which is similar to the results in Table 5. All the results indicated that miR-200a had an extensive role in milk fat synthesis (Table 5).
Sterol regulatory element-binding protein-1c (SREBP-1c), PPARγ, and liver X receptor α (LXRα) are key transcriptional factors that control the biological processes of lactation, including fatty acid synthesis (i.e., SREBP-1c targeting FASN and ACACA) (31), triglyceride synthesis [PPARγ targeting DGAT1 and ATP-binding cassette subfamily A member 1 (ABCA1)] (32), fat droplet formation (PPARγ targeting ADRP) (33), and cholesterol transport [LXRα targeting ATP-binding cassette subfamily G member 1 (ABCG1)] (34). When miR-200a was overexpressed, we found that the expression of PPARg (1.28-fold, p < 0.05) and its targets (i.e., DGAT1 and ABCA1) was upregulated, and the expression of SREBP-1c (1.15-fold, p < 0.05) and its targets (i.e., FASN and ACACA) was downregulated. Furthermore, expression of both LXRa (0.81-fold, p < 0.01) and its target ABCG1 was decreased. The parallel expression between transcription factors and their targets indicates that by regulating the mRNA expression of SREBP-1c, PPARg, and LXRa, elevated miR-200a expression can regulate the expression of genes associated with milk fat synthesis that are downstream of transcription factors during lactation.
Identification of Synergistic miRNAs
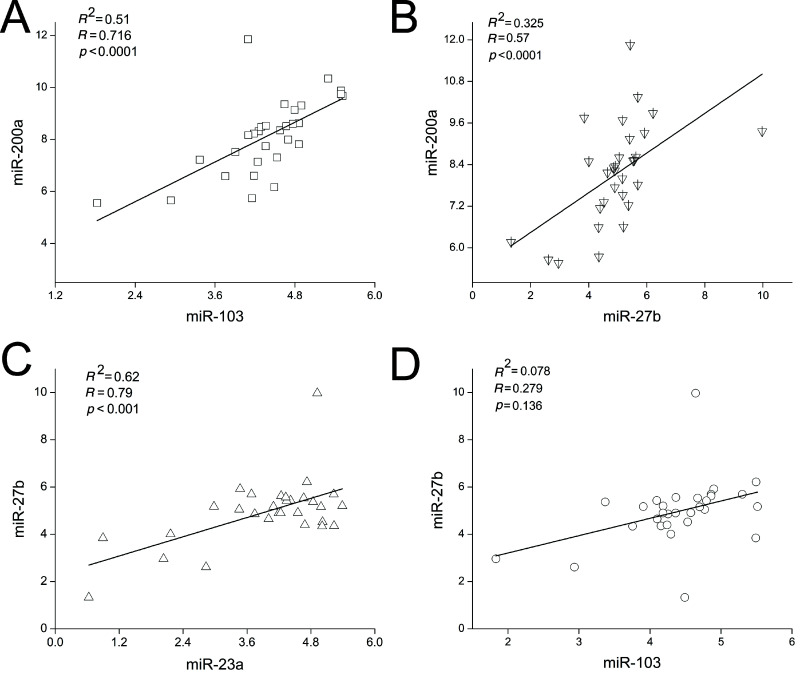
Under most conditions, miRNAs synergistically regulate complex biological processes and genes with the same or similar functions. For example, cardiac arrhythmogenesis is linked to miR-1 and miR-133, both of which act through the regulation of essential ion channel protein family (35). A limited number of miRNAs are thought to be able to control larger sets of genes through synergism. For example, Lin-4 and let-7 were the earliest miRNA pair to be experimentally verified and display cooperative regulation in Drosophila (36). During lactation of dairy goats, we wanted to determine whether miR-23a, miR-27b, miR-103, and miR-200a work simultaneously and synergistically to regulate milk fat synthesis. We selected 30 goats, sampled the mammary gland tissues at midlactation (120 days after parturition), and assessed miRNA expression in each goat by qRT-PCR. The Pearson correlation coefficient was used to determine any significant correlations in expression between the miRNAs. As a result, we found a strong correlation between miR-103 and miR-200a expression (Fig. 3A, R = 0.716, p < 0.0001), miR-27b and miR-200a expression (Fig. 3B, R = 0.57, p < 0.0001), and miR-23a and miR-27b expression (Fig. 3C, R = 0.79, p < 0.001). However, the expression of miR-23a did not correlate with either miR-200a or miR-103 expression (data not shown). In addition, miR-27b expression did not correlate with miR-103 expression (Fig. 3D, R = 0.279, p = 0.136). These correlations indicated that three miRNA pairs (i.e., miR-23a/miR-27b, miR-27b/miR-200a, and miR-200a/miR-103) are co-regulated during midlactation, suggesting that these miRNAs work in pairs to simultaneously regulate targets and induce the coexpression of their targets in the mammary gland of lactating goats.
Figure 3.
Correlation between miRNA expression in mammary gland of lactating goats. The expression levels of miR-23a, miR-27b, miR-103, and miR-200a were measured in each of the 30 goats (midlactation). Pearson analysis using the SPSS software was performed to identify the correlation coefficient. Intensity scatter plot shows comparison of two miRNA profiles in mammary gland. The expression of miRNAs was normalized to U6 (n = 30).
miRNAs can regulate miRNA expression (37,38). It is expected to determine whether mutual regulation exists between these miRNAs. We overexpressed miR-23a, miR-27b, miR-103, and miR-200a in mammary gland epithelial cells and assessed the expression of miRNAs in four Ad-miRNA-infected cells (Table 6). Ad-infected cells were used as a control, and we found that miRNAs mutually regulate expression in goat. Overexpression of miR-103 (2.93-fold, p < 0.05) downregulated miR-200a expression (0.88-fold, p < 0.05), and elevated miR-200a expression (2.44-fold, p < 0.01) decreased miR-103 expression (0.72-fold, p < 0.05). miR-200a overexpression (2.44-fold, p < 0.01) led to the upregulation of miR-27b expression (1.10-fold, p < 0.05), and miR-27b overexpression (1.93-fold, p < 0.05) resulted in an increased miR-200a expression (1.29-fold, p < 0.05). Additionally, mutual regulation was also observed between miR-27b and miR-23a, as the upregulation of one increased the expression of the other. However, no significant interaction was found between miR-23a and miR-103 or miR-103 and miR-27b. These results support our hypothesis that miRNAs with co-regulated expression can also mutually regulate expression in the mammary gland. Furthermore, the “co-regulation” and “mutual regulation” indicated the complexity of synergism of miRNA regulation.
Table 6.
miRNAs Mutually Regulate Expression in Goat Mammary Gland Epithelial Cells
| Cells Infected by Different Ad-miRNAs | miRNA Expression | |||
|---|---|---|---|---|
| miR-23a | miR-27b | miR-103 | miR-200a | |
| Ad-miR-23a | 3.52* | 1.17* | 0.89 | 1.08 |
| Ad-miR-27b | 2.92* | 1.93* | 1.04 | 1.29* |
| Ad-miR-103 | 0.87 | 1.77 | 2.93* | 0.88* |
| Ad-miR-200a | 0.91 | 1.10* | 0.72* | 2.44** |
miRNA expression was determined in epithelial cells at 72 h postinfection. Expression levels of miR-23a, miR-27b, miR-103, and miR-200a in four groups of Ad-miRNA-infected cells were measured by qRT-PCR, normalized to U2, and presented relative to Ad-infected cells (control) (n = 18). Bold numbers indicate the overexpressed miRNA that was mediated by its Ad-miRNA.
*p < 0.05, **p < 0.01.
DISCUSSION
miRNA Screening
Studies have shown that milk fat synthesis involves the synergistic regulation of genes during lactation (26,27). miRNAs are predicted to target one third of human genes (39). We speculated that miRNAs synergistically regulate milk fat synthesis in goats. Thus far, no miRNA function has been validated in goat mammary gland. We speculated that miRNAs that are reported to regulate fat synthesis in other tissues or cell lines may play similar roles in the goat mammary gland. To address our hypothesis, we performed four experiments: screening for differentially expressed miRNAs during the lactation cycle (Fig. 1A), analysis of miRNA sensitivity to prolactin (Fig. 1B), investigation of the role of miRNAs in milk fat synthesis (Fig. 2 and Table 5), and analysis of the relationship between miRNAs (Fig. 3). As expected, we obtained three pairs of miRNAs (miR-23a and miR-27b, miR-103 and miR-200a, miR-27b and miR-200a) that are synergistically expressed during lactation. In addition, elevated levels of miR-23a, miR-27b, miR-103, or miR-200a expression can affect gene mRNA expression associated with milk fat synthesis in goat mammary gland epithelial cells. Our results suggest that miR-23a, miR-27b, miR-103, and miR-200a participate in the regulation of lactation in goats, which supports our hypothesis.
Molecular Network of Milk Fat Synthesis
Hormone–miRNA Regulation. miRNA expression has been shown to be regulated by hormones (40,41). In this study, the expression levels of miR-23a, miR-27b, miR-103, and miR-200a were upregulated as the prolactin concentration increased. Prolactin acts through the STAT5/JAK2 pathway to increase gene mRNA expression associated with milk fat synthesis in mammary gland (42,43). When we used MicroCosm and PicTar to predict the targets of these four miRNAs (Table 4), we found that some targets are downstream of prolactin, such as STATB (miR-23) and STAT5A (miR-103), and some targets are downstream of STAT5/JAK2, including MGF (miR-200) and BTN1A1 (miR-27). When the prolactin concentration increased, the expression of these target mRNAs may be upregulated. However, epithelial cells generate much more these endogenous miRNAs (Fig. 1B). Therefore, the relationship between prolactin and miRNAs is complicated and needs further validation. From this point, milk fat synthesis may be involved in a hormone–miRNA regulatory network in mammary gland.
miRNA–miRNA Regulation. Some co-regulated miRNAs are cotranscribed (44). We speculated that the co-regulated miRNAs (i.e., miR-200a and miR-103, miR-23a and miR-27b, miR-103 and miR-27b) identified in this study have a common mechanism of miRNA transcription initiation. The transcription of miRNAs is controlled by RNA polymerase II (Pol II) or III (Pol III) (45,46). It has been shown that the miR-23a–miR-27b–miR-24-2 cluster is transcribed by Pol II in humans (47). This information supported our result that the expression levels of miR-23a and miR-27b are co-regulated (Fig. 3C). MiR-200a expression correlates with miR-27b expression (Fig. 3B), suggesting that miR-200a may also be transcribed by Pol II. Moreover, miR-103 is co-regulated with its host gene PANK, indicating they may be transcribed from the same mRNA, which is activated by Pol II (23). On the basis of this information, miR-23a, miR-27b, miR-103, and miR-200a may be all transcribed by Pol II. However, miR-27b expression does not correlate with miR-103 expression, and we have not found a reasonable explanation for this lack of correlation. Additionally, during pAd-miRNA construction, we cloned goat pri-miR-23a, pri-miR-27b, pri-miR-103, and pri-miR-200a according to the cow pri-miRNA sequences. When we analyzed these four pri-miRNA sequences in the mammal genome (human, cow, sheep, and goat), we found that the locations of these pri-miRNAs are conserved in the mammalian genome (Fig. 4). pri-miR-23a, pri-miR-27b, and pri-miR-200a are located in the intergenic region; miR-103-1 is located in intron 5 of pantothenate kinase 3 (PANK3). Although miR-23a and miR-27b are reported to be located in one cluster, the locations of pri-miR-23a and pri-miR-27b are distant (Fig. 4). We speculated that these two pri-miRNAs cloned in this study were located in the other region of the goat genome and not the miR-23a–miR-27b–miR-24-2 cluster. Moreover, transcription initiation is mediated mainly through transcription factor interactions and can be affected by different Pol enzymes (6,45). A difference in the activity of one factor can result in different miRNA transcription efficiency (47). Therefore, this complicated regulation of miRNA indicates that miRNAs control a complicated gene expression regulatory network during lactation.
Figure 4.
The location of pre-miRNAs in genome.
Interactions were found between three co-regulated miRNA pairs. Our data showed a mutual promotion of expression between miR-27b and miR-200a as well as miR-23a and miR-27b (Table 6). The interaction between miR-200a and miR-103 showed a mutual reduction in expression (Table 6). Through genome sequence analysis, miR-103-1 was found to be located in intron 5 of PANK3. We hypothesize that miR-103 and PANK3 are cotranscribed (23). Moreover, PANK3 is the predicted target of miR-200a (TargetScan, Release 6.2, 2012; Human). We speculate that elevated miR-200a expression decreased PANK3 expression, which can downregulate endogenous miR-103-1 expression. Combining our results (Fig. 3 and Table 6), we speculated that there is a universal miRNA–miRNA regulatory network in the mammary gland. For the other two miRNA pairs, we think that they mutually regulate expression in an indirect manner.
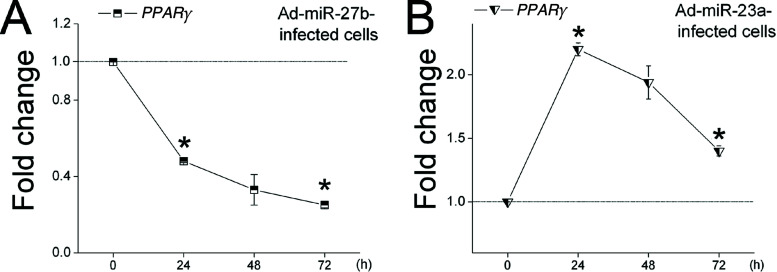
miRNA–mRNA Regulation. In this study, we demonstrated the role of miR-23a, miR-27b, miR-103, and miR-200a in the goat for the first time. Based on the significant effects of these four miRNAs on gene expression (i.e., miR-23a/CD36, miR-27b/ADRP, miR-103/LPL, miR-200a/ADRP and TIP47) (Fig. 2 and Table 5), we predicated that miR-23a and miR-103 may regulate raw material content for milk fat synthesis, and miR-27b and miR-200a may control milk fat droplet accumulation in epithelial cells. To further explore the function of miRNAs, we designed to investigate the mechanism of miRNA regulation in these genes. However, no miRNA binding sites are found in the 3′, 5′ or the coding region of these genes, indicating that the regulation of these miRNAs in mRNAs are indirect. A possible explanation is that miR-23a, miR-27b, miR-103, and miR-200a may target other genes that can affect the transcriptional activity of these genes. For example, overexpression of miR-27b decreased the mRNA level of ADRP (Fig. 2C). Previous studies have shown that PPARγ, participating in the transcriptional activation of lipogenic genes (i.e., ADRP), is a target of miR-27b in adipose tissue (13). We measured the mRNA levels of PPARg in Ad-miR-27b-infected cells at 0, 24, 48, and 72 h postinfection. We found that overexpression of miR-27b suppressed PPARg expression throughout the observation period (Fig. 5A). Moreover, the expression pattern of PPARg mRNA was consistent with the mRNA expression pattern of ADRP downstream of PPARγ. These findings can partially explain the decrease in ADRP expression as miR-27b overexpressed (Fig. 2C). However, we have not found underlying causes for the effects of miR-23a, miR-103, and miR-200a on gene expression (miR-23a/CD36, miR-103/LPL, miR-200a/ADRP and TIP47).
Figure 5.
miR-27b and miR-23a regulate the mRNA level of PPARg in goat mammary gland epithelial cells. (A) Overexpression of miR-27b decreases the mRNA level of PPARg. (B) Overexpression of miR-23a increases the mRNA level of PPARg. mRNA levels of PPARγ (A, B) were determined in epithelial cells 0, 24, 48, and 72 h after infecting with Ad-miRNAs or Ad (control). The mRNA levels were measured by qRT-PCR, normalized with GAPDH, and expressed relative to the mRNA amounts of Ad-infected cells (control). All data are presented as the mean ± SD (n = 18). *p < 0.05.
The transcription of miRNAs and their targets are usually negatively co-regulated. Upregulation of miRNA expression generally inhibits their targets’ mRNA expression. However, in some cases, the transcription of the miRNAs and their targets are positively co-regulated (i.e., in rat postnatal oligodendrocyte lineage cells) (38,48), and upregulation of expression of some miRNAs increases the mRNA levels of their targets. For instance, elevated miR-17-5p expression in human cancer cells by c-Myc transcriptionally upregulates E2F1, a target gene of miR-17-5p (48). Similarly, in the present study, overexpression of miR-23a upregulated the mRNA expression of ADRP, a predicted target of miR-23a (Fig. 2B). The repressive effect of miRNAs on target expression is often limited to the level of translation (4,50). Therefore, we wanted to ask if miR-23a acts in concert with other regulatory processes related to ADRP expression. We assessed the mRNA level of PPARg, a crucial transcription factor of ADRP (30), and we found that overexpression of miR-23a significantly upregulated PPARg expression (Fig. 5B). Furthermore, the expression patterns of PPARg and miR-23a were similar, which can partially explain the increased mRNA expression of ADRP.
The miR-200 family regulates the epithelial-to-mesenchymal transition by targeting ZEB1 (50). We performed an MTT test to investigate the effect of elevated miR-200a expression in epithelial cells. However, we have not observed a difference in the total numbers of Ad-miR-200a-infected cells and control cells. We speculate that the dominant function of miR-200a in normal cells may be different from that in cancer cells. Alternatively, the targets of miR-200a may vary in goats and other mammals. Therefore, the validation of miR-200a targets is important to us. Although overexpression of miR-200a had little effect on the mRNA expression of its predicted targets (i.e., BTN1A1) (Tables 4 and 5), we do not exclude the possibility that these predicted targets are true targets of miR-200a because the repressive effect of miRNAs on target expression is often limited to the level of protein (4,50). For example, overexpression of the miR-200 family in ovarian cancer cells suppressed the protein expression of its target E-cadherin repressors (ZEB1/2) but had no effect on the target’s mRNA expression (50). Thus far, we have used the MFOLD software to analyze the ΔG of the 80-bp flanking sequence of the miR-200a binding site on these predicted targets (51) and have constructed five miRNA sensors using the pGL3-control luciferase reporter vector inserted into the XbaI locus with 100–200 bp miRNA binding site of potential targets, including MGF, BTN1A1, GPR43, LPL, and SCD. However, we have not obtained any direct proof that one of these genes is the target of miR-200a. Target validation experiments for miR-23a, miR-27b, and miR-103 are also ongoing.
Our previous studies on the Saanen dairy goats showed that changes in milk fat fatty acid composition paralleled changes in different stages of lactation (52); additionally, some genes (i.e., FABP4 and ABCG2) also changed mRNA expression levels during the lactation cycle (53). MiR-23a, miR-27b, and miR-103 exhibit expression during the lactation cycle (Fig. 1A), and all these four miRNAs can regulate FASN expression (Fig. 2), a key regulator for milk fatty acid composition. We speculate that the changes in fatty acid composition of goat milk may be related to the co-regulation of miRNA and mRNA in mammary gland. Combined with our results (co-regulated and mutually regulated miRNAs) (Fig. 3 and Table 5), all the information suggests that the molecular network of milk fat synthesis is composed of many regulatory interactions, including miRNA to miRNA, miRNA to gene, gene to miRNA, and gene to gene. This complicated network can extend to all biological processes during the entire lactation process in the mammary gland, indicating the importance of molecular synergistic work.
Collectively, this study provides the first experimental evidence for the correlation of miRNAs in the regulation of lactation in goat mammary gland. For the first time, we have identified the role of miR-23a, miR-27b, miR-103, and miR-200a in the expression of mRNAs associated with milk fat synthesis. This work indicates that there must be a complicated molecular network of miRNA–miRNA and miRNA–mRNA regulatory interactions required for milk fat synthesis to occur in the mammary gland of lactating goats.
ACKNOWLEDGMENTS
We are grateful to Prof. Li Changan for biopsy sampling. This project was co-supported by Special Fund for Agro-scientific Research in the Public Interest of China (No. 201103038) and National Natural Science Foundation of China (No. 31072013).
REFERENCES
- 1. Heinlein GFW, Caccse R. Goat milk versus cow milk. Dairy Goat J 2003; 81:12–14. [Google Scholar]
- 2. Jenness R. Composition and characteristics of goat milk: Review 1968–1979. J Dairy Sci 1980; 63:1605–1630. [Google Scholar]
- 3. Haenlein GFW. Past, present, and future perspectives of small ruminant research. J Dairy Sci 2001; 84:2097–2115. [DOI] [PubMed] [Google Scholar]
- 4. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 5. Ambros V. The functions of animal microRNAs. Nature 2004; 431:350–355. [DOI] [PubMed] [Google Scholar]
- 6. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005; 6:376–385. [DOI] [PubMed] [Google Scholar]
- 7. Tanaka T, Haneda S, Imakawa K, Sakai S, Nagaoka K. A microRNA, miR-101a, controls mammary gland development by regulating cyclooxygenase-2 expression. Differentiation 2009; 77:181–187. [DOI] [PubMed] [Google Scholar]
- 8. Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003; 9:1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun L, Xie HM, Mori MA, Alexander R, Yuan BB, Hattangadi SM, et al. Mir193b–365 is essential for brown fat differentiation. Nat Cell Biol 2011; 13:958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao CC, Calado DP, Galler G, Thai T, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007; 131:146–159. [DOI] [PubMed] [Google Scholar]
- 11. Sacco J, Adeli K, Jennifer S, Khosrow A. MicroRNAs: Emerging roles in lipid and lipoprotein metabolism. Lipid Metab 2012; 23:220–225. [DOI] [PubMed] [Google Scholar]
- 12. Esau C, Davis S, Murray SF, Yu XX, Pandy SK, Pear M, et al. MiR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006; 3:87–98. [DOI] [PubMed] [Google Scholar]
- 13. Lin Q, Gao ZG, Alarcon RM, Ye JP, Yun Z. A role of miR-27 in the regulation of adipogenesis. FEBS J 2009; 276:2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011; 474:649–654. [DOI] [PubMed] [Google Scholar]
- 15. Ji ZB, Wang GZ, Xie ZJ, Zhang CL, Wang JM. Identification and characterization of microRNAs in the dairy goat (Capra hircus) mammary gland by Solexa deep-sequencing technology. Mol Biol Rep 2012; 39:9361–9371. [DOI] [PubMed] [Google Scholar]
- 16. Xu J, Li CX, Li YS, Lv JY, Ma Y, Shao TT, et al. MiRNA–miRNA synergistic network: Construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res 2011; 39:825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. German T, Barash I. Characterization of an epithelial cell line from bovine mammary gland. In Vitro Cell Dev Biol Anim 2012; 38:282–292. [DOI] [PubMed] [Google Scholar]
- 18. Folley SJ. Biochemical aspects of mammary gland function. Biol Rev 1949; 24:316–354. [DOI] [PubMed] [Google Scholar]
- 19. Hennighausen L, Robinson GW, Wagner KU, Liu XW. Prolactin signaling in mammary gland development. J Biol Chem 1997; 272:7567–7569. [DOI] [PubMed] [Google Scholar]
- 20. Hart IC, Bines JA, Morant SV, Ridley JL. Endocrine control of energy metabolism in the cow: Comparison of the levels of hormones (prolactin, growth hormone, thyroxine, and insulin) and metabolites in the plasma of high and low-yielding cattle at various stages of lactation. J Endocrinol 1978; 77:333–345. [DOI] [PubMed] [Google Scholar]
- 21. Koprowsk JA, Tucker HA. Serum prolactin during various physiological states and its relationship to milk production in the bovine. Endocrinology 1973; 92:1480. [DOI] [PubMed] [Google Scholar]
- 22. Frost RJA, Rooij EV. MiRNAs as therapeutic targets in ischemic heart disease. J Cardiovasc Translational Res 2010; 3:280–289. [DOI] [PubMed] [Google Scholar]
- 23. Xie HM, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are down regulated in obesity. Diabetes 2009; 58:1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teleman AA. MiR-200 De-FOGs insulin signaling. Cell Metab 2010; 11:8–9. [DOI] [PubMed] [Google Scholar]
- 25. Hansen HO, Grunnet I, Knudsen J. Triacylglycerol synthesis in goat mammary gland. The effect of ATP, Mg2+ and glycerol 3-phosphate on the esterification of fatty acids synthesized de novo. Biochem J 1984; 220:513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genomics 2008; 9:366–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bionaz M, Loor JJ. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform Biol Insights 2011; 5:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen JK, Knudsen J. Transacylation as a chain-termination mechanism in fatty acid synthesis by mammalian fatty acid synthetase: Synthesis of butyrate and hexanoate by lactating cow mammary gland fatty acid synthetase. Biochem J 1980; 186:287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kundsen J, Grunnet I. Transacylation as a chain-termination mechanism in fatty acid synthesis by mammalian fatty acid synthetase. Synthesis of medium-chain-length (C8-C12) acyl-CoA esters by goat mammary-gland fatty acid synthetase. Biochem J 1982; 202:139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McManaman JL, Russell TD, Schaack J, Orlicky DJ, Robenek H. Molecular determinants of milk lipid secretion. J Mammary Gland Biol Neoplasia 2007; 12:259–268. [DOI] [PubMed] [Google Scholar]
- 31. Damiano F, Alemanno S, Gnoni GV, Siculella L. Translational control of the sterol-regulatory transcription factor SREBP-1 mRNA in response to serum starvation or ER stress is mediated by an internal ribosome entry site. Biochem J 2010; 429:603–612. [DOI] [PubMed] [Google Scholar]
- 32. Kadegowda AKG, Bionaz M, Piperova LS, Erdman RA, Loor JJ. Peroxisome proliferator-activated receptor-γ activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J Dairy Sci 2009; 92:4276–4289. [DOI] [PubMed] [Google Scholar]
- 33. Yamazaki T, Shiraishi S, Kishimoto K, Miura SJ, Ezaki O. An increase in liver PPARγ2 is an initial event to induce fatty liver in response to a diet high in butter: PPARγ2 knockdown improves fatty liver induced by high-saturated fat. J Nutr Biochem 2011; 22:543–553. [DOI] [PubMed] [Google Scholar]
- 34. Cheng YX, Liu GB, Pan Q, Guo SF, Yang XH. Elevated expression of liver X receptor alpha (LXRα) in myocardium of streptozotocin-induced diabetic rats. Medicine 2011; 34:698–706. [DOI] [PubMed] [Google Scholar]
- 35. Luo XB, Lin HX, Pan ZW, Xiao JN, Zhang Y, Lu YJ, et al. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem 2008; 283:20045–20052. [DOI] [PubMed] [Google Scholar]
- 36. Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila . Genome Biol 2003; 5:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tuccoli A, Poliseno L, Rainaldi G. MiRNAs regulate miRNAs. Cell Cycle 2006; 5:2473–2476. [DOI] [PubMed] [Google Scholar]
- 38. Tsang J, Zhu J, Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 2007; 26:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Griffiths JS. The microRNA registry. Nucleic Acids Res 2004; 32:109–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheung L, Gustavsson C, Norstedt G, Tollet-Egnell P. Sex-different and growth hormone-regulated expression of microRNA in rat liver. BMC 2009; 10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cui J, Eldredge JB, Xu Y, Puett D. MicroRNA expression and regulation in human ovarian carcinoma cells by luteinizing hormone. PLoS ONE 2011; 6:e21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gouilleux F, Wakao H, Mundt M, Groner, B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J 1994; 13:4361–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hogan JC, Stephens JM. The regulation of fatty acid synthase by STAT5A. Diabetes 2005; 54:1968–1975. [DOI] [PubMed] [Google Scholar]
- 44. Gerin S, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem 2010; 285:33652–33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 2006; 13:1097–1101. [DOI] [PubMed] [Google Scholar]
- 46. Lee Y, Kim MJ, Han JJ, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004. 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roy A, Meisterernst M, Pognonec P, Roeder RG. Cooperative interaction of an initiator-binding transcription initiation factor and the helix–loop–helix activator USF. Nature 1991; 354:245–248. [DOI] [PubMed] [Google Scholar]
- 48. Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci 2008; 28:11720–11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. C-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005; 435:839–843. [DOI] [PubMed] [Google Scholar]
- 50. Park SM, Gaur AB, Lengyel E, Marcus EP. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008; 22:894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuhn DE, Martin MM, Feldman DS, Terry AV, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods 2007; 44:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luo J, Shan CY, Liu LP, Wang, HB, Sun XQ, Wang XQ, et al. Preliminary study on effect of parity on short- and medium-chain fatty acids composition in milk of Xinong Saanen goat. J Northwest Sci-Tech Univ Agric Forest 2005; 3:23–28. [Google Scholar]
- 53. Wu HJ, Luo J, Zhang LJ, Han XF, Yang BJ, Wang HB. Studies on differentially expressed genes in mammary gland of Xinong Saanen Goat at peak and early lactation stages. Chin J Anim Vet Sci 2008; 39:136-142. [Google Scholar]
- 54. Ramachandran D, Roy U, Garg S, Ghosh S, Pathak S, Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion pancreatic β-islets. FEBS J 2011; 278:1167–1174. [DOI] [PubMed] [Google Scholar]
- 55. Kim SY, Kim AY, Lee HW, Son YH, Lee GY, Lee JW, et al. MiR-27a is a negative regulator of adipocyte differentiation via suppressing PPARγexpression. Biochem Biophys Res Commun 2010; 392:323–328. [DOI] [PubMed] [Google Scholar]
- 56. He A, Zhu L, Gupta N, Chang Y, Fang, F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3 -L1 adipocytes. Mol Endocrinol 2007; 21:2785–2794. [DOI] [PubMed] [Google Scholar]
- 57. Davalos A, Goedeke L, Smibert P, Ramireza CM. MiR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA 2011; 108:9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, et al. MiR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptorγ expression. Mol Cell Biol 2011; 31:626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takanabe R, Ono K, Abe Y, Takaya T, Hore T, Wada H, et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun 2008; 376:728–732. [DOI] [PubMed] [Google Scholar]
- 60. Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res 2010; 86:410–420. [DOI] [PubMed] [Google Scholar]
- 61. Nakanishi N, Nakagawa Y, Tokushige N, Aoki N, Matsuzaka T, Ishii K, et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun 2009; 385:492–496. [DOI] [PubMed] [Google Scholar]