Abstract
The advent and persistence of the Severe Acute Respiratory Syndrome Coronavirus – 2 (SARS-CoV-2)-induced Coronavirus Disease (COVID-19) pandemic since December 2019 has created the largest public health emergency in over a century. Despite the administration of multiple vaccines across the globe, there continues to be a lack of approved efficacious non-prophylactic interventions for the disease. Flavonoids are a class of phytochemicals with historically established antiviral, anti-inflammatory and antioxidative properties that are effective against cancers, type 2 diabetes mellitus, and even other human coronaviruses. To identify the most promising bioactive flavonoids against the SARS-CoV-2, this article screened a virtual library of 46 bioactive flavonoids against three promising targets in the SARS-CoV-2 life cycle: human TMPRSS2 protein, 3CLpro, and PLpro. By examining the effects of glycosylation and other structural-activity relationships, the presence of sugar moiety in flavonoids significantly reduces its binding energy. It increases the solubility of flavonoids leading to reduced toxicity and higher bioavailability. Through protein-ligand contact profiling, it was concluded that naringin formed more hydrogen bonds with TMPRSS2 and 3CLpro.
In contrast, hesperidin formed a more significant number of hydrogen bonds with PLpro. These observations were complimented by the 100 ns molecular dynamics simulation and binding free energy analysis, which showed a considerable stability of docked bioflavonoids in the active site of SARS-CoV-2 target proteins. Finally, the binding affinity and stability of the selected docked complexes were compared with the reference ligands (camostat for TMPRSS2, GC376 for 3CLpro, and GRL0617 for PLpro) that strongly inhibit their respective SARS-COV-2 targets. Overall analysis revealed that the selected flavonoids could be potential therapeutic agents against SARS-CoV-2. Naringin showed better affinity and stability for TMPRSS2 and 3CLpro, whereas hesperidin showed a better binding relationship and stability for PLpro.
Keywords: Natural products, COVID-19, Computational studies, COVID-19 drug discovery, Glycosylated flavonoids, Hesperidin, Naringin, In silico, SARS-CoV-2, TMPRSS2, 3CLpro, PLpro
Graphical abstract
1. Introduction
In late December 2019, multiple cases of atypical pneumonia in Wuhan city, China, were reported to the World Health Organization (WHO); in less than three months, a global pandemic was declared [1]. The novel coronavirus (2019-nCoV, SARS-CoV-2) induced coronavirus disease (COVID-19) became the leading public health emergency worldwide, resulting in around 300 million infections and 5.5 million deaths as of January 2022 [2]. Clinical symptoms of this severe acute respiratory syndrome (SARS) manifest primarily in the upper and lower respiratory tracts. They include dry cough, fever, shortness of breath, sore throat, fatigue, nasal congestion, myalgia, chills, dizziness, and in some, loss of smell and taste [3]. The severity of these manifestations varies between individuals – in particular, the worsening prognosis was associated with an increase in age and/or the presence of underlying chronic or immunodeficiency disorders [4]. Despite the accelerated development and mass roll-out of multiple COVID-19 vaccines leading to the administration of over 9 billion doses worldwide, in addition to their widely reported possible resultant adverse effects, there continues to be a lack of approved and effective therapeutic drugs as treatment or cure [[5], [6], [7]]. Of the existing interventions, the most used are prophylaxis using antiviral or antiretroviral therapy, supporting treatment using corticosteroids and immunosuppressants, breathing support such as mechanical ventilation, adjunctive therapy, and convalescent plasma therapy [5,8]. The ever-increasing number of infections despite mass vaccination, the circulation of new and more infectious strains, and the long-term effects of COVID-19 infection, even on individuals with no prior comorbidities, urgently demand the expedited screening, investigation, and development of target-specific anti-SARS-CoV-2 drug candidates to treat active cases [9].
Characterized by crown-like spike proteins on the virion surface, coronaviruses (CoVs) refer to a group of enveloped viruses with positive-sense single-stranded RNA ((+)-ssRNA) belonging to the taxonomical subfamily Orthocoronavirinae/Coronavirinae, under the family Coronaviridae, and the order Nidovirales [10]. They are classified into the genera of alpha-, beta-, gamma- and delta coronaviruses based on their respective hosts: while alpha- and beta coronaviruses infect mammals (e.g., porcine epidemic diarrhea virus and SARS-CoV), the latter two cause diseases in birds (e.g., avian coronavirus and bulbul coronavirus HKU11) [11]. Six species of coronaviruses were identified as pathogens in humans, of whom three beta-CoVs have been responsible for significant outbreaks in the 21st century: the severe acute respiratory syndrome (SARS) epidemic in 2003, the Middle East respiratory syndrome (MERS) epidemic in 2012, and recently, the SARS-CoV-2-induced COVID-19. Multiple SARS-CoV-2 variants of concern are in circulation as the virus spreads, generating new mutations. Many such variants question the real-world effectiveness of existing vaccines and emphasize the importance of drug discovery efforts for this viral respiratory disease.
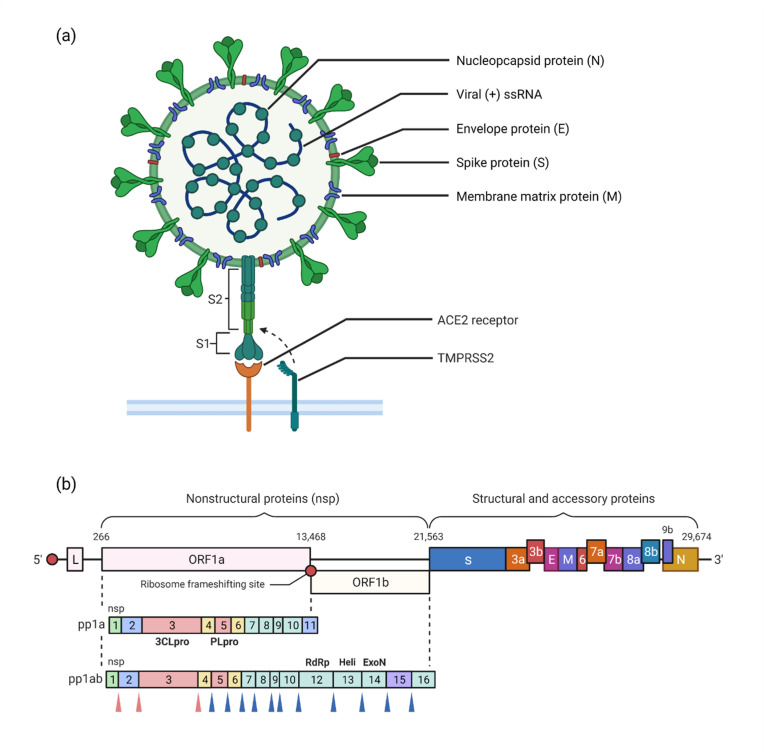
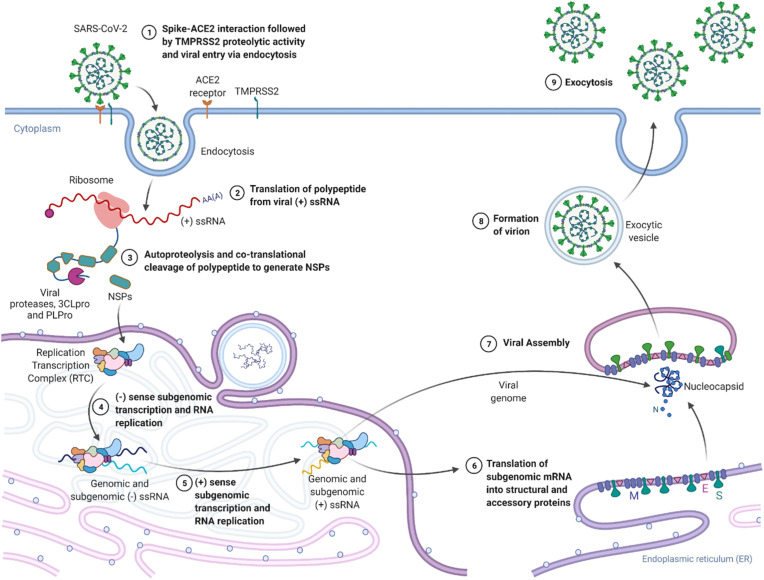
SARS-CoV-2 is characterized by multiple structural proteins including the Nucleocapsid (N), Envelope (E), Membrane matrix (M), and Spike (S) proteins (Fig. 1 a), in addition to various other significant non-structural proteins (NSPs), including its RNA-dependent RNA polymerase (RdRp, nsp12), helicase (nsp13), exonuclease (nsp14), as well as two major cysteine proteases: the main protease (3CLpro or Mpro, nsp3) and the Papain-like protease (PLpro, nsp5) [12]. Following binding of the S1 subunit of spike protein to the Angiotensin-converting enzyme-2 (ACE2) receptor in human cells, the Transmembrane Protease Serine 2 (TMPRSS2) protease in human cell membranes cleaves the S protein. It activates the S2 domain, ultimately leading to cell fusion and viral entry. Following spike fusion through endocytosis and unloading of the viral genome into the cytoplasm, the released viral (+) ssRNA uses host ribosomal machinery to translate open reading frames (ORFs) 1a and 1b into polypeptides pp1a and pp1ab [13]. These proteins are post-translationally cleaved and modified by the proteases 3CLpro and PLpro into 16 non-structural proteins (NSPs) that give rise to proteins of the viral replication and transcription complex (RTC), which leads to viral genome replication and structural protein synthesis, followed by exocytosis of the newly formed virion progeny (Fig. 2 ) [[14], [15], [16], [17]].
Fig. 1.
Schematic diagram of major SARS-CoV-2 structural and functional components and their genes. (a) A single-stranded positive sense (+) viral RNA (ssRNA) is encapsulated regularly with Nucleocapsid (N) proteins. The viral lipid bilayer membrane consists of both the Membrane matrix (M) and the Envelope (E) proteins, from where the Spike (S) protein (consisting of subunits S1 and S2) protrudes and interacts with human Angiotensin Converting Enzyme 2 (ACE2) receptor, leading to activation of the S protein by Transmembrane Protease Serine 2 (TMPRSS2), ultimately facilitating viral entry and cell fusion. (b) Overview of the most important SARS-COV-2 protein-coding genes: Open Reading Frames 1a & 1b code for polypeptides pp1a and pp1ab, which constitute multiple Non-Structural Proteins (NSPs) such as Papain-like Protease (PLpro), 3-Chyomtrypsin-like protease (3CLpro or Mpro), RNA-dependent RNA-Polymerase (RdRp), Helicase (Heli) and Exonuclease (ExoN). The genome's structural proteins encoded include the S, E, M, and N proteins. During post-translational stages, the corresponding PLpro and 3CLpro cleavage sites (indicated by red and blue arrowheads, respectively) are demarcated. The entire SARS-CoV-2 viral genome consists of ∼29.9 kb. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
The SARS-CoV-2 viral life cycle and potential sites for drug-action.
1.1. SARS-CoV-2 3CLpro
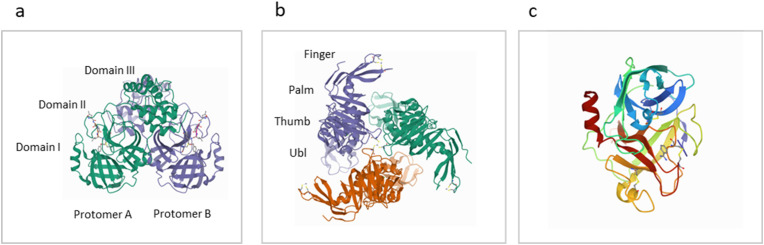
The SARS-CoV-2 3CLpro is a 306-amino acid, 33.8 kDa, a hydrophilic proteolytic enzyme belonging to the PA class of proteases. Its sequence identity is conserved with those of SARS-CoV (96.08%), MERS-CoV (87.00%), Human-CoV (90.00%), and Bovine-CoV (90.00%) homologs [18]. The enzyme cleaves the functional polypeptides at 11 specific sites (Fig. 1b). It is a homodimer consisting of two chains (protomers A & B) connected by an N-finger. Each chain consists of three domains (I, II & III) and an active catalytic dyad (Cys-145 and His-41) that cleaves at ↓: X‐(L/F/M)‐Q↓(G/A/S (Fig. 3 a) [19]. Due to its vital role in the viral life cycle and the absence of alternative agents for proteolytic processing in human cells, this enzyme is a desirable target for drug development and subsequent trials.
Fig. 3.
The X-ray crystal structures and domains of potential SARS-CoV-2 targets. (a): 3CLpro complexed with N3 inhibitor (PDB: 6LU7). (b) PLpro (PDB: 6W9C). (c) DESC1, a member of the TMPRSS2 family (PDB: 2OQ5).
1.2. SARS-CoV-2 PLpro
PLpro is a slightly basic, ∼37 kDa, 315 residue enzyme of the more prominent 1945-amino acid, 212 kDa, Nsp3. It lies between the SARS unique domain (SUD/HVR) and a nucleic acid-binding domain (NAB). The enzyme cleaves the functional polypeptides at 3 different sites, but along with 3CLpro, it is considered a significant player in the viral life cycle (Fig. 1b). It acts at recognition sequence consensus LXGG ↓ XX. Like SARS-CoV-2 3CLpro, the PLpro sequence is conserved among other human beta coronaviruses, being 83% identical and 90% similar to SARS-CoV, and 31% identical and 49% similar to MERS-CoV [19]. It is a monomeric enzyme consisting of two domains, the Ubiquitin-like domain (Ubl) and the catalytic domain, which subdomains, thumb, palm, and fingers (Fig. 3b) define its characteristics. The active site, consisting of the catalytic triad Cys111, His272, and Asp286, is located between the thumb and palm subdomains [17]. In addition to its role in polypeptide processing, the enzyme also cleaves the antiviral response-regulating ubiquitin and ubiquitin-like interferon-stimulated gene 15 protein (ISG15) from host proteins, subsequently leading to the reversal of post-translational modifications [20].
1.3. Human TMPRSS2 protein
Human TMPRSS2 proteins are 32–70 kDa, up to 530 amino acid transmembrane serine proteases coded for by the TMPRSS2 gene(s). They activate various endogenous substrates such as pro-hepatocyte growth factor, PAR-2, matriptase/ST14, and currently of interest, ACE2 [19,20]. TMPRSS2 cleaves the viral S protein at the S1/S2 site (RAR685↓) following interaction with the ACE2 receptor, ultimately leading to cell fusion and viral entry. The human DESC1 protease belongs to the type II transmembrane serine proteinase family. It consists of a 20 aa cytoplasmic region, a 14 aa transmembrane domain, and a 120 aa extracellular SEA domain, followed by a C-terminal trypsin-like serine proteinase domain, with the characteristic catalytic triad of Ser195, His57, and Asp102 (Fig. 3c) [[21], [22], [23]].
There is scope for blocking multiple stages of the viral life cycle via drug-induced inhibition. However, we focus on 3CLpro, PLpro, and TMPRSS2 given their undisputed roles in crucial stages of the viral machinery: viral protein processing, construction of RTC, disruption of host antiviral responses, and viral entry. De novo development of antiviral drugs is a long, expensive, and strenuous task, requiring interdisciplinary teams of researchers and precise safety considerations [24,25]. Phytochemicals are hailed for their effectiveness in combatting major public health concerns, including cancer and diabetes. Within the last two decades, coronaviruses such as those causing SARS and MERS [1,[26], [27], [28]].
Given that the current treatment against COVID-19 is limited to prophylactic interventions such as monoclonal antibodies, peptides, ventilation support, and interferon therapies, we favor the exploration of natural, plant-based compounds, such as flavonoids, for their anti-SARS-CoV-2 potential. The role of the flavanone hesperidin against COVID-19 was investigated since the early phases of the pandemic [29]. Cheng et al. recently reported that the flavanone hesperidin and its aglycone, hesperetin, bind to hTMPRSS2 and hACE2. This leads to downstream cascade inhibition and reduces viral entry and infection [30]. A minor attraction was reported for the proteases PLpro and 3CLpro. They were using VeroE6 cells to investigate the high cytotoxic IC50 values of 1491 μM (hesperetin) and 1435 μM (hesperidin). Hesperetin also reduces the binding activity between hACE2 and the Spike protein, while both hesperetin and hesperidin, slightly decreased the enzyme activity of TMPRSS2. Interestingly, Cheng et al. report that both these flavanones possess the ability to downregulate the protein expression of hACE2 and hTMPRSS2 in normal (Beas 2B cell) and malignant lung (H460) cells post-transcriptionally through their interaction with heat-shock proteins (HSP70/90). Clinical trials such as NCT04452799 were launched to investigate the flavonoid's role in prophylactic and treatment mechanisms against the disease; this, along with other trials such as those reviewed by Kaul et al. [28] represent what clinicians must closely follow to discover possible therapeutics for the increasing number of patients.
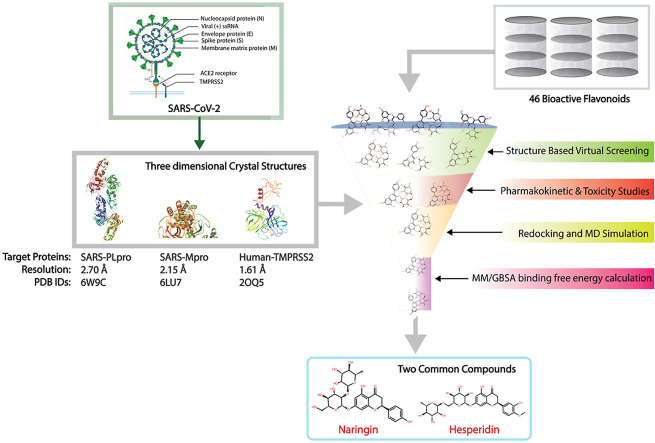
In this study, a database containing 46 bioactive flavonoids was screened virtually via structure-based virtual screening to identify common potential therapeutic drugs which are potentially effective against SARS-CoV-2 3CLpro, SARS-CoV-2 PLpro, and human TMPRSS2 protein(s). Selected compounds were also assessed for their drug-likeness and toxicity by SwissADME and ProTox-II online tools, respectively. In addition, the selected docked protein-ligand complexes were analyzed for their intermolecular interactions followed and their dynamic stability via molecular dynamics simulation. The pharmacokinetics and toxicity of the compounds chosen as well as the binding affinity and stability of the selected docked complexes were compared with the reference ligands reported inhibiting the three SARS-CoV-2 targets [[31], [32], [33], [34]].
2. Methodology
2.1. Structure-based virtual screening
The three-dimensional crystal structure of TMPRSS2 (PDB ID: 2OQ5) [23], 3CLpro (PDB ID: 6LU7) [35], and PLpro (PDB ID: 6W9C) [19] solved at 1.61 Å, 2.15 Å, and 2.70 Å resolution respectively by x-ray crystallography were selected as the target proteins. Flavonoids were screened against the selected target proteins via SBVS at the MTI-OpenScreen web server [36]. In this study, a total of 46 bioactive flavonoids (Table S1), previously reported as active against different infectious diseases. Their three-dimensional structures were retrieved from the PubChem online database (https://pubchem.ncbi.nlm.nih.gov/) [37]. All three target proteins were prepared for SBVS by removing the water molecules and heteroatoms, assigning the Gasteiger charges, adding the polar hydrogen atoms, and removing non-polar hydrogen atoms using the Dock Prep tool in UCSF Chimera-1.14 with default parameters [38]. Following protein preparation, the MTi-OpenScreen webserver was used to screen the collected bioflavonoids against the active site of the target proteins TMPRSS2, 3CLpro, and PLpro. As a result, two common bioflavonoids, hesperidin and naringin, which exhibited the highest negative docking scores respective to all three proteins were selected for ADMET (absorption, distribution, metabolism, excretion, and toxicity) analysis using the SwissADME (http://www.swissadme.ch/index.php) online tool (Fig. S1) [39].
2.2. Pharmacokinetic studies
The SwissADME online tool was used to evaluate predictions of the pharmacokinetic data of the newly suggested compounds, hesperidin, and naringin [39]. The canonical SMILES of these flavones plus three controls were extracted from PubMed (https://pubmed.ncbi.nlm.nih.gov/) and inserted into the software, which in turn returned the structures of the compounds and several data values used to evaluate their physicochemical properties, lipophilicity, water-solubility, pharmacokinetics, drug-likeliness, and medicinal chemistry (Table S2). “BOILED-Egg” analysis representing the GI and BBB (blood-brain barrier) permeability of the control compounds is also reported (Fig. S2). At the same time, radar charts estimate their respective oral bioavailability's based on physicochemical factors (Fig. S3).
2.3. Toxicity studies
All compounds were further analyzed for toxicity using another online software tool known as ProTox-II (https://tox-new.charite.de/protox_II/index.php?site=compound_input) [40]. The software returned several values, including the predicted median lethal dose (LD50), predicted toxicity class, average similarity, prediction accuracy, and toxicity against various targets represented in Tables 1, S3, and S4. The flavones naringin and hesperidin analyses were then compared to approved (control) molecules.
2.4. Redocking simulation and intermolecular interaction analysis
Molecular docking simulations of TMPRSS2, 3CLpro, and PLpro were performed with the selected bioflavonoids hesperidin and naringin using the Chimera-AutoDock Vina plugin setup to understand and decipher the active site residues that are involved in the interaction. The compounds camostat, GC376, and GRL0617 are known to inhibit TMPRSS2 [31,33], 3CLpro [32], and PLpro [34] respectively were used as the reference ligands in this study. Using default parameters in Chimera, the target proteins and the selected compounds were prepared for docking by removing the native ligands, thereby adding the charges and polar hydrogen atoms to the crystal structure. Docking was followed by the docking simulation in AutoDock Vina as a plugin under the default setting by repositioning and adjusting the grid size of (−5.82 × 22.094 x 23.24) at the binding site of TMPRSS2 protein, the grid size of (−13.09 × 13.99 x 69.30) at the binding site of 3CLpro protein, and grid size of (−27.34 × 23.41 x 37.45) at the binding site of PLpro protein, along the X, Y, and Z-axis to cover all essential residues. The grid adjustment is required to provide sufficient space for the different conformations generated by ligands during the docking process. As a result of docking and docking simulation, at least 10 docked poses were collected based on the highest negative docking score, and least root mean square deviation (RMSD) (by default zero in AutoDock Vina). These were extracted to analyze the binding poses using the 2D interaction diagram in the free academic Maestro v12.9 package (Schrödinger Release 2021-3: Maestro, Schrödinger, LLC, New York, NY, 2021). Eventually, all the three-dimensional and two-dimensional interaction images were generated in the free academic Maestro v12.9 package, and similar steps were performed for the docking of reference inhibitors with the target proteins for the comparative analysis of binding pose against the selected bioflavonoid compounds.
2.5. Molecular dynamics simulation
The dynamic stability and intermolecular interactions of the docked complexes acquired from the docking experiments were analyzed by the molecular dynamics (MD) simulation, which was performed under the Linux system running on a HP Z2 Microwater workstation using the Desmond v5.6 [41] module of Schrödinger-Maestro v11.8 (Schrödinger Release 2018-4: Maestro, Schrödinger, LLC, New York, NY, 2018). Using the system builder module in Desmond, the MD system for all the docked complexes was constructed as an orthorhombic grid box (10 Å × 10 Å × 10 Å buffer) and then minimized by adding solvent as TIP4P (transferable intermolecular potential 4 points). The neutralization of the complete system followed the system minimization by adding the counterions (Na+ or Cl−) and further re-minimization using the minimization tool with default settings. Finally, the complete system was forced to simulate at 300K temperature and 1.01325 bar pressure. Along with the reference complex, all the prepared and minimized MD systems were analyzed with the 100 ns MD simulation with OPLS-2005 (force field) and 2000 frames collected at 50 ps interval during the stimulation interval. The trajectories generated via MD simulation were further analyzed for the RMSD, root mean square function (RMSF), and protein-ligand profiling using the simulation interaction diagram tool Desmond v5.6 module of Schrödinger-Maestro v11.8 [41].
2.6. Molecular mechanics/generalized born surface area (MM/GBSA) calculation
To calculate the binding free energy, MM/GBSA was performed on the last ten poses extracted at regular interval of 10 ns from the respective MD simulation trajectory under default parameters of Prime MMGBSA module in Schrödinger suite (Schrödinger Release 2018-3: Prime, Schrödinger, LLC, New York, NY, 2018). The solvent molecules and the ions were deleted for the refinement of extracted poses required for the MMGBSA calculation [42]. The net binding free energy () was calculated by the following equation:
In the above equation, binding free energy is denoted by , the free energy of complex is denoted by And the energy for the receptor and ligand is denoted by and , respectively.
3. Results and discussion
3.1. Structure-based virtual screening
All the 46 compounds selected for structure-based virtual screening (SBVS) against the ligand-binding site of the three target proteins TMPRSS2, 3CLpro, and PLpro, belong to the flavonoids class. The binding energies of these 46 compounds were recorded in the considerable range of −5.0 to −10 kcal/mol against the active site residues of all three proteins but the common compounds, hesperidin and naringin, showed the highest docking scores (lower binding energies) against all three target proteins (Table S1). Based on this, they were selected for redocking and intermolecular interaction analysis (IMI analysis) with TMPRSS2, 3CLpro, and PLpro, compared with the reference ligands camostat, GC376, and GRL0617.
3.2. Role of glycosylation and structure-activity relationships
We observed a pattern between the glycosylation status of the most studied flavonoids retrieved from structure-based virtual screening and their corresponding binding energies to each target protein. Specifically, the top ten flavonoids with the lowest binding energies for TMPRSS2 were all glycosylated; similarly, eight out of the top ten flavonoids for 3CLpro were glycosylated, as were seven of the top ten flavonoids for PLpro. Therefore, we propose that glycosylation plays an important inhibitory role in the binding mechanism of flavonoids against these targets (Table S1). Such flavonoids could mechanistically block attachment and disrupt the entry of virions into host cells while interfering with various stages of viral replication, translation, and polyprotein processing in order to prevent the release of the viruses [43].
3.2.1. Properties of glycosylated flavonoids
Glycosylated flavonoids consist of a base flavan structure and sub-class specific –R groups in addition to a sugar moiety such as arabinose, rhamnose, rutinose, galactose, or glucose; which impacts their respective biological activities [43]. Moreover, flavonoids without sugar moieties are seen to be largely insoluble in water. Thus clinical applications of such compounds have always been in doubt due to their inherent tendency to form polymers. However, glycosylation of flavonoids leads to higher bioavailability and reduces the potential for toxicity due to their newfound solubility provided by the sugar moieties [43]. The sugar moiety block and thereby protects the phenolic group from oxidative degradation. Biological activities of such flavonoids are enhanced against HIV and rotavirus, compared to their non-sugar precursors [44].
For decades, flavonoids were lauded for their antioxidant and anti-inflammatory properties, and glycosylation complements this effect. For example, hesperidin, one of the molecules of interest in this study, has demonstrated high antioxidant activity through radical scavenging and an ability to reduce tissue damage via antioxidant cellular defenses using the ERK/Nrf2 signaling pathway [43,45]. The same glycosylated flavonoid downregulates inducible Nitric oxide synthase (iNOS) and Cyclooxygenase-2 (COX-2). Wherein lies its anti-inflammatory properties [46]. It modulates inflammatory markers such as (Interleukin) IL-6, IL-1β, and TNF-α, in addition to pro-inflammatory cytokines such as IFN-γ and IL-2, in various vital organs in animal models [47]. Moreover, by downregulating the abnormal levels of Angiotensin 2 in hypertensive rodents, hesperidin has been shown to attenuate inflammation. This model has the potential to be replicated in human SARS-CoV-2 patients, given the extensive role of the receptor in viral entry and pathogenesis of associated symptoms such as cytokine storm [43].
Similarly, naringin, a glycosylated flavonoid that is richly present in citrus fruits, is also known to have wide-ranging anti-inflammatory and antiviral activities that are potentially applicable for COVID-19 related therapy. By inhibiting the expression of COX-2, IL-1, IL-6, and iNOS via the suppression of the high mobility group box 1 (HMGB1), it offers anti-inflammatory activities against COVID-19 pathogenesis [45]. Perhaps even more promisingly, naringin is widely investigated through live human clinical trials for its anti-hypercholesteraemic and anti-hypertensive activities. One study found a higher dosage of naringin reduces diastolic blood pressure significantly [48]. In addition to these two glycosylated flavonoids, others like baicalin and silymarin showed simultaneous anti-neuroinflammatory and antiviral potentials for combating SARS-CoV-2 neural complications [45].
3.2.2. Structure-activity relationships of glycosylated flavonoids
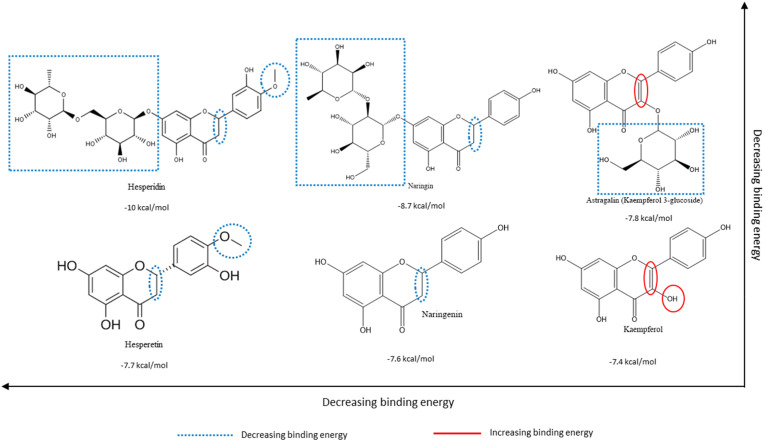
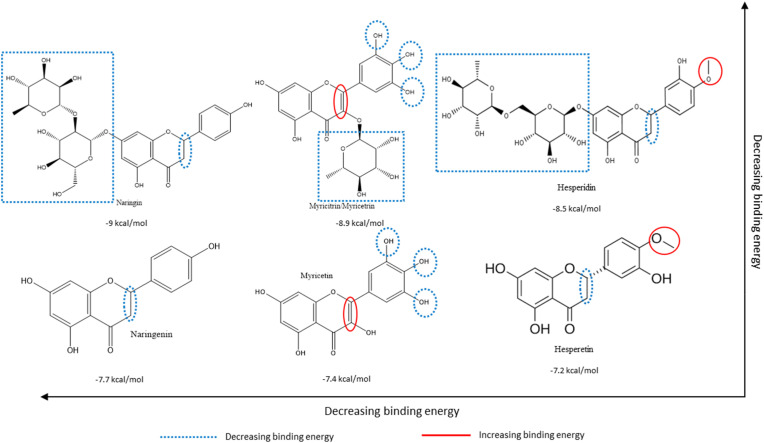
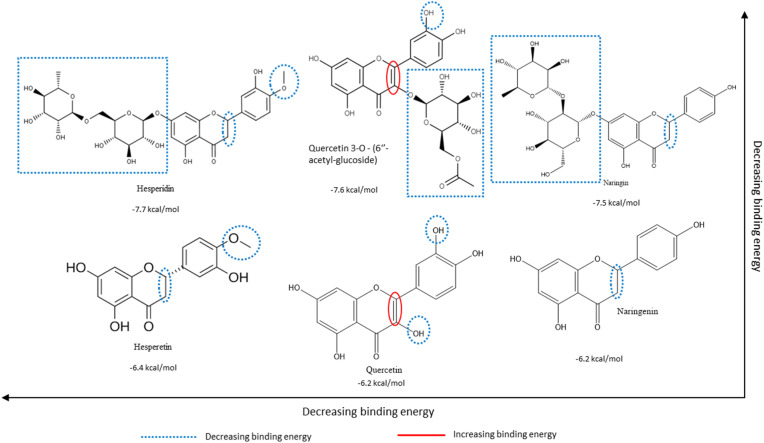
From the multiple flavonoids presented from structure-based virtual screening in Table S1, the structure-activity relationships of hesperidin, naringin, and a third glycosylated flavonoid were compared to their respective aglycones. From Fig. 4, Fig. 5, Fig. 6 , it is evident that glycosylation generally leads to higher binding affinities (translated to lower binding energies) to all three targets presented in this study.
Fig. 4.
Structure-Activity Relationships of select glycosylated flavonoids (top row) and their corresponding aglycones (bottom row) based on their relative binding energies with respect to human TMPRSS2 protein derived from structure-based virtual screening. Substitutions and groups that increase/decrease binding affinities (decrease binding energy) are blue and red, respectively. Lower binding energy represents higher binding affinity. Refer to Table S1 for complete data of structure-based virtual screening. Structures adapted from PubChem.org. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Structure-Activity Relationships of select glycosylated flavonoids (top row) and their corresponding aglycones (bottom row) based on their relative binding energies for SARS-CoV-2 3CLpro, as derived from structure-based virtual screening. Substitutions and groups that increase/decrease binding affinities (decrease binding energy) are blue and red, respectively. Lower binding energy represents higher binding affinity. Refer to Table S1 for complete data of structure-based virtual screening. Structures adapted from PubChem.org. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
Structure-Activity relationships of select glycosylated flavonoids (top row) and their corresponding aglycones (bottom row) based on their relative binding energies for SARS-CoV-2 PLpro, as derived from structure-based virtual screening. Substitutions and groups that increase/decrease binding affinities (decrease binding energy) are blue and red, respectively. Lower binding energy represents higher binding affinity. Refer to Table S1 for complete data of structure-based virtual screening. Structures adapted from PubChem.org.(For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4 exhibits how the affinity for the human TMPRSS2 protein follows the order hesperidin > naringin > astragalin based on the relative presence/absence and positioning of hydroxyl groups on the backbone, in addition to the presence/absence of the C-ring C2–C3 double bond, which leads to lower binding energies when absent, as seen in hesperidin and naringin. However, the difference in affinity due to glycosylation can only be appreciated when flavonoids with sugar moieties are compared to their respective aglycones. We observe a staggering increase of up to 2.3 kcal/mol (hesperidin vs. hesperetin) due to the absence of glycosylation, highlighting its significant role in the binding mechanism.
Fig. 5, Fig. 6 portray similar structure-activity relationships. The glycosylated and aglycosylated forms of myricetin and quercetin (under 3CLpro and PLpro, respectively) have much lower binding energies for the target proteins, further supporting the critical role of glycosylation of flavonoids in the SARS-CoV-2 protein binding mechanisms.
Our previous work supports the position that, overall, the presence of multiple hydroxyl groups on the B-ring of flavonoids reflects positively on the binding affinities of respective flavonoids to the viral targets [28]. Hesperidin and naringin are promising compounds against all the targets examined in this study, but especially against the human TMPRSS2 protein as per the structure-function tests. Glycosylated flavonoids were generally shown to bind more effectively to each of the targets than their aglycone counterparts. The analysis of structure-activity relationships plays a vital role in the initial stages of drug discovery and synthesis. It allows for the identification of particular structural aspects of compounds that enable efficacious binding and, later, activity.
3.3. Pharmacokinetic studies and toxicity studies
Hesperidin and naringin show several favorable biochemical properties, including good water solubility, no BBB permeability, and are substrates for P glycoprotein (Table S2). Solubility is one of the significant factors affecting the absorption of a drug into cells: being soluble is a requirement for drugs meant for parenteral usage, and enough of the drug for treatment can be delivered in a lower dosage. Hesperidin and naringin show even better solubility than the chosen control molecules [39]. Like the approved controls GC376 and camostat, they are not BBB permeable; also, like controls GRL0617 and GC376, they are P-glycoprotein (p-gp) substrates, suggesting good elimination from the body, reducing the potential for drug-induced toxicity (Fig. S2). The overall pharmacokinetic properties of the drugs are represented in the bioavailability radar in (Fig. S3).
SwissADME is a helpful tool to ascertain the drug-likeliness of molecules before they proceed to pre-clinical trials. The drug-likeliness of a compound assesses the probability for a molecule to become an orally administered drug based on bioavailability [39]. The Lipinski Rule of Five is a parameter that is commonly used to evaluate the drug-likeliness of orally bioavailable drugs and states certain parameters that a drug must meet for the compound to be orally bioavailable. These predicted conditions include molecular mass less than 500 Dalton, high lipophilicity (expressed as consensus LogP <5), less than 5 hydrogen bond donors, less than 10 hydrogen bond acceptors, and a molar refractivity between 40 and 130 (Fig. S3).
As Table S2 shows, naringin and hesperidin, both have violations against the Lipinski Rule of Five. However, the Lipinski Rule of Five is only a tool that can help to predict the possibility of drug-likeliness based on orally bioavailable drugs – it is not a parameter that must always be met to obtain approval standards [49]. This is illustrated by our control molecule GC376, an approved prodrug that readily converts to peptide aldehyde GC373 in cats and has activity against SARS-CoV-2 in humans [32,50]. Additionally, naringin and hesperidin's bioavailability score (defined as F>10% in rats) is 0.17, as shown in Table S2. This is the same as the bioavailability score for the approved drug GC376.
Additionally, while most flavonoids exhibit poor oral absorption and bioavailability due to the hydrophilic nature of the flavonoid glycoside group, several approaches improve the bioavailability of flavonoids including nanoformulations, encapsulation, and microemulsions [50]. Previous literature states that these molecules still display antiviral activities despite these problems. For example, naringin displays anti-adsorption effects against dengue virus type 2 [51], and hesperidin has also shown antiviral activity against rotavirus [51].
Additionally, several sources reported that the aglycones of these molecules had increased bioavailability. The aglycone of naringin is naringenin, which exerts therapeutic effects against SARS-CoV-2 through its action against the 3CLpro, main protease, and by reduction of ACE receptors [51]. Naringenin also displays anti-inflammatory activity against SARS-CoV-2 by reducing viral replication and cytokine production [51]. Naringenin and hesperetin (the aglycone of hesperidin) additionally show antiviral activity against the 17D strain of Yellow Fever Virus [52]. Generally, these flavonoids exhibit a high safety profile, with all three compounds having LD50 values of >2000 mg/kg to a minimum of 70% accuracy. Hesperidin appears to be the safest compound with a predicted LD50 of approximately 12,000 mg/kg, followed by naringin at 2300 mg/kg. All these values are higher or comparable to the LD50 for the approved control drugs (Table 1 ).
Table 1.
Toxicity Model Reports for selected compounds hesperidin and naringin and selected control compounds GL376, GLR0617 and Camostat.
| Molecule/Compound | Naringin | Hesperidin | GRL0617 | GC376 | Camostat |
|---|---|---|---|---|---|
| Predicted LD50 (mg/kg) | 2300 | 12,000 | 2800 | 300 | 3000 |
| Predicted Toxicity Class | 5 | 6 | 5 | 3 | 5 |
| Average Similarity | 80.21% | 99.30% | 69% | 46.24% | 62.15% |
| Prediction Accuracy | 70.97% | 72.90% | 68.07% | 54.26% | 68.07% |
3.4. Redocking simulation and intermolecular interaction analysis
Through SBVS, the compounds with the best conformation and stability to target protein were predicted using the scoring function, which needed to be validated further via redocking and IMI analysis. To get the most appropriate binding poses of hesperidin and naringin in the binding pocket of TMPRSS2, 3CLpro, and PLpro, redockings were executed for each compound against all three proteins using AutoDock Vina with a minimum RMSD value (zero) and lesser docking score (highly negative). Both hesperidin and naringin exhibited binding energies in the acceptable range of −7.7 to −10 kcal/mol, and −7.5 to −9 kcal/mol respectively against TMPRSS2, 3CLpro, and PLpro.
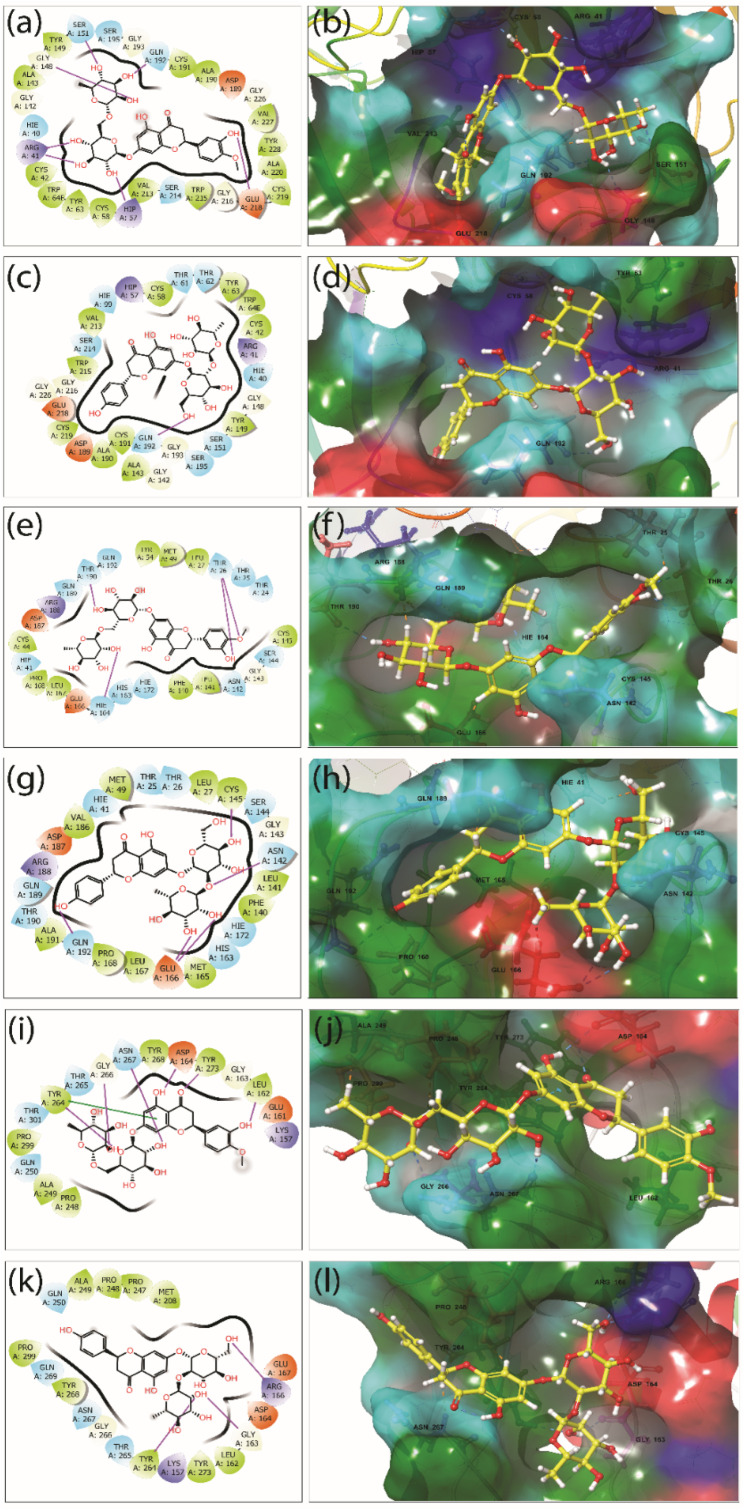
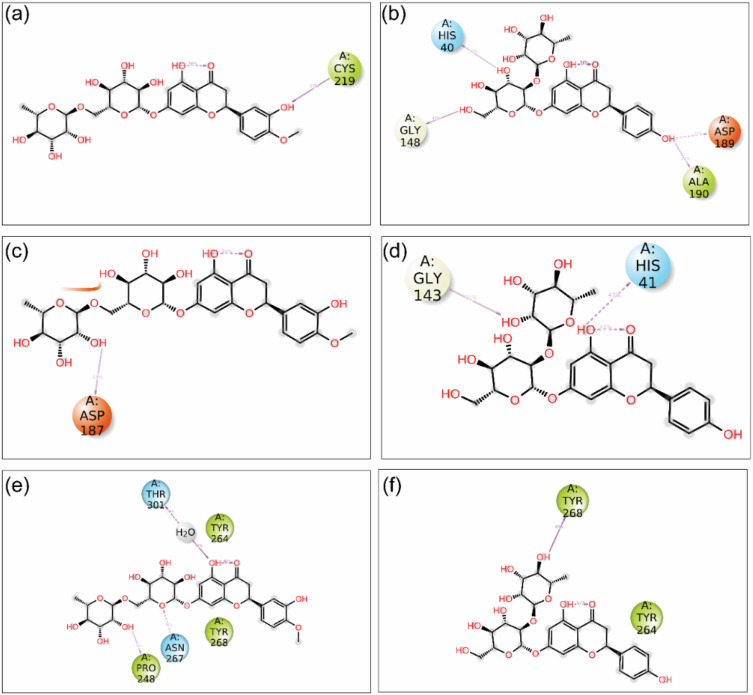
To understand the interaction profile between the ligands (hesperidin and naringin) and the target proteins (TMPRSS2, 3CLpro, and PLpro), IMI analysis was performed compared to the reference inhibitors camostat, GC376, and GRL0617. The TMPRSS2-hesperidin docked complex exhibited a binding energy of −10 kcal/mol. This involved seven hydrogen-bonds (H-bonds) with Arg41, Hip57, Gly148, Ser151, Gln192, and Glu218 residues, and additional interactions such as hydrophobic (Cys58, Tyr63, Ala143, Tyr149, Ala190, Cys191, Val213, Trp215, Cys219, Ala220, Val227, and Tyr228 residues), polar (Ser151, Der195, Gln192, and Ser214 residues), negative (Asp189, and Glu218 residues), positive (Arg41, and Hip57 residues), and glycine (Gly142, Gly148, Gly193, Gly216, and Gly226 residues) (Fig. 7 a and b). In contrast, the TMPRSS2-naringin complex involved only one H-bond (Gly192) with binding energy −8.7 kcal/mol; other intermolecular interaction included hydrophobic (Cys42, Cys58, Tyr63, Tyr149, Ala143, Ala190, Cys191, Val213, Trp215, and Cys219 residues), polar (Thr61, Thr62, Ser151, Ser195, Gln192, Ser214, and residues), positive (Arg41 residue), and negative (Asp189, and Glu218 residues) (Fig. 7c and d). The conformation of the binding pocket and the interacting residues of the protein TMPRSS2 are the same for hesperidin, naringin, and the reference compound camostat (Figs. S4a and S4b).
Fig. 7.
2D and 3D interaction profiles for the docked complexes; (a–b) TMPRSS2-hesperidin, (c–d) TMPRSS2-naringin, (e–f) 3CLpro-hesperidin, (g–h) 3CLpro-naringin, (i–j) PLpro-hesperidin, (k–l) PLpro-naringin, representing the active site residue around 4 Å region in the binding site of TMPRSS2, 3CLpro, and PLpro proteins. 3D and 2D structures were rendered from Maestro 12.9. In 3D structure, the ligand and interacting amino acid residues are represented by ball and stick model, and binding region of protein under 4 Å are shown by solid surface. In 2D interaction profiles, hydrogen bonds (pink arrows), hydrophobic (green), polar (blue), negative (red), positive (violet), and glycine (grey) interactions are shown to be involved between the docked protein-ligand complexes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
IMI analysis of 3CLpro-hesperidin found that this complex accommodates four H-bonds (Thr26, His164, and Thr190 residues) (Fig. 7e and f). In comparison, 3CLpro-naringin accommodates five H-bonds (Asn142, Cys145, Glu166, and Gln192 residues) (Fig. 7g and h) with binding energies of −8.5 kcal/mol and −9 kcal/mol, respectively. Other than the H-bonds, both the complexes involved the same hydrophobic interactions (Leu27, Met49, Phe140, Leu141, Cys145, Leu167, Pro168), polar (Thr25, Thr26, Asn142, Ser144, His163, His172, Gln189, Thr190, and Gln192 residues), positive (Arg188), negative (Glu166, and Asp 187 residues), and glycine (Gly143), except for a few amino acid residues. The similarity of interacting residues in both of these interactions (except for the hydrogen bonding) suggests that both hesperidin and naringin interact with the 3CLpro with almost the same affinity: this is also apparent from their binding scores (Fig. 7e–h). The conformation of the binding pocket and the interacting residues of protein 3CLpro are the same for hesperidin, naringin, and the reference compound GC376 (Figs. S4c and S4d).
The binding energies for PLpro-hesperidin and PLpro-naringin are −7.7 kcal/mol and −7.5 kcal/mol, comparatively less than the other docked complexes but in the acceptable range. The PLpro-hesperidin docked complex contains six H-bonds (Leu162, Asp164, Tyr273, Tyr264, Gln266, and Asn267 residues) along with hydrophobic interactions (Leu162, Tyr264, Tyr268, Tyr273, Pro299, Pro248, and Ala249 residues), polar (Gln250, Thr265, Asn267, and Thr301 residues), positive (Lys157), negative (Asp167, and Glu161 residues), and glycine (Gly163, and Gly266) (Fig. 7iandj). In comparison to PLpro-hesperidin, the PLpro-naringin docked complex involves only three H-bonds (Gly163, Arg166, and Tyr264 residues) along with the hydrophobic interactions (Leu162, Met208, Pro248, Ala249, Tyr264, Tyr268, Tyr273, and Pro299 residues), polar (Gln250, Thr265, Asn267, and Gln269 residues), positive (Lys157, and Arg166 residues), negative Asp164, and Glu167 residues), and glycine (Gly163). Due to the presence of more H-bonds, the PLpro-hesperidin complex can be considered more stable than the PLpro-naringin docked complex (Fig. 7k–l). Hesperidin and naringin interact with a high number of amino acid residues compared to the reference compound GRL0617 (Figs. S4e and S4f).
Analyzing the intermolecular interactions of all the docked complexes, the TMPRSS2-hesperidin docked complex may be considered a more stable complex due to the higher number of H-bonds than with TMPRSS2-naringin. Similarly, of the PLpro-hesperidin and PLpro-naringin complexes, the former docked complex may be considered more stable. In the case of 3CLpro, the docked complexes 3CLpro-hesperidin and 3CLpro-naringin appear to accommodate almost the same number of H-bonds, while the identical residues are involved in various other intermolecular interactions. These two complexes are considered to be almost equally stable.
As discussed in section 3.2.2, the glycosylation of the selected bioflavonoids reduces the binding energy and plays a significant role in the binding with the SRAS-CoV-2 target proteins (Fig. 4, Fig. 5, Fig. 6). The affinity between the bioflavonoids and the target proteins increases due to the sugar moiety's presence, which actively participates in the intermolecular interaction with the active site residues seen in the two-dimensional interaction diagrams (Fig. 7). In TMPRSS2-hesperidin docked complex, six out of seven hydrogen bonds were formed by the sugar moiety of hesperidin (Fig. 7a), and in TMPRSS2-narigin docked complex, only one hydrogen bond was seen, and it was formed by the sugar moiety of naringin (Fig. 7c). In the 3CLpro-hesperidin docked complex, two out of four hydrogen bonds were formed by the sugar moiety of hesperidin (Fig. 7e). In the 3CLpro-naringin docked complex, four out of five hydrogen bonds were created by the sugar moiety of naringin (Fig. 7g). Similarly, in PLpro-hesperidin docked complex, three out of six hydrogen bonds were formed by the sugar moiety of hesperidin (Fig. 7i). In contrast, all three hydrogen bonds were formed by the sugar moiety of naringin in the PLpro-naringin docked complex (Fig. 7k). Therefore, by analyzing the result of SAR and redocking altogether, it is clear that glycosylation plays a crucial role in the interaction between the selected bioflavonoids and SARS-CoV-2 target proteins.
3.5. Molecular dynamics simulation
Molecular dynamics (MD) simulation provides insights into a protein's dynamic behavior and the conformational changes that occur on binding with ligands by predicting the stability and intermolecular interactions of docked complexes with respect to time [52,53] Here, to check the stability of docked protein-ligand complexes, various parameters such as root mean square deviation (RMSD), root mean square fluctuation (RMSF), and protein-ligand contact profiling was recorded and tracked during the entire 100 ns simulation trajectories. Usually, the RMSD is used to monitor the structural fluctuations at the global level. It gives information about the deviation and conformational changes in the protein and ligand structure in the docked complex. In contrast, the RMSF monitors the fluctuation at the local level and measures the mean deviation of each residue and atom of protein and ligand, respectively, in each other's presence. Additionally, protein-ligand contact profiling assesses the interactions between the protein and ligand in the docked complexes during the simulation with respect to time, further exploring the stability of the docked ligand in the protein binding pocket.
3.5.1. RMSD and RMSF analysis
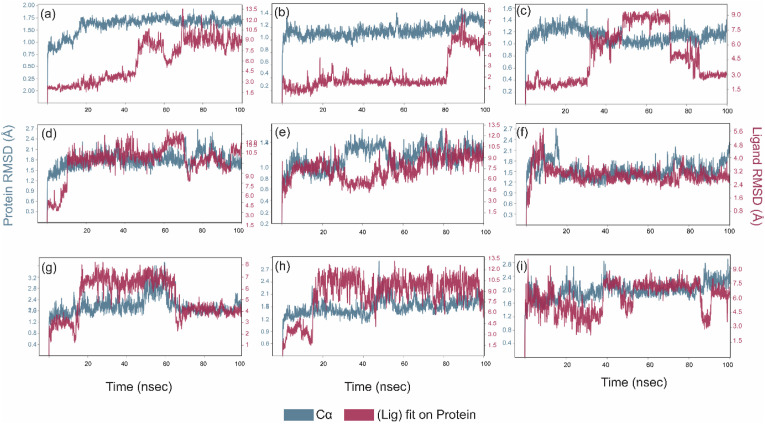
Primarily, the RMSD analysis of protein and ligand for each of the docked protein-ligand complexes was recorded with respect to the initial pose taken as a reference frame (Fig. 8 ). The RMSD for TMPRSS2 deviated <2 Å in both the TMPRSS2-hesperidin and TMPRSS2-naringin docked complexes. This indicated that the protein did not experience conformational changes when bound either to hesperidin or naringin. In contrast, the ligands showed significant fluctuations in both the docked complexes but remained in the binding pocket. The RMSD for hesperidin showed a very high deviation (−9.0 Å) (Fig. 8a) that was observed in the case of the reference ligand camostat (Fig. 8c). This is supported by the RMSF values, where several atoms showed fluctuations close to 8 Å (Figs. S5a and S5c). However, unlike camostat, hesperidin remained buried inside the binding pocket. The RMSD for the naringin showed less deviation (−5.0 Å) as compared to the reference ligand (−9.0 Å) and remained inside the binding pocket of TMPRSS2 (Fig. 8b and c). Therefore, it is concluded that both the docked complexes exhibit higher stability and rigidity as compared to the reference TMPRSS-camostat complex. Additionally, the TMPRSS2-naringin docked complex showed higher dynamic stability with less deviation than the TMPRSS2-hesperidin docked complex (Fig. 8a-c).
Fig. 8.
RMSD plot for the α-carbon atoms of proteins and selected bioflavonoid compounds: (a) TMPRSS2-hesperidin, (b) TMPRSS2-naringin, (c) TMPRSS2-camostat, (d) 3CLpro-hesperidin, (e) 3CLpro-naringin, (f) 3CLpro- GC376 (g) PLpro-hesperidin, (h) PLpro-naringin, (i) PLpro- GRL0617 were plotted for the 100 ns MD simulation time.
Similarly, the RMSD for the protein 3CLpro in the docked complexes 3CLpro-hesperidin and 3CLpro-naringin showed a mean deviation of <2.5 Å and <1.5 Å, respectively, considered highly acceptable. If we compare the RMSD of hesperidin and naringin in comparison to the reference ligand GC376 when bound to 3CLpro, both showed a higher mean deviation (>10 Å) than GC376 (<3.5 Å) (Fig. 8d-f). Their RMSF values support this observation, wherein hesperidin few atoms exhibited an internal fluctuation ≥8.0 Å, and in naringin the fluctuation was <3 Å (Figs. S5d and S5e). Additionally, hesperidin tended to leave the binding pocket between 60 and 70 ns of the 100 ns trajectory and again after 90 ns. This was not observed in the case of naringin, which remained buried inside the binding pocket during the entire trajectory of the simulation (100ns). Finally, although hesperidin and naringin showed more fluctuations than the reference ligand GC376, they remained associated with the protein trajectory (Fig. 8d-f). Therefore, both bioflavonoids showed considerable dynamic stability with the 3CLpro, but naringin is a more suitable ligand than hesperidin due to less deviation in RMSD.
The RMSD for the protein PLpro in the docked complexes PLpro-hesperidin and PLpro-naringin showed mean deviation over a considerable range (<3.2 Å and <2.4 Å, respectively) throughout the 100 ns simulation. In comparison to the RMSD value of the reference ligand GRL0617 (∼7.5 Å), naringin showed a higher deviation (∼3.0 Å to ∼11.0 Å) after 15 ns followed by the exhibition of a stable trajectory at 10.5 Å until ∼95 ns and then seemed to fall again. A longer simulation would give a better understanding of the dynamic stability of naringin. Hesperidin, also showed deviation from ∼3.0 Å to 7.0 Å after exactly 15 ns. Still, in contrast to naringin it returned to 4.0 Å after 60 ns and then exhibited equilibrium until the end of the 100 ns simulation (Fig. 8g-h). RMSF analysis showed that atoms in both the ligands showed maximum fluctuations of up to ∼6.0 Å (Figs. S5g and S5h), suggesting that the naringin's high RMSD is not due to internal fluctuations but occurs upon interaction with PLpro. Overall, hesperidin is a more suitable ligand than naringin, as it deviated for only a short time before attaining a stable trajectory, which is then exhibited until the end of the 100 ns simulation.
3.5.2. Protein-ligand interaction profiling
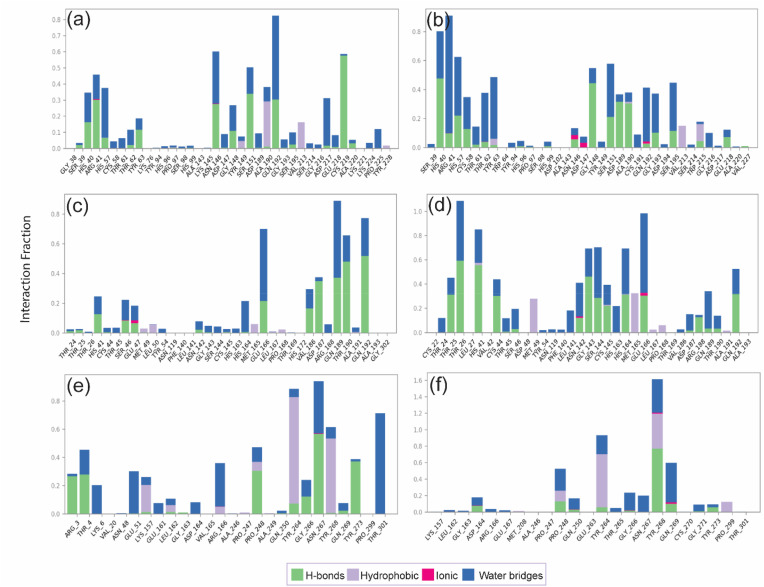
The protein-ligand interaction profiling includes several interactions (H-bonds, hydrophobic interactions, ionic interactions, and salt bridges). It is a critical parameter to understand the modification in interactions between protein and ligand during the 100 ns MD simulation. The interaction of hesperidin and naringin with the active site residues of TMPRSS2, 3CLpro, and PLpro during the 100 ns simulation was further analyzed for various interactions occurring at the atomic and intermolecular level for time (Fig. 9 ).
Fig. 9.
Protein-ligand interaction profiling for the docked complexes: (a) TMPRSS2-hesperidin, (b) TMPRSS2-naringin, (c) 3CLpro-hesperidin, (d) 3CLpro-naringin, (e) PLpro-hesperidin, (f) PLpro-naringin, were extracted at 30% of the total MD simulation interaction interval.
In the TMPRSS2-hesperidin docked complex, mainly H-bonds and hydrophobic interactions were observed during the simulation. The residues Arg41, Asn146, Gly148, and Gln192 formed H-bonds more than 25% of the simulation time, and Cys219 created the H-bond for more than 50% of the time. The residues involved in hydrophobic interactions were Tyr149, Ala190, and Val213, but only Ala190 contributed to more than 30% of the simulation time. The residues His57, Asn146, Gln192 formed a water bridge for more than 30% of the simulation time in addition to developing H-bonds (Fig. 9a). In the TMPRSS2-naringin docked complex, the residues His40 and Gly148 formed H-bonds for more than 40% of the simulation time, and Asp189 and Ala190 formed H-bond for more than 30% of the simulation time. Hydrophobic interaction was not significantly involved in this complex. A water bridge was formed by His57, Thr62, Tyr63, Ser151, Gln192, and Ser195 residues for more than 35% of the simulation time and by Arg41 for more than 80% of the simulation time. Ionic interactions were formed by Asn146, Asp147, and Gln192 but only for a brief period of the simulation (Fig. 9b). The smaller number of interactions indicates the stability of the TMPRSS2-naringin docked complex is not suitable, as was also observed in the analysis of MD simulation data (Fig. 9c).
In the 3CLpro-hesperidin docked complex, H-bonds by Asp187, Gln189, Thr190, and Gln192, and water bridging by Glu166, Gln189 were involved in the interaction for more than 30% of the simulation. Comparatively, the 3CLpro-naringin docked complex showed a more significant number of H-bonds and hydrophobic interactions, suggesting the complex is dynamically more stable than the 3CLpro-hesperidin docked complex. H-bonds were formed by Thr26, His41, and Gly143 for more than 40% of the simulation, and by Thr25, Cys44, and His164, Glu166, and Gln192 for more than 30% of the simulation (Fig. 9d).
Among the PLpro-ligand docked complexes, the PLpro-hesperidin complex involved more H-bonds, hydrophobic interactions, and water bridges than PLpro-naringin and can be considered more stable. In the PLpro-hesperidin complex, H-bonds were formed by Arg3, Thr4, Pro248, and Tyr273 for more than 20% of the simulation. By Asn267 for more than 50% of the simulation, the hydrophobic interactions formed by Tyr264, and Tyr 267 for more than 50% of the simulation, and water bridges were formed by Thr301 for more than 60% of the simulation and by Glu51, Arg166, and Asn267 for more than 25% of the simulation (Fig. 9e and f). A similar observation was obtained through the MD simulation analysis of both the complexes where hesperidin was more stable in the binding pocket of PLpro.
The interactions between the three proteins and hesperidin and naringin were determined at a total 30% interval of 100 ns MD simulation. Interestingly, all the docked complexes exhibited the formation of H-bonds and water bridges, contributing to the dynamic stability of these bioflavonoids within the active site of target proteins. Consequently, combining MD simulation and protein-ligand interaction profiling to assess the stability of the docked complexes, we can conclude that: (i) the TMPRSS2-naringin docked complex seems to be more stable than TMPRSS2-hesperidin; (ii) 3CLpro-naringin is more stable than 3CLpro-hesperidin; and (iii) PLpro-hesperidin is more stable than PLpro-naringin (Fig. 10 ).
Fig. 10.
Schematic representation for the interaction profile of proteins docked with the bioflavonoids: (a) TMPRSS2-hesperidin, (b) TMPRSS2-naringin, (c) 3CLpro-hesperidin, (d) 3CLpro-naringin, (e) PLpro-hesperidin, (f) PLpro-naringin, extracted at 30% of total ns simulation interval.
3.6. Binding free energy analysis
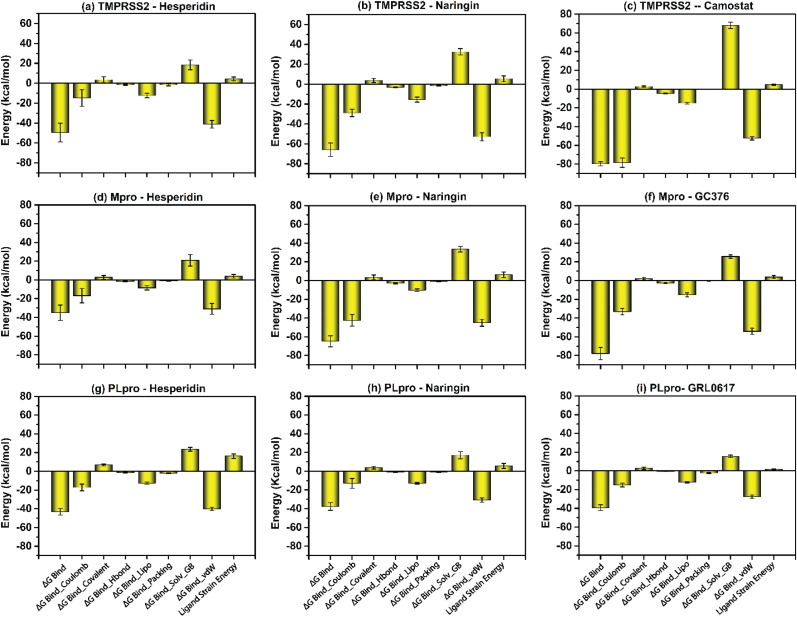
Using molecular mechanics/generalized born surface area calculation (MM/GBSA) approach, the binding free energy and the binding affinity of the selected protein-ligand complexes were evaluated. Followed by the MD simulation, the last ten poses of each docked complexes i.e., TMPRSS2-hesperidin, TMPRSS2-naringin, 3CLpro-hesperidin, 3CLpro-naringin, PLpro-hesperidin, and PLpro-naringin, were extracted from 100 ns MD simulation trajectory at a regular interval of 10 ns to calculate the average binding free energy against the reference complexes, viz. TMPRSS2- Camostat, 3CLpro- GC376, and PLpro- GRL0617 (Supplementary Table 5). MMGBSA calculation showed that naringin exhibited significant binding free energy with TMPRSS2 (−65.83854 kcal/mol) and 3CLpro (−64.76481) in comparison to the reference inhibitors i.e., camostat (−79.84702 kcal/mol) and GC376 (−77.88006 kcal/mol) respectively. In contrast, hesperidin exhibited lesser binding energy (−43.14713) with the PLpro protein in comparison to the reference inhibitor i.e., GRL0617 (−39.32349) (Table 2 ). In addition, parameters related with the dissociation energy were also calculated for each of the docked complexes and exhibited substantial support of Van der Waals' interaction energy (), and Lipophilic energy () in the stability whereas Generalized Born electrostatic solvation energy () was found to contribute in the instability of the respective docked complexes (Fig. 11 ). Coulomb energy (Δ ) also contributed significantly to the stability of all the docked complexes except TMPRSS2-flavonoid complexes. Overall, the MMGBSA result suggested that the selected bioflavonoids exhibit significant binding free energy and substantial stability by forming Van der Waals’ interactions with the essential residues in the active site of SARS-CoV-2 target proteins.
Table 2.
Binding free energy calculation of the selected bioflavonoids against the SARS-CoV-2 target proteins in comparison to the reference inhibitors.
| S. No. | Compounds | MM/GBSA Binding free energy calculation (kcal/mol) |
||
|---|---|---|---|---|
| TMPRSS2 | 3CLpro | PLpro | ||
| 1. | Hesperidin | −49.50252 | −35.01595 | −43.14713 |
| 2. | Naringin | −65.83854 | −64.76481 | −37.7361 |
| 3. | Camostat (Control for TMPRSS2) | −79.84702 | – | – |
| 4. | GC376 (Control for 3CLpro) | – | −77.88006 | – |
| 5. | GRL0617 (Control for PLpro) | – | – | −39.32349 |
Fig. 11.
Binding free energy and individual dissociation energy components calculation performed for SARS-CoV-2 target proteins docked with the bioflavonoids (a) TMPRSS2-hesperidin, (b) TMPRSS2-naringin, (c) TMPRSS2-camostat, (d) 3CLpro-hesperidin, (e) 3CLpro-naringin, (f) 3CLpro- GC376 (g) PLpro-hesperidin, (h) PLpro-naringin, (i) PLpro- GRL0617, on extracted poses from total 100 ns simulation interval.
4. Conclusion
This study adopted a molecular docking and simulation approach to identify common bioactive flavonoids potentially active against the SARS-CoV-2 targets 3CLpro, PLpro, and TMPRSS2. Two potent flavonoids, hesperidin, and naringin, were identified through virtual screening followed by evaluation for drug likeliness, toxicity, and dynamic stability in comparison to the reference ligands camostat (for TMPRSS2), GC376 (for 3CLpro), and GRL0617 (for PLpro). The importance of glycosylation to the anti-inflammatory and antiviral activities and its contribution to the structure-activity relationship was discussed. Molecular docking simulation showed that the binding energies of hesperidin and naringin were indicative of strong complex formation with the target proteins. Molecular dynamics simulation for 100 ns showed that the deviation in dynamic stability for all the docked complexes lies in an acceptable range. Therefore, these complexes can be considered stable due to the formation of multiple intermolecular interactions. Based on our analysis of the drug likeness, toxicity, binding energy, MD simulation trajectory, and protein-ligand interaction profiles, we conclude that the bioflavonoids hesperidin and naringin are potential therapeutic agents against SARS-CoV-2. Of the two, naringin showed better affinity and stability with TMPRSS2 and 3CLpro, whereas hesperidin showed better binding affinity and stability with PLpro. The binding free energy calculation further supported the stability and affinity of the docked flavonoids through MM/GBSA where naringin showed a significant binding affinity with TMPRSS2 and 3CL pro.
In contrast, hesperidin exhibited significant binding affinity with the PLpro protein of SARS-CoV-2. The screened flavonoids should be further validated through in vitro inhibition and neutralization assays to develop a therapeutic drug molecule against SARS-CoV-2. Our results add to existing knowledge concerning the antiviral effects of flavonoids and set the stage for potentially further studies in the field. Hopefully, they invite in vitro, in vivo, and clinical trials to investigate the effects of the currently mentioned flavonoids and other phytochemicals in this global pandemic against a common foe.
Author contributions
A.C. designed the study, critically supervised the project, revised, and reviewed the manuscript with the help of S.K.J and SSM; S.K., and P.Y. performed the experiments; S.K., and A.C. analyzed the data and wrote the manuscript; P.P. and R.K. wrote parts of the manuscript and generated some of the tables and the graphs. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Data availability statement
No new data were created or analyzed in this study.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We acknowledge the help of Dr. Dietrich Büsselberg. Professor of Physiology and Biophysics at Weill Cornell Medicine-Qatar and Dr. Ross S. MacDonald, Weill Cornell Medicine-Qatar Distributed eLibrary, for editing the manuscript. We Thank Hidenori Miyagawa, Weill Cornell Medicine-Qatar Distributed eLibrary, for helping and advising in producing some figures. Open Access funding was provided by the Qatar National Library.
Sanjay Kumar is recipient of Council of Scientific and Industrial Research (CSIR), Govt. of India fellowship.Multiple figures have been created using icons and templates retrieved from BioRender.com with appropriate publication licenses. Few chemical structures have been created using ChemDraw and adapted from PubChem.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105231.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Russo M., Moccia S., Spagnuolo C., Tedesco I., Russo G.L. Roles of flavonoids against coronavirus infection. Chem. Biol. Interact. 2020;328 doi: 10.1016/j.cbi.2020.109211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alimohamadi Y., Sepandi M., Taghdir M., Hosamirudsari H. Determine the most common clinical symptoms in COVID-19 patients: a systematic review and meta-analysis. J. Prev. Med. Hyg. 2020;61:E304–E312. doi: 10.15167/2421-4248/jpmh2020.61.3.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saed Aldien A., Ganesan G.S., Wahbeh F., Al-Nassr N., Altarawneh H., Al Theyab L., Saed Aldien S., Tomerak S., Naveed H., Elshazly M.B., Zakaria D. Systemic inflammation may induce cardiac injury in COVID-19 patients including children and adolescents without underlying cardiovascular diseases: a systematic review. Cardiovasc. Revascularization Med. 2021 doi: 10.1016/J.CARREV.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umakanthan S., Chattu V.K., VRanade A., Das D., Basavarajegowda A., Bukelo M. A rapid review of recent advances in diagnosis, treatment and vaccination for COVID-19. AIMS Publ. Health. 2021;8:137. doi: 10.3934/PUBLICHEALTH.2021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Ali D., Elshafeey A., Mushannen M., Kawas H., Shafiq A., Mhaimeed N., Mhaimeed O., Mhaimeed N., Zeghlache R., Salameh M., Paul P., Homssi M., Mohammed I., Narangoli A., Yagan L., Khanjar B., Laws S., Elshazly M.B., Zakaria D. Cardiovascular and haematological events post COVID-19 vaccination: a systematic review. J. Cell Mol. Med. 2021 doi: 10.1111/JCMM.17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ameena Shafiq A., Salameh M.A., Laswi I., Mohammed I., Mhaimeed O., Mhaimeed N., Mhaimeed N., Paul P., Mushannen M., Elshafeey A., Fares A., Holroyd S., Zakaria D. Neurological immune related adverse events post-COVID-19 vaccination: a systematic review. J. Clin. Pharmacol. 2021 doi: 10.1002/JCPH.2017. [DOI] [PubMed] [Google Scholar]
- 8.Rudrapal M., Issahaku A.R., Agoni C., Bendale A.R., Nagar A., Soliman M.E.S., Lokwani D. 2021. In silico screening of phytopolyphenolics for the identification of bioactive compounds as novel protease inhibitors effective against SARS-CoV-2. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary S., Silakari O. Scaffold morphing of arbidol (umifenovir) in search of multi-targeting therapy halting the interaction of SARS-CoV-2 with ACE2 and other proteases involved in COVID-19. Virus Res. 2020;289:198146. doi: 10.1016/J.VIRUSRES.2020.198146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11:210. doi: 10.3390/V11030210. 11 (2019) 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wertheim J.O., Chu D.K.W., Peiris J.S.M., Kosakovsky Pond S.L., Poon L.L.M. A case for the ancient origin of coronaviruses. J. Virol. 2013;87:7039–7045. doi: 10.1128/JVI.03273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mody V., Ho J., Wills S., Mawri A., Lawson L., Ebert M.C.C.J.C., Fortin G.M., Rayalam S., Taval S. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun. Biol. 2021. 2021;41(4):1–10. doi: 10.1038/s42003-020-01577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamid S., Mir M.Y., Rohela G.K. Novel coronavirus disease (COVID-19): a pandemic (epidemiology, pathogenesis and potential therapeutics) New Microbes New Infect. 2020;35 doi: 10.1016/j.nmni.2020.100679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Sen Tan K., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- A n update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Grunewald M., Perlman S. Methods Mol. Biol. Humana Press Inc.; 2020. Coronaviruses: an updated overview of their replication and pathogenesis; pp. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahir ul Qamar M., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osipiuk J., Azizi S.A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A., Kang S., Kathayat R.S., Kim Y., Lisnyak V.G., Maki S.L., Nicolaescu V., Taylor C.A., Tesar C., Zhang Y.A., Zhou Z., Randall G., Michalska K., Snyder S.A., Dickinson B.C., Joachimiak A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12:1–9. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stasiulewicz A., Maksymiuk A.W., Nguyen M.L., Bełza B., Sulkowska J.I. SARS-CoV-2 papain-like protease potential inhibitors—in silico quantitative assessment. Int. J. Mol. Sci. 2021;22:3957. doi: 10.3390/IJMS22083957. 22 (2021) 3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y.W., Lee M.S., Lucht A., Chou F.P., Huang W., Havighurst T.C., Kim K., Wang J.K., Antalis T.M., Johnson M.D., Lin C.Y. TMPRSS2, a serine protease expressed in the prostate on the apical surface of luminal epithelial cells and released into semen in prostasomes, is misregulated in prostate cancer cells. Am. J. Pathol. 2010;176:2986–2996. doi: 10.2353/AJPATH.2010.090665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monticelli M., Mele B.H., Benetti E., Fallerini C., Baldassarri M., Furini S., Frullanti E., Mari F., Andreotti G., Cubellis M.V., Renieri A. Protective role of a tmprss2 variant on severe covid-19 outcome in young males and elderly women. Genes. 2021;12 doi: 10.3390/GENES12040596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyrieleis O.J.P., Huber R., Ong E., Oehler R., Hunter M., Madison E.L., Jacob U. Crystal structure of the catalytic domain of DESC1, a new member of the type II transmembrane serine proteinase family. FEBS J. 2007;274:2148–2160. doi: 10.1111/j.1742-4658.2007.05756.x. [DOI] [PubMed] [Google Scholar]
- 24.Jeong G.U., Song H., Yoon G.Y., Kim D., Kwon Y.C. Therapeutic strategies against COVID-19 and structural characterization of SARS-CoV-2: a review. Front. Microbiol. 2020;11:1723. doi: 10.3389/fmicb.2020.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das G., Ghosh S., Garg S., Ghosh S., Jana A., Samat R., Mukherjee N., Roy R., Ghosh S. An overview of key potential therapeutic strategies for combat in the COVID-19 battle. RSC Adv. 2020;10:28243–28266. doi: 10.1039/d0ra05434h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Chen Y., Chen Y., Zhou B., Shan X., Yang G. Eriodictyol inhibits IL-1β-induced inflammatory response in human osteoarthritis chondrocytes. Biomed. Pharmacother. 2018;107:1128–1134. doi: 10.1016/j.biopha.2018.08.103. [DOI] [PubMed] [Google Scholar]
- 27.Lee J.S., Bukhari S.N.A., Fauzi N.M. Effects of chalcone derivatives on players of the immune system. Drug Des. Dev. Ther. 2015;9:4761–4778. doi: 10.2147/DDDT.S86242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaul R., Paul P., Kumar S., Büsselberg D., Dwivedi V.D., Chaari A. Promising antiviral activities of natural flavonoids against SARS-CoV-2 targets: systematic review. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haggag Y.A., El-Ashmawy N.E., Okasha K.M. Is hesperidin essential for prophylaxis and treatment of COVID-19 Infection? Med. Hypotheses. 2020;144:109957. doi: 10.1016/J.MEHY.2020.109957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng F.J., Huynh T.K., Yang C.S., Hu D.W., Shen Y.C., Tu C.Y., Wu Y.C., Tang C.H., Huang W.C., Chen Y., Ho C.Y. Hesperidin is a potential inhibitor against sars-cov-2 infection. Nutrients. 2021;13 doi: 10.3390/NU13082800/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann M., Hofmann-Winkler H., Smith J.C., Krüger N., Sørensen L.K., Søgaard O.S., Hasselstrøm J.B., Winkler M., Hempel T., Raich L., Olsson S., Yamazoe T., Yamatsuta K., Mizuno H., Ludwig S., Noé F., Sheltzer J.M., Kjolby M., Pöhlmann S. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. BioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.08.05.237651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:1–8. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S K., Tai Young Kim C.J.L., Jeon Sangeun, Jang Youngho, Gotina Lizaveta, Won Joungha, Ju Yeon Ha, Kim Sunpil, Jang Minwoo Wendy, Won Woojin, Gordon Park Mingu, Pae Ae Nim, Sunkyu Han, Platycodin D. 2021. A Natural Component of Platycodon Grandiflorum, Prevents Both Lysosome- and TMPRSS2-Driven SARS-CoV-2 Infection by Hindering Membrane Fusion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Z., Huang B., Tang J., Liu S., Liu M., Ye Y., Liu Z., Xiong Y., Zhu W., Cao D., Li J., Niu X., Zhou H., Zhao Y.J., Zhang G., Huang H. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-020-20718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 36.Labbé C.M., Rey J., Lagorce D., Vavruša M., Becot J., Sperandio O., Villoutreix B.O., Tufféry P., Miteva M.A. MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Res. 2015;43:W448–W454. doi: 10.1093/nar/gkv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., Zaslavsky L., Zhang J., Bolton E.E. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25 doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/SREP42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46:W257–W263. doi: 10.1093/NAR/GKY318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowers K.J., Chow E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., Salmon J.K., Shan Y., Shaw D.E. Proc. 2006 ACM/IEEE Conf. Supercomput. Association for Computing Machinery; New York, NY, USA: 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. 84–es. [DOI] [Google Scholar]
- 42.Shiv Bharadwaj S.G.K., Lee Kyung Eun, Dhar Dwivedi Vivek, Yadava Umesh, Nees Matthias. Density functional theory and molecular dynamics simulation support Ganoderma lucidum triterpenoids as broad range antagonist of matrix metalloproteinases. J. Mol. Liq. 2020;311:10. doi: 10.1016/j.molliq.2020.113322. [DOI] [Google Scholar]
- 43.Godinho P.I.C., Soengas R.G., Silva V.L.M. Therapeutic potential of glycosyl flavonoids as anti-coronaviral agents. Pharmaceuticals. 2021;14 doi: 10.3390/ph14060546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao J. Dietary flavonoid aglycones and their glycosides: which show better biological significance? Crit. Rev. Food Sci. Nutr. 2017;57:1874–1905. doi: 10.1080/10408398.2015.1032400. [DOI] [PubMed] [Google Scholar]
- 45.Fakhri S., Piri S., Majnooni M.B., Farzaei M.H., Echeverría J. Targeting neurological manifestations of coronaviruses by candidate phytochemicals: a mechanistic approach. Front. Pharmacol. 2020;11:621099. doi: 10.3389/fphar.2020.621099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi S.-J., Tai B.H., Cuong N.M., Kim Y.-H., Jang H.-D. Antioxidative and anti-inflammatory effect of quercetin and its glycosides isolated from mampat (Cratoxylum formosum) Food Sci. Biotechnol. 2012;212(21):587–595. doi: 10.1007/S10068-012-0075-4. 2012. [DOI] [Google Scholar]
- 47.Agrawal P.K., Agrawal C., Blunden G. Pharmacological significance of hesperidin and hesperetin, two citrus flavonoids, as promising antiviral compounds for prophylaxis Against and combating COVID-19. Nat. Prod. Commun. 2021;16 doi: 10.1177/1934578X211042540. [DOI] [Google Scholar]
- 48.Salehi B., Fokou P.V.T., Sharifi-Rad M., Zucca P., Pezzani R., Martins N., Sharifi-Rad J. The therapeutic potential of naringenin: a review of clinical trials. Pharm. Times. 2019;12:11–12. doi: 10.3390/PH12010011. (2019) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neidle S. 9 - Design Principles for Quadruplex-Binding Small Molecules. Academic Press; Boston: 2012. pp. 151–174. [DOI] [Google Scholar]
- 50.Alzaabi M.M., Hamdy R., Ashmawy N.S., Hamoda A.M., Alkhayat F., Khademi N.N., Al Joud S.M.A., El-Keblawy A.A., Soliman S.S.M. Flavonoids are promising safe therapy against COVID-19. Phytochemistry Rev. 2021:1. doi: 10.1007/S11101-021-09759-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zandi K., Teoh B.-T., Sam S.-S., Wong P.-F., Mustafa M.R., AbuBakar S. Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol. J. 2011. 2011;81(8):1–11. doi: 10.1186/1743-422X-8-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bharadwaj S., Rao A.K., Dwivedi V.D., Mishra S.K., Yadava U. Structure-based screening and validation of bioactive compounds as Zika virus methyltransferase (MTase) inhibitors through first-principle density functional theory, classical molecular simulation and QM/MM affinity estimation. J. Biomol. Struct. Dyn. 2021;39:2338–2351. doi: 10.1080/07391102.2020.1747545. [DOI] [PubMed] [Google Scholar]
- 53.Bharadwaj S., Lee K.E., Dwivedi V.D., Yadava U., Panwar A., Lucas S.J., Pandey A., Kang S.G. Discovery of Ganoderma lucidum triterpenoids as potential inhibitors against Dengue virus NS2B-NS3 protease. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-55723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were created or analyzed in this study.